Introduction
The Maelstrom (MAEL) gene was first identified in
Drosophila(1). It plays a
role in the establishment of oocyte polarity by acting in the
positioning of mRNA and the microtubule-organizing center (MTOC) in
early oocytes (1,2). Drosophila maelstrom protein
co-localizes with Vasa and Aubergine in the nuage (3), a germline-unique perinuclear structure
that serves as a platform for PIWI-interacting RNA (piRNA)
biogenesis and piRNA-dependent silencing of transposons and some
other harmful selfish elements (4,5). The
mouse homolog of MAEL is also found to localize in the nuage
(6,7) and is essential for spermatogenesis and
transposon repression (7). Further
study by Aravin et al(8)
demonstrated that MAEL co-localizes with MIWI2/PIWIL4 in a type of
nuage named piP-body, which harbors both piRNA pathway proteins
(MIWI2, TDRD9 and MAEL) and P-body (processing body) components
(DDX6, DCP1a, XRN1 and GW182), and that loss of MAEL disrupts
normal MIWI2 localization and piRNA production leading to
transposon activation. In rat spermatogenic cells, MAEL protein is
detected in all types of nuage except the cluster of 30-nm
particles (i.e. 70- to 90-nm particles, satellite body,
intermitochondrial cement, cluster of 60- to 90-nm particles,
chromatoid body) (9). MAEL protein
is also found in non-nuage compartments (10) and the nucleus (3,9). In
the nucleus, MAEL binds the miR-7 promoter and represses its
expression (11).
Our previous study demonstrated that MAEL is a
cancer-testis (CT) gene (12). The
CT genes, also known as cancer-germline (CG) genes, are
predominantly expressed in germ cells of the testis or/and ovary
and have no or little expression in somatic adult tissues, but are
aberrantly expressed in various types of cancers (13,14).
We found that the human MAEL gene is only expressed in germ cells
of the testis among normal tissues, but is expressed in various
human cancer cell lines such as breast cancer MDA-MB-231 cells and
colorectal cancer SW480 cells. Like most of the CT genes (15,16),
the expression of the MAEL gene is activated by hypomethylation in
cancer (12). Kim et
al(17) analyzed the
methylation profile of 27,578 CpG sites spanning >14,000 genes
in colorectal cancer and adjacent normal mucosa, and found that
among the genes tested, MAEL and SFT2D3 showed the lowest
methylation level in tumor tissues when compared to the normal
mucosa (17). However, the precise
subcellular localization and function of MAEL in cancer cells are
not clear.
In the present study, we identified the interacting
partners of MAEL in breast cancer MDA-MB-231 and colorectal cancer
SW480 cells using mass spectrometric method, and demonstrated that
MAEL localizes in stress granules (SGs) in cancer cells and
interacts with several components of SGs.
Materials and methods
Plasmids
The Myc-tagged YBX1 expression plasmid pDESTmycYBX1
[Addgene plasmid 19878, deposited by Landthaler (18)] was obtained from Addgene (Cambridge,
MA, USA). The other expression plasmids (pHA-MAEL, pMyc-DDX39,
pMyc-EIF4A1, pMyc-EIF3F, pMyc-ELAV1, pMyc-PABPC1, pMyc-SYNCRIP and
pMyc-KHSRP) were constructed as follows. The coding sequence of
each gene was amplified from the first strand cDNA of MDA-MB-231 or
SW480 cells with the primers described in Table I. The amplified products were cloned
into the pMD18-T vector (Takara, Dalian, China) by TA cloning
method, and subcloned into the pCMV-HA or pCMV-Myc vector (Clontech
Laboratories, Inc., Mountain View, CA, USA) with restriction enzyme
sites in the primer (Table I).
 | Table IPrimers used for amplifying the
coding region of the genes. |
Table I
Primers used for amplifying the
coding region of the genes.
| Gene name | Sequence
(5′→3′) | Restriction
site |
|---|
| MAEL-F |
GAATTCCCATGCCGAACCGTAAGG | EcoRI |
| MAEL-R |
GGTACCGGGCCTGTTACTGTTTTCAGAA | KpnI |
| DDX39-F |
GAATTCTCATGGCAGAACAGGATGTG | EcoRI |
| DDX39-R |
CTCGAGTGGTGGTTACCGGCTCTG | XhoI |
| EIF4A1-F |
GAATTCTCATGTCTGCGAGCCAGG | EcoRI |
| EIF4A1-R |
GTCGACGCTGGGTGGCAGGACAG | SalI |
| EIF3F-F |
GAATTCACAAGATGGCCACACCG | EcoRI |
| EIF3F-R |
CTCGAGCCATTCACAGGTTTACAAGTTT | XhoI |
| ELAV1-F |
GTCGACAATGTCTAATGGTTATGAAGACC | SalI |
| ELAV1-R |
GGTACCGCATGAGCGAGTTATTTGTG | KpnI |
| PABPC1-F |
GTCGACCGAGATGAACCCCAGTGC | SalI |
| PABPC1-R |
GGTACCTTTAAACAGTTGGAACACCG | KpnI |
| SYNCRIP-F |
GTCGACTGGAAACATGGCTACAGAACA | SalI |
| SYNCRIP-R |
GGTACCCTACTTCCACTGTTGCCCAA | KpnI |
| KHSRP-F |
GAATTCCCATGTCGGACTACAGCAC | EcoRI |
| KHSRP-R |
GGTACCGATTCATTGAGCCTGCTGC | KpnI |
Immunoprecipitation (IP) of the MAEL
protein complex
The human breast cancer MDA-MB-231 and colorectal
cancer SW480 cells [American Tissue Culture Collection (ATCC),
Manassas, VA, USA] were cultured in L-15 media that was
supplemented with glutamine, antibiotics and 10% fetal bovine serum
(FBS) at 37°C in 5% CO2. The cells were washed with
phosphate-buffered saline (PBS) and incubated with 0.5 mM
cross-linker DSP (Pierce, Rockford, IL, USA) for 30 min. After
cross-linking, cells were sequentially washed with PBS, PBS
containing 25 mM Tris-HCl buffer and PBS. Then, cells were lysed in
RIPA buffer by sonication, and subjected to centrifugation at
12,000 × g for 15 min. The supernatants were transferred to a new
tube for immunoprecipitation.
Immunoprecipitation was performed as described by
Lin et al(19). In each
1.5-ml tube, 2.5 mg of lysates was mixed with 2 μg of polyclonal
antibody against MAEL or preimmune rabbit IgG (Signalway Antibody
Co., Ltd., Nanjing, China), and incubated overnight at 4°C with
gentle shaking. Immune complexes were collected on protein A/G
agarose beads and washed three times to remove non-specific binding
using the same buffer as for the cell lyses. Proteins were eluted
from the beads by boiling the beads in loading buffer for 10 min,
and were then separated on SDS-PAGE gel and visualized with silver
staining.
Nano-LC-MS/MS
Each excised protein gel band was destained and
digested with trypsin as described by Lin et al(19). Nano-LC MS/MS experiment was
performed on an HPLC system composed of two LC-20AD nano-flow LC
pumps, an SIL-20AC autosampler and an LC-20AB micro-flow LC pump
(Shimadzu, Tokyo, Japan) connected to an LTQ Orbitrap mass
spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Sample
was loaded on a CapTrap column (0.5 × 2 mm; Michrom Bioresources,
Inc., Auburn, CA, USA) for 6 min at a flow rate of 25 μl/min. The
sample was subsequently separated by a C18 reverse-phase column
(0.10 × 150 mm, packed with 3 μm Magic C18AQ particles; Michrom
Bioresources) at a flow rate of 500 nl/min. The mobile phases were
2% acetonitrile with 0.1% formic acid (phase A and the loading
phase) and 95% acenitrile with 0.1% formic acid (phase B). To
achieve proper separation, a 60-min linear gradient from 0 to 80%
phase B was employed. The separated sample was introduced into the
mass spectrometer via an Advance 30-μm silica tip (Michrom
Bioresources). The spray voltage was set at 1.6 kV and the heated
capillary at 180°C. The mass spectrometer was operated in the
data-dependent mode, and each cycle of duty consisted of one
full-MS survey scan at the mass range 385–2000 Da with resolution
power of 100,000 using the Orbitrap section, followed by MS/MS
experiments for 10 strongest peaks using the LTQ section. The AGC
expectation during full-MS and MS/MS were 1,000,000 and 10,000,
respectively. Peptides were fragmented in the LTQ section using
collision-induced dissociation with helium and the normalized
collision energy value set at 35%. Previously fragmented peptides
were excluded for 60 sec.
Database search
Tandem mass spectra were extracted by BioWorks
version 3.3.1 SP1 (Thermo Fisher Scientific). All MS/MS samples
were analyzed using Sequest (Thermo Fisher Scientific; version 28).
Sequest was set up to search the human proteome database from
UniProt release 2010_08 (downloaded from the official website of
UniProtKB following this link: http://www.uniprot.org/uniprot/?query=organism:9606+keyword:181&format=*&compress=yes,
downloaded at 2010-07-13) assuming the digestion enzyme trypsin.
Sequest was searched with a fragment ion mass tolerance of 1.00 Da
and a parent ion tolerance of 20 ppm. Oxidation of methionine
(+15.99492 Da) and acetylation of lysine (+42.01057 Da) were
specified as variable modifications. Trans-Proteomic Pipeline
(20) was used to validate MS/MS
based peptide and protein identifications. Peptide probability was
specified by the Peptide Prophet algorithm (21). Protein probabilities were assigned
by the Protein Prophet algorithm (22). Proteins that contained similar
peptides and could not be differentiated based on MS/MS analysis
alone were grouped to satisfy the principles of parsimony.
Gene Ontology analysis
Gene Ontology (GO) enrichment analysis was performed
using the Database for Annotation, Visualization and Integrated
Discovery (DAVID) functional annotation tool (http://david.abcc.ncifcrf.gov) (23). UniProt accession numbers of
MAEL-associated proteins were submitted to DAVID system and
categorized based on biological process (BP), molecular function
(MF) and cellular component (CC) using the software.
Anti-tag co-immunoprecipitation
Transfection and co-IP were performed as previously
described (24). Hek293 cells were
co-transfected with HA-tagged MAEL expression plasmids and
expression plasmids of each Myc-tagged SG gene. At 24 h after
transfection, cell lysates were prepared and immunoprecipitated
with rabbit anti-HA polyclonal antibody or preimmune rabbit IgG,
and the precipitated protein was detected by western blot analysis
using the anti-Myc monoclonal antibody. The anti-HA, anti-Myc and
IgG antibodies were purchased from Santa Cruz Biotechnology (Santa
Cruz, CA, USA).
Immunofluorescence staining of SGs
SW480 cells were grown on glass coverslips, and
treated with 0.5 mM hydrogen peroxide (H2O2)
for 1 h to induce the formation of SGs. Treated cells and control
cells were fixed with 4% paraformaldehyde and permeabilized in 0.1%
Triton X-100, respectively. To reduce the unspecific binding, the
cells were incubated in 1% BSA overnight the 4°C. Then, cells were
incubated with anti-MAEL polyclonal antibody and anti-PABPC1
monoclonal antibody (Pierce, Rockford, IL, USA) for 2 h at room
temperature, followed by incubation with Texas Red-conjugated
anti-rabbit IgG (red) and FITC-conjugated anti-mouse IgG (green)
for 1 h. The nuclei were labeled by DAPI. Images were obtained
using a Zeiss LSM 510 confocal microscope (Carl Zeiss Imaging,
Oberkochen, Germany). Texas Red-conjugated anti-rabbit IgG and
FITC-conjugated anti-mouse IgG were purchased from Santa Cruz
Biotechnology.
Results
RNA-binding proteins are enriched in the
MAEL protein complex
To identify the potential interacting partners of
MAEL protein, we performed the co-IP experiments to isolate the
MAEL complex in human breast cancer MDA-MB-231 and colorectal
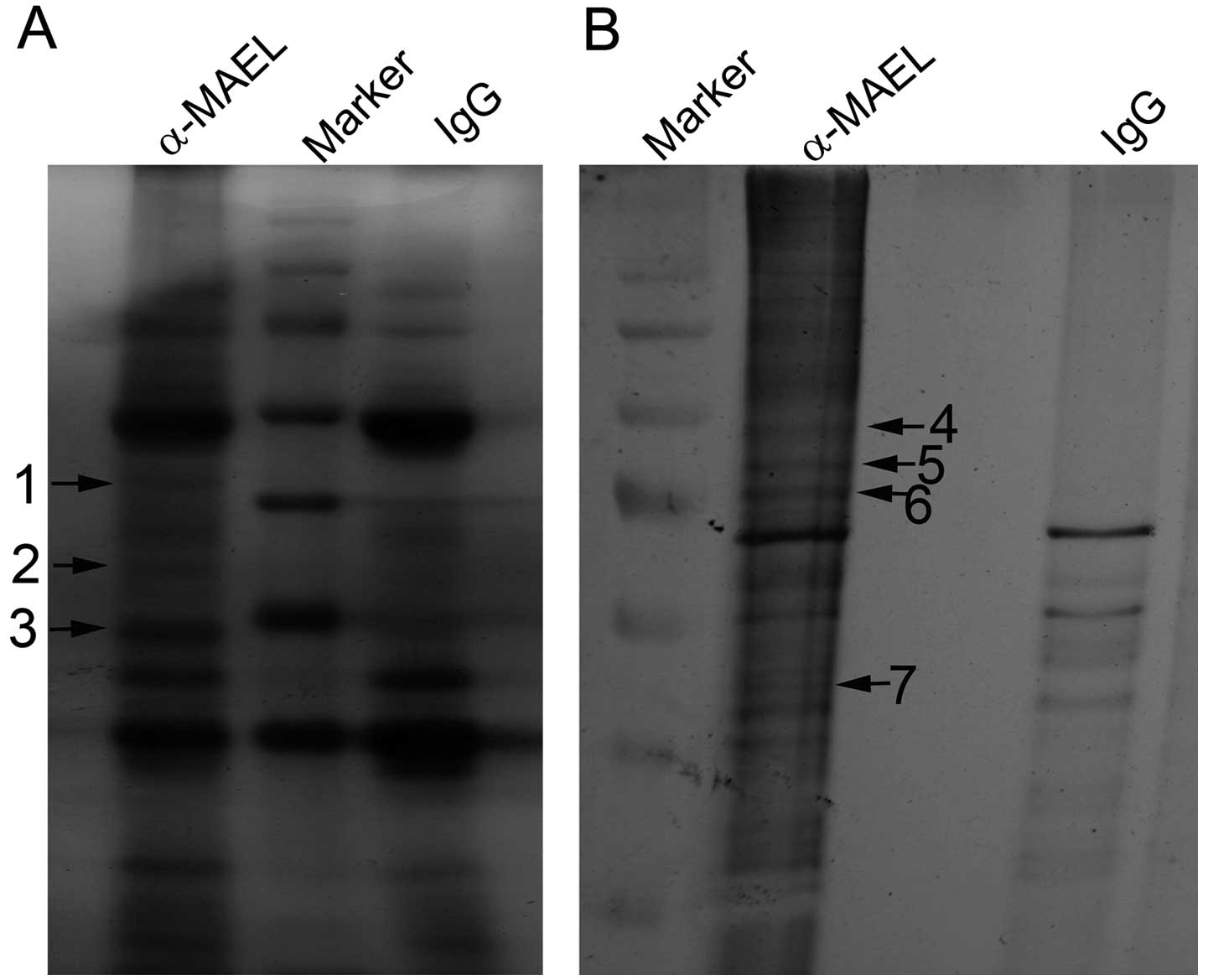
cancer SW480 cells. The immune complexes of the anti-MAEL antibody
and IgG were resolved on SDS-PAGE gel followed by silver staining.
The protein bands which only existed in the complex of the
anti-MAEL antibody were excised (Fig.
1), digested with trypsin and analyzed by Nano-LC MS/MS method.
All MS/MS samples were analyzed using Sequest. According to the
database search results, we identified 178 non-redundant proteins
in the three bands from MDA-MB-231 cells and 167 proteins in the
four bands from SW480 cells. Among these proteins, 78 proteins were
common to both MDA-MB-231 and SW480 cells.
To find whether these 78 proteins share some
particular features, we performed GO enrichment analysis with DAVID
(23). Table II shows the top 5 significantly
enriched terms in the MAEL-associated proteins. In the molecular
function (MF) category, the RNA binding term is the most
significant term (with the lowest P-value) and contains the largest
number of proteins (32.1% of the 77 proteins assigned to MF
category). In the cellular component (CC) category, the term with
the lowest P-value was the non-membrane-bounded organelle, and that
with the second lowest P-value was the ribonucleoprotein complex
term. Additionally, a large number of MAEL-associated proteins were
enriched in the nuclear lumen, consisting of 23 proteins (29.5% of
the 70 proteins assigned to CC category). In the biological process
(BP) category, the top three terms with the lowest P-value were RNA
process, chromatin assembly and macromolecular complex assembly.
When considering all three categories, we found that most
MAEL-associated proteins were RNA-binding proteins existing in the
ribonucleoprotein complex.
 | Table IITop 5 significantly enriched GO terms
in the MAEL-associated proteins. |
Table II
Top 5 significantly enriched GO terms
in the MAEL-associated proteins.
| Category | Term | Count | Percentage | Value |
|---|
| MF | RNA binding | 25 | 32.1 | 1.12E-12 |
| Structural
constituent of the cytoskeleton | 10 | 12.8 | 4.04E-10 |
| Structural molecule
activity | 15 | 19.2 | 1.57E-05 |
| Transcription
corepressor activity | 6 | 7.7 | 1.60E-03 |
| NAD or NADH
binding | 4 | 5.1 | 2.60E-03 |
| CC | Non-membrane-bound
organelle | 38 | 48.7 | 8.98E-10 |
| Ribonucleoprotein
complex | 18 | 23.1 | 1.28E-09 |
| Spliceosome | 11 | 14.1 | 1.98E-09 |
| Intermediate
filament | 10 | 12.8 | 5.62E-07 |
| Nuclear lumen | 23 | 29.5 | 4.10E-06 |
| BP | RNA splicing | 17 | 21.8 | 2.63E-12 |
| Chromatin
assembly | 7 | 8.9 | 6.84E-06 |
| Macromolecular
complex assembly | 11 | 14.1 | 9.58E-06 |
| Ectoderm
development | 8 | 10.3 | 1.14E-04 |
| Intermediate
filament-based process | 4 | 5.1 | 2.34E-04 |
MAEL interacts with stress granule
proteins
MAEL protein is a component of nuage (3,6,7), a
germline-unique ribonucleoprotein complex particle. According to
the above results (Table II), MAEL
associates with RNA-binding proteins, possibly a component of the
ribonucleoprotein complex in cancer cells. However, we did not find
other nuage components in the MAEL-associated proteins. Notably,
the MAEL-associated proteins contained 14 components of SGs: PABPC1
(25), FUBP1/FBP1 (26), KHSRP/FBP2 (26), YBX1/YB-1 (27), SYNCRIP/hnRNP Q (28), EIF3F (29), EIF4A1 (30), EIF4B (31), ELAVL1 (32), HNRNPA1 (33), HNRNPA2B1 (34), DDX1 (27), DDX3X (35) and DDX39 (36). Among these, 8 proteins (PABPC1,
FUBP1, KHSRP, YBX1, SYNCRIP, EIF4B, HNRNPA1 and HNRNPA2B1) were
identified from both MDA-MB-231 and SW480 cells (Table III); 6 proteins were found only in
MDA-MB-231 (EIF4A1, EIF3F and DDX3X) or SW480 cells (DDX39, ELAVL1
and DDX1) (data not shown).
 | Table IIIThe SG proteins identified in the
MAEL complexes from both MDA-MB-231 and SW480 cells. |
Table III
The SG proteins identified in the
MAEL complexes from both MDA-MB-231 and SW480 cells.
| | | MDA-MB-231
cells | SW480 cells |
|---|
| | |
|
|
|---|
| Accession no. | Protein | Official
symbol | Peptide
sequence | Percent
coverage | Peptide
sequence | Percent
coverage |
|---|
| P11940 | PABP1 | PABPC1 | YQGVNLYVK | 8.5 | NLDDGIDDER | 10.2 |
| | | GFGFVSFER | | PAAAAAAATPAVR | |
| | | ALDTM.NFDVIK | | NFGEDM.DDER | |
| | | SGVGNIFIK | | FGPALSVK | |
| | | EFSPFGTITSAK | | VDEAVAVLQAH | |
| Q96AE4 | FUBP1 | FUBP1 | IGGNEGIDVPIPR | 5.2 | GTPQQIDYAR | 17.6 |
| | | IAQITGPPDR | | IGGNEGIDVPIPR | |
| | | | |
SVQAGNPGGPGPGGR | |
| | | | |
QQAAYYAQTSPQGM.PQHPPAPQGQ | |
| | | | | NPPPNADPNMK | |
| | | | | IQIAPDSGGLPER | |
| | | | |
AQTSPQGMPQHPPAPQGQ | |
| | | | |
TSPQGM.PQHPPAPQGQ | |
| | | | | IAQITGPPDR | |
| | | | | LLDQIVEK | |
| | | | | ITGDPYK | |
| Q92945 | FUBP2 | KHSRP | IGGGIDVPVPR | 7.6 | IGGGIDVPVPR | 13.5 |
| | | M.M.LDDIVSR | | IINDLLQSLR | |
| | | VPDGM.VGLIIGR | | M.M.LDDIVSR | |
| | | SVSLTGAPESVQK | | SVSLTGAPESVQK | |
| | | DAFADAVQR | | VQISPDSGGLPER | |
| | | | | VPDGM.VGLIIGR | |
| | | | | DAFADAVQR | |
| | | | | MM.LDDIVSR | |
| | | | | M.LDDIVSR | |
| | | | | KDAFADAVQR | |
| | | | | TSM.TEEYR | |
| | | | | MMLDDIVSR | |
| P67809 | YBOX1 | YBX1 |
AADPPAENSSAPEAEQGGAE | 40.7 |
AADPPAENSSAPEAEQGGAE | 28.4 |
| | | EDVFVHQTAIK | |
GAEAANVTGPGGVPVQGSK | |
| | |
GAEAANVTGPGGVPVQGSK | | NGYGFINR | |
| | |
NDTKEDVFVHQTAIK | | EDVFVHQTAIK | |
| | | NGYGFINR | |
NDTKEDVFVHQTAIK | |
| | | NYQQNYQNSESGEK | | FPPYYM.R | |
| | |
GAEAANVTGPGGVPVQGSK | |
EDGNEEDKENQGDETQGQQPPQR | |
| | |
PGTTGSGAGSGGPGGLTSAAPAGGDK | | | |
| | | FPPYYM.R | | | |
| O60506 | HNRPQ | SYNCRIP | LM.M.DPLTGLNR | 7.1 | TGYTLDVTTGQR | 6.8 |
| | | TGYTLDVTTGQR | 7.1 | LM.M.DPLTGLNR | |
| | | | | LFVGSIPK | |
| P23588 | IF4B | EIF4B | VAPAQPSEEGPGR | 3.6 |
AASIFGGAKPVDTAAR | 6.3 |
| | | | | VAPAQPSEEGPGR | |
| | | | | GLNISAVR | |
| | | | |
AASIFGGAKPVDTAAR | |
| P09651 | ROA1 | HNRNPA1 | IEVIEIM.TDR | 3.7 | DYFEQYGK | 39.8 |
| | | | |
GFAFVTFDDHDSVDK | |
| | | | |
GFGFVTYATVEEVDAAM.NAR | |
| | | | | IEVIEIM.TDR | |
| | | | |
LFIGGLSFETTDESLR | |
| | | | |
NQGGYGGSSSSSSYGSGR | |
| | | | |
SSGPYGGGGQYFAKPR | |
| | | | | IGGLSFETTDESLR | |
| | | | | GGGFGGNDNFGR | |
| | | | | SFETTDESLR | |
| | | | | SESPKEPEQLR | |
| | | | | EDSQRPGAHLTVK | |
| | | | | LTDCVVM.R | |
| P22626 | ROA2 | HNRNPA2B1 | IDTIEIITDR | 11 |
GFGFVTFDDHDPVDK | 37.5 |
| | |
GGGGNFGPGPGSNFR | | IDTIEIITDR | |
| | | GGNFGFGDSR | |
LFIGGLSFETTEESLR | |
| | | | |
NM.GGPYGGGNYGPGGSGGSGGYGGR | |
| | | | | YHTINGHNAEVR | |
| | | | | DYFEEYGK | |
| | | | |
GGGGNFGPGPGSNFR | |
| | | | | GGNFGFGDSR | |
| | | | | YHTINGHNAEVR | |
| | | | | SFETTEESLR | |
| | | | | LTDCVVM.R | |
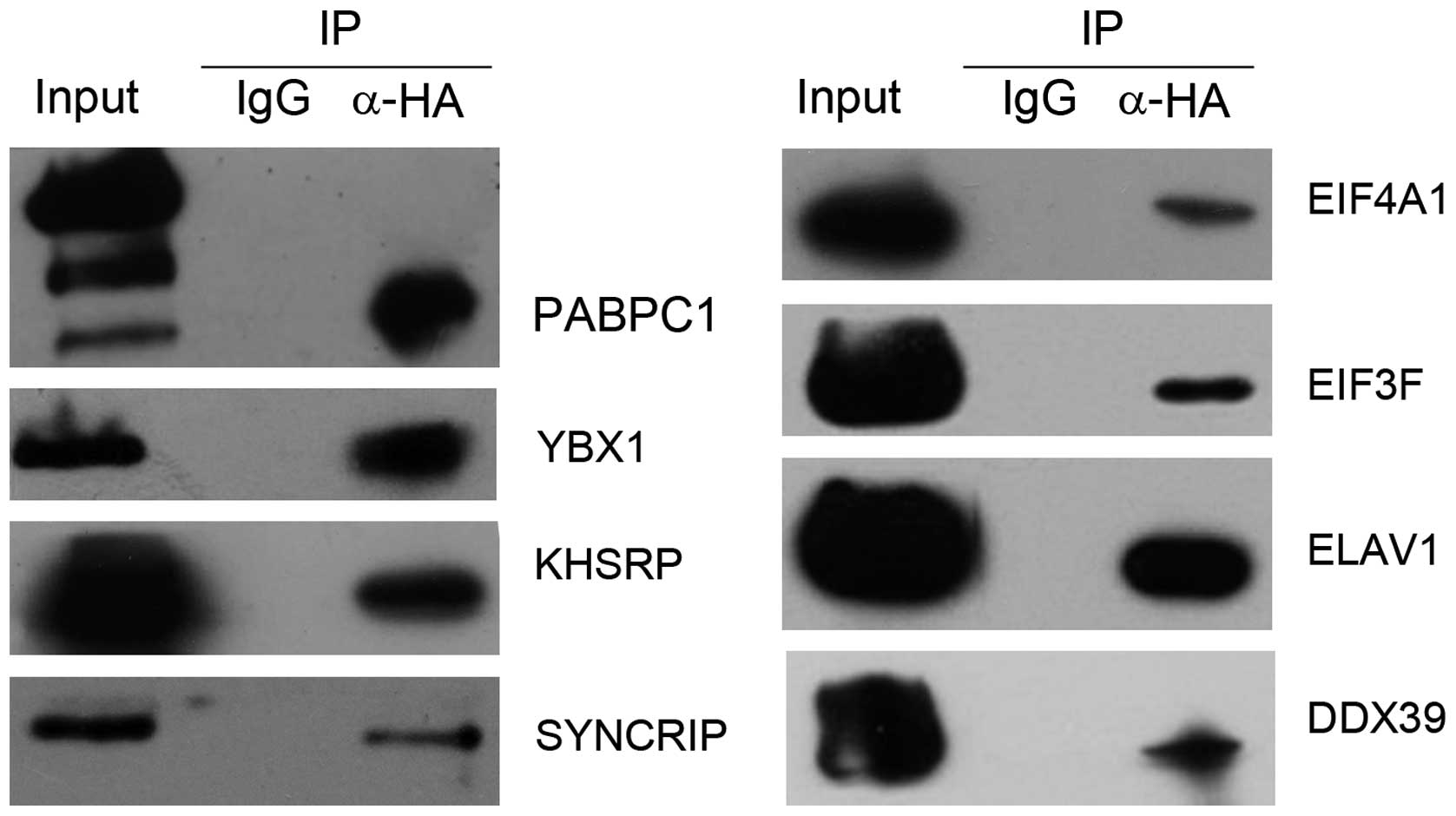
Then, 8 (PABPC1, KHSRP, YBX1, SYNCRIP, EIF4A1,
DDX39, ELAVL1 and EIF3F) of the identified SG proteins were
selected for further confirmation of their interactions with MAEL
protein by co-IP experiments. The coding regions of these 8 genes
were cloned into mammalian expression vector pCMV-Myc for
constructing Myc-tagged expression plasmids. The Myc-tagged
expression plasmid of each SG protein was co-transfected with
HA-tagged MAEL expression plasmid into Hek293 cells, respectively.
The anti-tag immunoprecipitation assays showed that all these 8 SG
proteins were able to interact with the MAEL protein (Fig. 2).
MAEL localizes to SGs
As stated above, MAEL protein interacts with SG
proteins. Therefore, we performed immunofluorescence analysis to
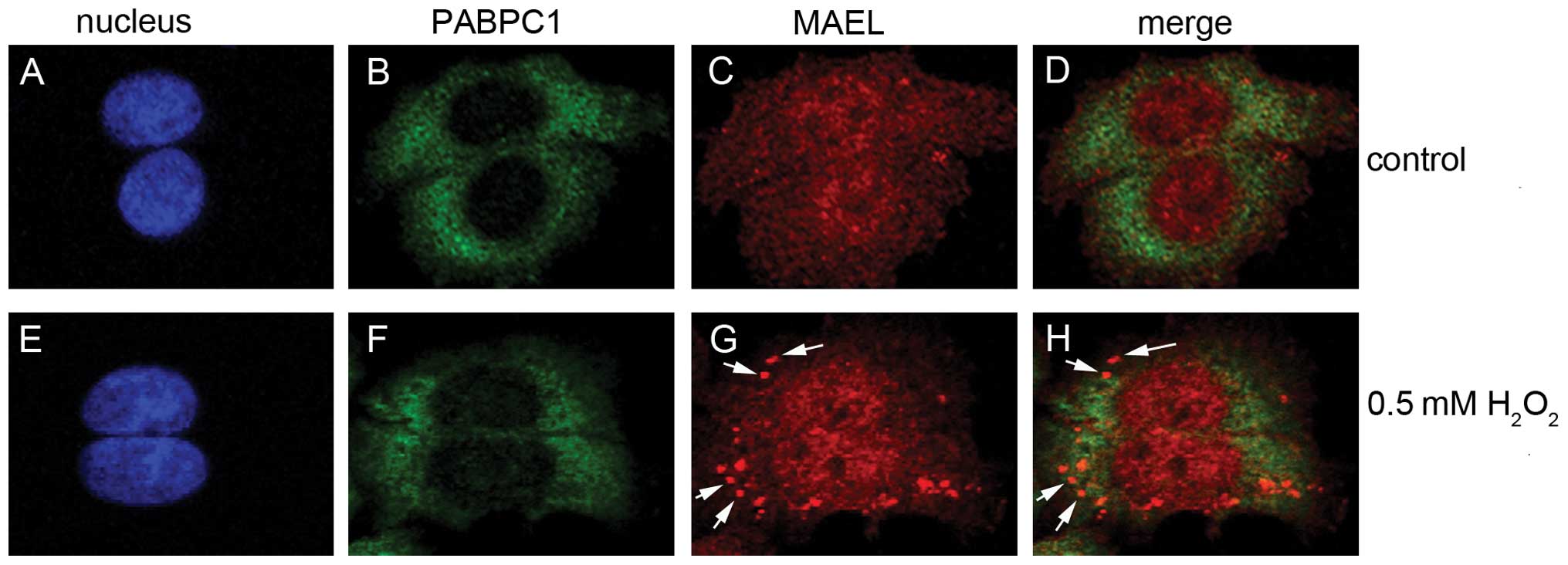
investigate whether the MAEL protein localizes in SGs. In the
untreated SW480 cells, MAEL proteins were distributed homogeneously
in both the cytoplasm and the nucleus (Fig. 3). After induction with
H2O2, some cytoplasmic MAEL proteins
displayed a distribution pattern of distinct speckles, and
co-localization with SG marker PABPC1 (25) in the speckle (Fig. 3), suggesting that MAEL proteins
localize to SGs. Strangely, after treatment with
H2O2, PABPC1 did not condense into apparent
speckles as it did in the cells treated with arsenite (37).
Discussion
Our previous study demonstrated that the human MAEL
gene is only expressed in the spermatocytes and spermatids of the
testis under normal condition, but is aberrantly expressed in
various types of human cancer cells (12). The role of MAEL in spermatogenesis
has been studied thoroughly, but no investigation on its function
in cancer has been performed. In the present study, we identified
178 MAEL-associated proteins in breast cancer MDA-MB-231 cells and
167 in colorectal cancer SW480 cells by immunoprecipitation and
Nano-LC-MS/MS analysis. In these MAEL-associated proteins, we found
14 components of SGs, but none of the nuage. The protein
interactions between MAEL and these stress granule proteins were
confirmed by anti-tag immunoprecipitation. Immunofluorescence
analysis showed that MAEL co-localizes with PABPC1 in SGs during
oxidative stress. This is not surprising, given that both SGs and
nuage are cytoplasmic ribonucleoprotein complex particles (RNA
granules). Nuage is a germline-unique structure (4), whereas SGs are somatic RNA granules
and are assembled when cells are exposed to stress (25,38,39).
However, both nuage and SGs have similar composition, and serve as
sites for small RNA-mediated gene silencing (40). MAEL has been found predominantly in
the nuage of germline cells, where it plays an indispensable role
in piRNA biogenesis and piRNA-mediated silencing of transposons
(3,6,7).
Therefore, we hypothesized that MAEL plays a role in miRNA-mediated
gene silencing in SGs, as it does in the nuage (7).
Additionally, we demonstrated that in cancer cells,
the MAEL protein is distributed in both the cytoplasm and the
nucleus, similar to its localization in germline cells (3,7). In
the nucleus, MAEL functions as a regulator of gene expression. For
example, mouse MAEL was found to interact with chromatin-remodeling
factors SNF5 and SIN3B in the testis (6); Drosophila MAEL represses the
transcription of miR-7 (11). In
accordance with the nuclear function of MAEL in germline cells,
Gene Ontology annotation showed that some MAEL-associated proteins
in cancer cells are involved in chromatin remodeling and
transcription repression.
Our results suggest that MAEL plays a similar role
in germline and cancer cells, similar to most CT genes. For
example, GAGE functions as a regulator of chromatin reorganization
in both germ and cancer cells (41). To date, 156 families of CT genes
have been collected in the CT database (http://www.cta.lncc.br). The discovery of CT genes led
to the hypothesis that gametogenesis and carcinogenesis share a
similar mechanism; the expression of germline genes in cancer
reflects the activation of the silenced gametogenesis programme in
somatic cells; this programme might contribute characteristic
features to the neoplastic phenotype, including immortality,
invasiveness, hypomethylation and metastatic capacity (42). Costa et al(43) believed that CT genes play a role in
stem cell self-renewal; the expression of CT genes in tumor tissues
contributes to maintaining stem cell properties and favors tumor
proliferation. This hypothesis was supported by Yamada et
al(44) who found that
considerable numbers of CT genes showed preferential expression in
cancer stem-like cells. Previous studies have shown that MAEL is
necessary for proper germline stem cell lineage differentiation
(11) and knockdown of MAEL
expression disrupts the differentiation of mouse embryonic stem
cells into germ cells (45).
Therefore, whether MAEL also plays a role in cancer stem cells
warrants further investigation.
In summary, we used proteomic approaches for
isolating the interacting partners of MAEL, and demonstrated that
MAEL, a component of nuage in germline cells, localizes in SGs and
interacts with several components of SGs, suggesting that MAEL
plays a role in carcinogenesis by post-transcriptionally regulating
gene expression, as it does in gametogenesis. This finding should
be valuable toward understanding the function of MAEL in
carcinogenesis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81272318 and
81071656).
References
|
1
|
Clegg NJ, Frost DM, Larkin MK,
Subrahmanyan L, Bryant Z and Ruohola-Baker H: maelstrom is
required for an early step in the establishment of
Drosophila oocyte polarity: posterior localization of
grk mRNA. Development. 124:4661–4671. 1997.PubMed/NCBI
|
|
2
|
Clegg NJ, Findley SD, Mahowald AP and
Ruohola-Baker H: Maelstrom is required to position the MTOC in
stage 2–6 Drosophila oocytes. Dev Genes Evol. 211:44–48.
2001.PubMed/NCBI
|
|
3
|
Findley SD, Tamanaha M, Clegg NJ and
Ruohola-Baker H: Maelstrom, a Drosophila
spindle-class gene, encodes a protein that colocalizes with
Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development.
130:859–871. 2003. View Article : Google Scholar
|
|
4
|
Pek JW, Patil VS and Kai T: piRNA pathway
and the potential processing site, the nuage, in the
Drosophila germline. Dev Growth Differ. 54:66–77. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lim AK and Kai T: Unique germ-line
organelle, nuage, functions to repress selfish genetic elements in
Drosophila melanogaster. Proc Natl Acad Sci USA.
104:6714–6719. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa Y, Speed RM, Gautier P, et al: Mouse
MAELSTROM: the link between meiotic silencing of unsynapsed
chromatin and microRNA pathway? Hum Mol Genet. 15:2324–2334. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Soper SF, van der Heijden GW, Hardiman TC,
et al: Mouse maelstrom, a component of nuage, is essential for
spermatogenesis and transposon repression in meiosis. Dev Cell.
15:285–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aravin AA, van der Heijden GW, Castaneda
J, Vagin VV, Hannon GJ and Bortvin A: Cytoplasmic
compartmentalization of the fetal piRNA pathway in mice. PLoS
Genet. 5:e10007642009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yokota S: Nuage proteins: their
localization in subcellular structures of spermatogenic cells as
revealed by immunoelectron microscopy. Histochem Cell Biol.
138:1–11. 2012. View Article : Google Scholar
|
|
10
|
Takebe M, Onohara Y and Yokota S:
Expression of MAEL in nuage and non-nuage compartments of rat
spermatogenic cells and colocalization with DDX4, DDX25 and MIWI.
Histochem Cell Biol. 140:169–181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pek JW, Lim AK and Kai T:
Drosophila maelstrom ensures proper germline stem cell
lineage differentiation by repressing microRNA-7. Dev Cell.
17:417–424. 2009. View Article : Google Scholar
|
|
12
|
Xiao L, Wang Y, Zhou Y, et al:
Identification of a novel human cancer/testis gene MAEL that is
regulated by DNA methylation. Mol Biol Rep. 37:2355–2360. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Janic A, Mendizabal L, Llamazares S,
Rossell D and Gonzalez C: Ectopic expression of germline genes
drives malignant brain tumor growth in Drosophila. Science.
330:1824–1827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hofmann O, Caballero OL, Stevenson BJ, et
al: Genome-wide analysis of cancer/testis gene expression. Proc
Natl Acad Sci USA. 105:20422–20427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De Smet C and Loriot A: DNA
hypomethylation and activation of germline-specific genes in
cancer. Adv Exp Med Biol. 754:149–166. 2013.PubMed/NCBI
|
|
16
|
Kim R, Kulkarni P and Hannenhalli S:
Derepression of cancer/testis antigens in cancer is associated with
distinct patterns of DNA hypomethylation. BMC Cancer. 13:1442013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim YH, Lee HC, Kim SY, et al: Epigenomic
analysis of aberrantly methylated genes in colorectal cancer
identifies genes commonly affected by epigenetic alterations. Ann
Surg Oncol. 18:2338–2347. 2011. View Article : Google Scholar
|
|
18
|
Landthaler M, Gaidatzis D, Rothballer A,
et al: Molecular characterization of human Argonaute-containing
ribonucleoprotein complexes and their bound target mRNAs. RNA.
14:2580–2596. 2008. View Article : Google Scholar
|
|
19
|
Lin Z, Crockett DK, Lim MS and
Elenitoba-Johnson KS: High-throughput analysis of protein/peptide
complexes by immunoprecipitation and automated LC-MS/MS. J Biomol
Tech. 14:149–155. 2003.PubMed/NCBI
|
|
20
|
Keller A, Eng J, Zhang N, Li XJ and
Aebersold R: A uniform proteomics MS/MS analysis platform utilizing
open XML file formats. Mol Syst Biol. 1:2005.0017. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Keller A, Nesvizhskii AI, Kolker E and
Aebersold R: Empirical statistical model to estimate the accuracy
of peptide identifications made by MS/MS and database search. Anal
Chem. 74:5383–5392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nesvizhskii AI, Keller A, Kolker E and
Aebersold R: A statistical model for identifying proteins by tandem
mass spectrometry. Anal Chem. 75:4646–4658. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
24
|
Zhou J, Qiao X, Xiao L, et al:
Identification and characterization of the novel protein CCDC106
that interacts with p53 and promotes its degradation. FEBS Lett.
584:1085–1090. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kedersha N, Stoecklin G, Ayodele M, et al:
Stress granules and processing bodies are dynamically linked sites
of mRNP remodeling. J Cell Biol. 169:871–884. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rothe F, Gueydan C, Bellefroid E, Huez G
and Kruys V: Identification of FUSE-binding proteins as interacting
partners of TIA proteins. Biochem Biophys Res Commun. 343:57–68.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Onishi H, Kino Y, Morita T, Futai E,
Sasagawa N and Ishiura S: MBNL1 associates with YB-1 in cytoplasmic
stress granules. J Neurosci Res. 86:1994–2002. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Quaresma AJ, Bressan GC, Gava LM, Lanza
DC, Ramos CH and Kobarg J: Human hnRNP Q re-localizes to
cytoplasmic granules upon PMA, thapsigargin, arsenite and
heat-shock treatments. Exp Cell Res. 315:968–980. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wen F, Zhou R, Shen A, Choi A, Uribe D and
Shi J: The tumor suppressive role of eIF3f and its function in
translation inhibition and rRNA degradation. PLoS One.
7:e341942012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wolf A, Krause-Gruszczynska M, Birkenmeier
O, Ostareck-Lederer A, Huttelmaier S and Hatzfeld M: Plakophilin 1
stimulates translation by promoting eIF4A1 activity. J Cell Biol.
188:463–471. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Buchan JR, Yoon JH and Parker R:
Stress-specific composition, assembly and kinetics of stress
granules in Saccharomyces cerevisiae. J Cell Sci.
124:228–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gallouzi IE, Brennan CM, Stenberg MG, et
al: HuR binding to cytoplasmic mRNA is perturbed by heat shock.
Proc Natl Acad Sci USA. 97:3073–3078. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guil S, Long JC and Caceres JF: hnRNP A1
relocalization to the stress granules reflects a role in the stress
response. Mol Cell Biol. 26:5744–5758. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim HJ, Kim NC, Wang YD, et al: Mutations
in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem
proteinopathy and ALS. Nature. 495:467–473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK
and Wu Lee YH: Critical roles of RNA helicase DDX3 and its
interactions with eIF4E/PABP1 in stress granule assembly and stress
response. Biochem J. 441:119–129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goodier JL, Zhang L, Vetter MR and
Kazazian HH Jr: LINE-1 ORF1 protein localizes in stress granules
with other RNA-binding proteins, including components of RNA
interference RNA-induced silencing complex. Mol Cell Biol.
27:6469–6483. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wippich F, Bodenmiller B, Trajkovska MG,
Wanka S, Aebersold R and Pelkmans L: Dual specificity kinase DYRK3
couples stress granule condensation/dissolution to mTORC1
signaling. Cell. 152:791–805. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Anderson P and Kedersha N: Stress
granules: the Tao of RNA triage. Trends Biochem Sci. 33:141–150.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Decker CJ and Parker R: P-bodies and
stress granules: possible roles in the control of translation and
mRNA degradation. Cold Spring Harb Perspect Biol. 4:a0122862012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Anderson P and Kedersha N: RNA granules:
post-transcriptional and epigenetic modulators of gene expression.
Nat Rev Mol Cell Biol. 10:430–436. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gjerstorff MF, Rosner HI, Pedersen CB, et
al: GAGE cancer-germline antigens are recruited to the nuclear
envelope by germ cell-less (GCL). PLoS One. 7:e458192012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Simpson AJ, Caballero OL, Jungbluth A,
Chen YT and Old LJ: Cancer/testis antigens, gametogenesis and
cancer. Nat Rev Cancer. 5:615–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Costa FF, Le Blanc K and Brodin B: Concise
review: cancer/testis antigens, stem cells, and cancer. Stem Cells.
25:707–711. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamada R, Takahashi A, Torigoe T, et al:
Preferential expression of cancer/testis genes in cancer stem-like
cells: proposal of a novel sub-category, cancer/testis/stem gene.
Tissue Antigens. 81:428–434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bahena I, Xu E, Betancourt M, et al: Role
of Mael in early oogenesis and during germ-cell
differentiation from embryonic stem cells in mice in vitro.
Zygote. 1–8. 2013.[Epub ahead of print].
|