Introduction
According to global cancer statistics, the morbidity
and mortality rates of digestive tumors rank first in both male and
female populations (1). The
etiology and pathogenesis of digestive tumors have a lot in common.
To prevent digestive tumors, it is necessary to understand the
related predisposing factors. Studies have shown that environmental
factors, diet, intake of non-steroidal and anti-inflammatory drugs,
and endogenous factors can significantly affect the individual
susceptibility to digestive tumors (2,3).
MicroRNAs (miRNAs) are a group of small non-coding
RNAs that are ~22 (18–25) nucleotides (nt) long. They have been
found to be associated with a variety of diseases, including
cancer. An increasing number of findings confirm that miRNAs play
essential roles in the development, but also the diagnosis,
treatment and prognosis of a variety of tumors. The value of using
miRNAs as biomarkers for diagnosis and as target molecules for
treatment of cancer is increasingly being recognized (4).
Numerous studies have shown that miR-146a is
involved in the development, apoptosis, invasion and metastasis of
digestive tumors; studies have reported that miR-146a is
downregulated in gastric cancer or gastric cancer cells and that it
regulates cell proliferation and apoptosis of gastric cancer cells
(5). In addition, Vinci et al
(6) studied the distribution of
sequence variants of miR-146a in colorectal cancer and the effects
of miRNA expression. He et al(7) reported that miR-146a modulated
TGF-β1-induced hepatic stellate cell proliferation by targeting
SMAD4. Tomokuni et al(8)
demonstrated that miR-146a inhibited the anticancer effect of IFN-α
in hepatocellular cancer (HCC) cells, and that this effect was
mediated by SMAD4. Another study found that the expression of
miR-146a inhibited the invasive capacity of pancreatic cancer cells
with concomitant downregulation of EGFR and the NF-κB regulatory
kinase interleukin 1 receptor-associated kinase 1 (9). Therefore, miR-146a appears to play a
crucial role in the properties of digestive tumors.
Studies have shown that single nucleotide
polymorphisms (SNPs) in human miRNAs constitute one of the main
forms of genetic variation in human genomic DNA sequences and that
they might play central roles in the susceptibility to human
disease. miRNA SNPs exhibit interindividual differences that are
relevant to disease diagnosis, treatment and prognosis.
A number of recent studies have suggested that the
miR-146a expression is deregulated in numerous solid tumors, and it
has become evident that miR-146a might act as a tumor-suppressor
(5,10,11).
The miR-146a rs2910164 G>C polymorphism is caused by the
miR-146a leader sequence G:U and C:U base pair mismatching. Studies
have shown that the miR-146a gene polymorphism rs2910164 is
associated with the occurrence of a variety of cancer types, such
as prostate, breast and cervical cancer (12–14).
From 2008 to 2013, researchers have repeatedly reported
associations between the miR-146a rs2910164 polymorphism and the
risk for digestive tumors, but the results were mixed or even
conflicting. Therefore, we performed a meta-analysis to derive a
more precise estimation of the association between the miR-146a G/C
SNP and the risks of developing cancer in the digestive system.
Materials and methods
Screening and identification of relevant
studies
Identification and eligibility of relevant studies
was performed using the search terms ‘miR/microRNA-146a’,
‘digestive cancer’, ‘biliary cancer’, ‘hepatocellular cancer’,
‘esophageal squamous cell carcinoma’, ‘gastric cancer’, ‘colorectal
cancer’, ‘pancreatic cancer’, ‘rs2910164’, ‘genotype’,
‘polymorphism’ and ‘variant’ in the PubMed, Ovid and Embase
databases and in the Cochrane Library (last search update: 17
April, 2013). The search was limited to English language articles
and only published studies with full-text articles were included.
We evaluated potentially relevant publications by manually
examining their title and abstract.
Inclusion and exclusion criteria
Inclusion criteria were: i) assessment of miR-146a
rs2910164 polymorphism and the risk of suffering from digestive
cancer; ii) a separate case-control study on humans; iii)
statistically sound genotype data with odds ratio (OR) values and
95% confidence intervals (CI); iv) full-text search. Exclusion
criteria were: i) lack of controls in the studies; ii) repetition
of previous results; iii) summary, comment, review and editorial
articles; iv) a focus on benign tumors of the digestive tract.
Data extraction and study
characteristics
Two researchers (Xiaohui Xu and Xiaodong Yang)
independently extracted all data that met the above inclusion
criteria and the existing differences in the resulting datasets
were resolved by team discussions. From each study, the following
information was extracted: last name of the first author, year of
publication, ethnicity, tumor type, source of research, research
methods, the number of cases and controls, the number of various
genotypes of cases and controls. If a study did not provide
complete data, we sent requests for this information to the
corresponding author. A total of 21 eligible studies, comprising
10,318 cases and 12,478 controls met the inclusion criteria
(Table I). The studies were
published in the period from 2008 to 2013, and all were
case-control studies. The case groups were only suffering from one
type of cancer (gastrointestinal), while the control groups did not
present with any tumor. From these 21 studies, 16 involved
individuals of Asian ethnicity and 5 of Caucasian. In addition, the
21 studies used different detection methods, with 14 studies using
the traditional method of polymerase chain reaction-restriction
fragment length polymorphism (PCR-RFLP), 1 using the PCR
confronting two-pair primer (PCR-CTPP) method, 4 using the
TaqMan-polymerase chain reaction (TaqMan-PCR) and another 2 using a
SNP assay. All the control groups were selected from a healthy
population and the age and gender were consistent with cases. All
the above data complied with the Hardy-Weinberg equilibrium (HWE)
principles (Table I).
 | Table ICharacteristics of the 21 studies
included in the meta-analysis. |
Table I
Characteristics of the 21 studies
included in the meta-analysis.
| Author (ref.) | Year | Ethnicity | Tumor type | Study design | Genotyping
method | No. of cases | No. of
controls | Cases | Controls | HWE p-value |
|---|
|
|
|---|
| CC | CG | GG | CC | CG | GG |
|---|
| Xu et
al(16) | 2008 | Asian | HCC | PB | PCR-RFLP | 479 | 504 | 158 | 241 | 80 | 197 | 245 | 58 | 0.12 |
| Ye et
al(17) | 2008 | Caucasian | ESCC | PB | SNPlex assay | 346 | 346 | 231 | 101 | 14 | 229 | 105 | 12 | 0.71 |
| Srivastava et
al(18) | 2010 | Asian | GBC | PB | PCR-RFLP | 230 | 224 | 11 | 90 | 129 | 5 | 81 | 138 | 0.08 |
| Guo et
al(19) | 2010 | Asian | ESCC | PB | SNPshot | 444 | 468 | 20 | 190 | 234 | 42 | 220 | 206 | 0.12 |
| Zeng et
al(20) | 2010 | Asian | GC | HB | PCR-RFLP | 304 | 304 | 89 | 153 | 62 | 119 | 132 | 53 | 0.12 |
| Min et
al(21) | 2012 | Asian | CRC | PB | PCR-RFLP | 446 | 502 | 151 | 233 | 62 | 188 | 245 | 69 | 0.443 |
| Akkiz et
al(22) | 2011 | Caucasian | HCC | PB | PCR-RFLP | 222 | 222 | 10 | 75 | 137 | 11 | 67 | 144 | 0.38 |
| Hishida et
al(23) | 2011 | Asian | GC | HB | PCR-CTPP | 583 | 1,637 | 230 | 271 | 82 | 633 | 825 | 229 | 0.12 |
| Okubo et
al(24) | 2010 | Asian | GC | HB | PCR-RFLP | 552 | 697 | 236 | 243 | 73 | 322 | 254 | 121 | 0.28 |
| Zhang et
al(25) | 2011 | Asian | HCC | HB | PCR-RFLP | 925 | 840 | 319 | 450 | 156 | 303 | 386 | 151 | 0.149 |
| Xiang et
al(26) | 2012 | Asian | HCC | PB | PCR-RFLP | 100 | 100 | 28 | 45 | 27 | 33 | 46 | 21 | 0.51 |
| Kim et
al(27) | 2012 | Asian | HCC | PB | PCR-RFLP | 159 | 201 | 57 | 88 | 14 | 74 | 103 | 24 | 0.19 |
| Zhou et
al(28) | 2012 | Asian | GC | HB | TaqMan | 1,686 | 1,895 | 286 | 822 | 578 | 393 | 951 | 551 | 0.64 |
| Ahn et
al(29) | 2012 | Asian | GC | HB | PCR-RFLP | 461 | 477 | 159 | 231 | 71 | 164 | 221 | 62 | 0.36 |
| Hezova et
al(30) | 2012 | Caucasian | CRC | HB | TaqMan | 212 | 197 | 9 | 79 | 124 | 12 | 70 | 115 | 0.41 |
| Mihalache et
al(31) | 2012 | Caucasian | CCA | HB | TaqMan | 182 | 350 | 11 | 53 | 118 | 17 | 122 | 211 | 0.91 |
| Zhou et
al(32) | 2012 | Asian | HCC | HB | PCR-RFLP | 186 | 483 | 67 | 86 | 33 | 158 | 254 | 71 | 0.056 |
| Song et
al(33) | 2013 | Asian | GC | HB | PCR-RFLP | 1,208 | 1,166 | 423 | 586 | 199 | 344 | 615 | 207 | 0.87 |
| Ma et
al(34) | 2013 | Asian | CRC | HB | TaqMan | 1,147 | 1,203 | 169 | 534 | 444 | 192 | 614 | 397 | 0.075 |
| Lv et
al(35) | 2013 | Asian | CRC | HB | PCR-RFLP | 353 | 540 | 47 | 230 | 54 | 143 | 274 | 96 | 0.08 |
| Pavlakis et
al(36) | 2013 | Caucasian | PC | HB | PCR-RFLP | 93 | 122 | 51 | 38 | 4 | 79 | 39 | 4 | 0.76 |
Statistical analysis
According to the case and control genotype
frequencies, the correlation between the miR-146a rs2910164
polymorphism and the risk of gastrointestinal cancer was assessed
via OR values with 95% CI. In addition, we analyzed whether tumor
type and ethnicity may affect the relationship between the miR-146a
rs2910164 polymorphism and the risk of gastrointestinal cancer.
Statistical analysis was performed on OR values (with 95% CI) for 5
distinct genotypic comparisons: the allelic one (G vs. C),
comparison to the dominant genetic model (GC + GG vs. CC),
comparison to the recessive genetic model (GG vs. GC + CC), the
homozygote (GG vs. CC) and the heterozygote comparison (GC vs. CC).
The Chi-square-based Q statistic was used to assess heterogeneity
between studies, with a p-value (Pheterogeneity)
<0.05 considered to indicate statistically significant
heterogeneity between studies. The I2 index, expressed
as a percentage, quantified the degree of heterogeneity throughout
the study, with I2 values of 25, 50 and 75% referring to
low, medium and high heterogeneity, respectively. Funnel plots were
used to assess the publication bias. When the effects were assumed
to be homogenous, the fixed-effects model was used (Mantel-Haenszel
method). If inter-study heterogeneity was detected, the
random-effects model was applied (DerSimonian and Laird method)
(15). All data analyses were
performed using the software Stata 11.0 and all the p-values are
derived two-sided tests.
Results
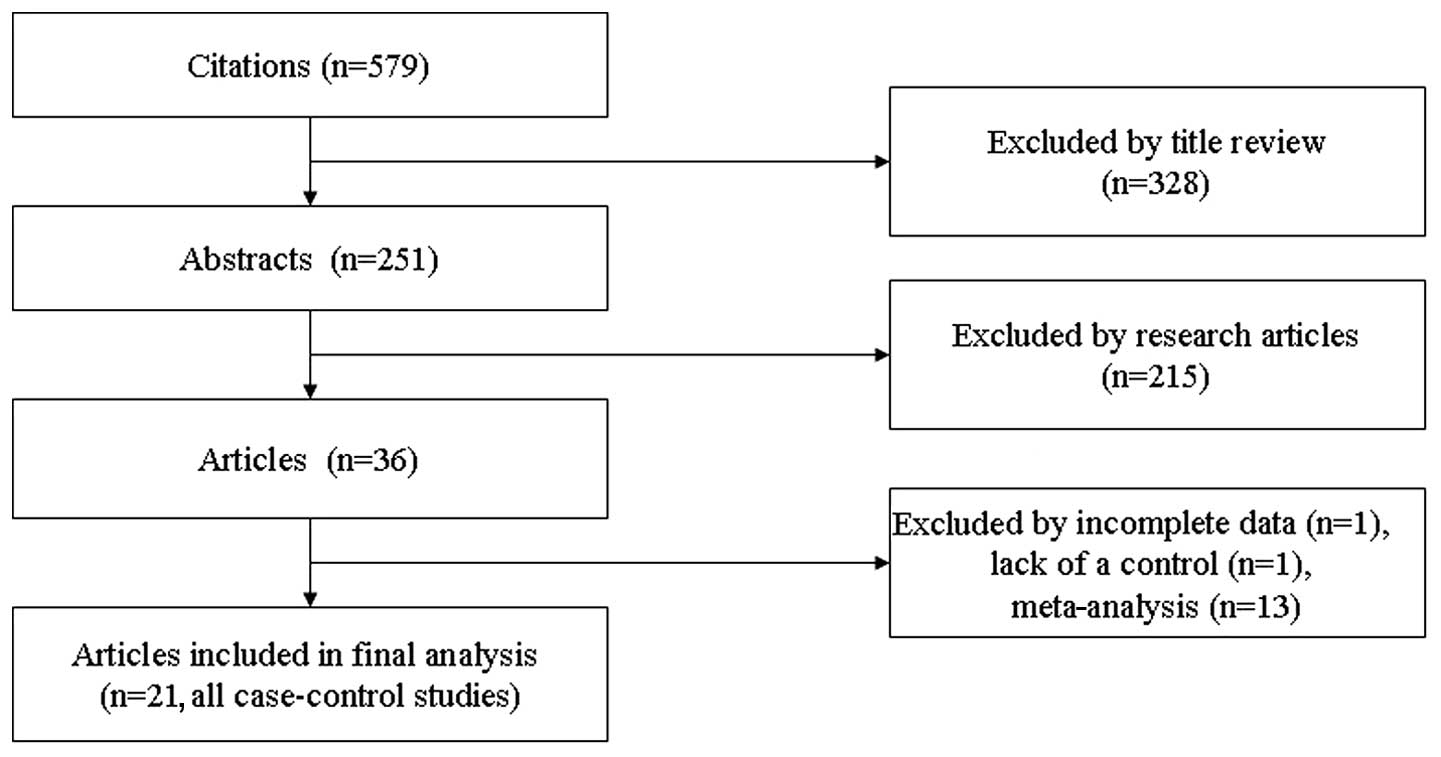
Study characteristics
A total of 545 articles relevant to the used search
terms were identified, and only 32 studies concerned the
association between digestive cancer and the miR-146a rs2910164
polymorphism. According to the inclusion and exclusion criteria
described in Materials and methods, 21 publications (16–36)
were included in the final meta-analysis, 8 using population-based
controls and 13 using hospital-based controls (Fig. 1). From the 21 publications, 2
concerned esophageal, 4 colorectal, 6 gastric and 6 hepatocellular
cancer. The main characteristics of the studies included in the
meta-analysis are summarized in Table
I.
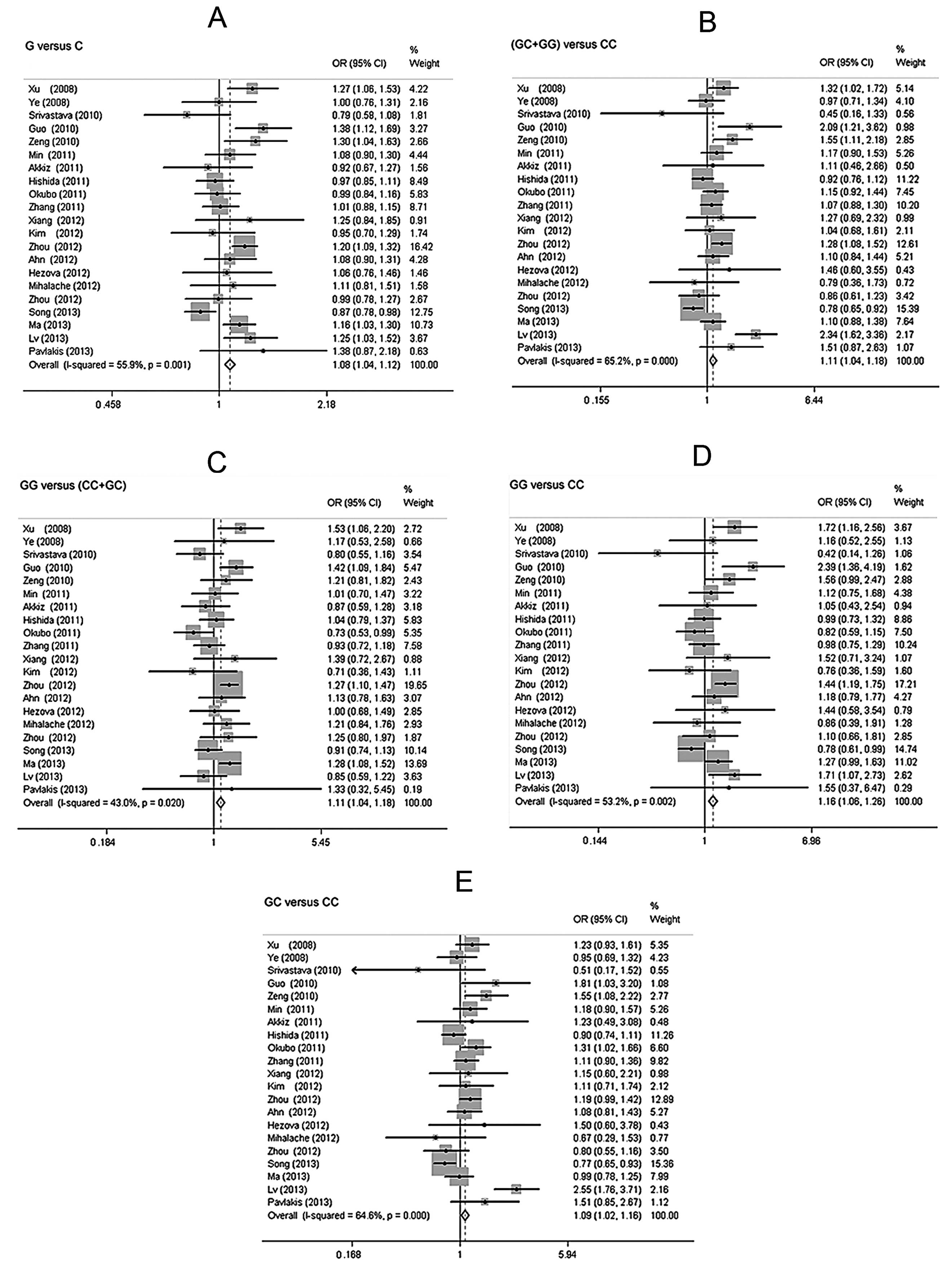
Overall analyses
The overall analysis of all cases revealed a
statistically significant positive association between the miR-146a
rs2910164 polymorphism and the risk of developing digestive tumors
(Fig. 2).
Allele G vs. C: OR=1.08, 95% CI: 1.04–1.12,
Pheterogeneity=0.001 (Fig.
2A). The results suggested that individuals with the G allele
were more susceptible to digestive cancer than those with the C
allele.
Dominant genetic model GC + GG vs. CC: OR=1.11, 95%
CI: 1.04–1.18, Pheterogeneity<0.001 (Fig. 2B). The results suggested that
individuals following the dominant genetic model GC + GG may show
an increased susceptibility to digestive cancer.
Recessive genetic model GG vs. CC + GC: OR=1.11, 95%
CI: 1.04–1.18, Pheterogeneity=0.020 (Fig. 2C). The results suggested that
compared to CC + GC, individuals following the recessive genetic
model GG were more susceptible to digestive cancer.
Homozygous GG vs. CC: OR=1.16, 95% CI: 1.06–1.26,
Pheterogeneity=0.002 (Fig.
2D). The results suggested that individuals following the
homozygous GG model showed no significant difference compared to
individuals with the CC allele with regards to developing digestive
cancer.
Heterozygous GC vs. CC: OR=1.09, 95% CI: 1.02–1.16,
Pheterogeneity<0.001 (Fig. 2E). The results suggested that
individuals following the heterozygous model GC were more
susceptible to digestive cancer than those with the CC allele.
Furthermore, in the stratified analysis exploring
the contributions from the type of digestive cancer (Table II), significantly increased risks
for esophageal squamous cell carcinoma were found [(for G vs. C:
1.23 (1.04–1.44), Pheterogeneity=0.054; for GG vs. CC +
GC: 1.39 (1.09–1.78), Pheterogeneity=0.655; for GG vs.
CC: 1.88 (1.19–2.97), Pheterogeneity=0.145)], as well as
risks for colorectal cancer [(for G vs. C: 1.15 (1.06–1.25),
Pheterogeneity=0.697; for (GC + GG) vs. CC: 1.31
(1.12–1.52), Pheterogeneity=0.005, for GG vs. CC + GC:
1.14 (1.00–1.31), Pheterogeneity=0.16; for GG vs. CC:
1.30 (1.08–1.57), Pheterogeneity=0.587, for GC vs. CC:
1.28 (1.09–1.50), Pheterogeneity<0.001)], but not for
biliary, hepatocellular, gastric and pancreatic cancer.
 | Table IISubgroup analysis of the association
between the miR-146a rs2910164 polymorphism and the risk of
digestive cancer. |
Table II
Subgroup analysis of the association
between the miR-146a rs2910164 polymorphism and the risk of
digestive cancer.
| Comparison | Subgroup | No.a | OR (95% CI) | I2
(%) | P-valueb |
|---|
| G vs. C | Ethnicity |
| Asian | 16 | 1.08
(1.04–1.13) | 65.1 | <0.001 |
| Caucasian | 5 | 1.05
(0.91–1.21) | 0.0 | 0.701 |
| Digestive cancer
type |
| Biliary
cancer | 2 | 0.94
(0.75–1.17) | | |
| Hepatocellular
cancer | 6 | 1.06
(0.97–1.16) | 23.4 | 0.258 |
| Esophageal
squamous cell carcinoma | 2 | 1.23
(1.04–1.44) | 70.8 | 0.054 |
| Gastric
cancer | 6 | 1.05
(0.99–1.11) | 78.6 | <0.001 |
| Colorectal
cancer | 4 | 1.15
(1.06–1.25) | 0.0 | 0.697 |
| Pancreatic
cancer | 1 | 1.38
(0.87–2.18) | | |
| GC + GG vs. CC | Ethnicity |
| Asian | 16 | 1.11
(1.04–1.18) | 72.5 | <0.001 |
| Caucasian | 5 | 1.08
(0.85–1.37) | 0.0 | 0.576 |
| Digestive cancer
type |
| Biliary
cancer | 2 | 0.65
(0.35–1.23) | | |
| Hepatocellular
cancer | 6 | 1.10
(0.97–1.26) | 0.0 | 0.549 |
| Esophageal
squamous cell carcinoma | 2 | 1.19
(0.91–1.56) | 82.1 | 0.018 |
| Gastric
cancer | 6 | 1.04
(0.96–1.14) | 79.7 | <0.001 |
| Colorectal
cancer | 4 | 1.31
(1.12–1.52) | 76.5 | 0.005 |
| Pancreatic
cancer | 1 | 1.51
(0.87–2.63) | | |
| GG vs. CC + GC | Ethnicity |
| Asian | 16 | 1.12
(1.04–1.19) | 54.6 | 0.005 |
| Caucasian | 5 | 1.04
(0.84–1.29) | 0.0 | 0.792 |
| Digestive cancer
type |
| Biliary
cancer | 2 | 0.98
(0.76–1.28) | | |
| Hepatocellular
cancer | 6 | 1.06
(0.90–1.24) | 40.8 | 0.133 |
| Esophageal
squamous cell carcinoma | 2 | 1.39
(1.09–1.78) | 0.0 | 0.655 |
| Gastric
cancer | 6 | 1.09
(0.99–1.20) | 64.5 | 0.015 |
| Colorectal
cancer | 4 | 1.14
(1.00–1.31) | 42 | 0.16 |
| Pancreatic
cancer | 1 | 1.33
(0.32–5.45) | | |
| GG vs. CC | Ethnicity |
| Asian | 16 | 1.16
(1.06–1.26) | 64.1 | <0.001 |
| Caucasian | 5 | 1.12
(0.75–1.68) | 0.0 | 0.92 |
| Digestive cancer
type |
| Biliary
cancer | 2 | 0.68
(0.36–1.28) | | |
| Hepatocellular
cancer | 6 | 1.14
(0.95–1.38) | 28.8 | 0.219 |
| Esophageal
squamous cell carcinoma | 2 | 1.88
(1.19–2.97) | 53 | 0.145 |
| Gastric
cancer | 6 | 1.10
(0.98–1.23) | 76.3 | 0.001 |
| Colorectal
cancer | 4 | 1.30
(1.08–1.57) | 0.0 | 0.587 |
| Pancreatic
cancer | 1 | 1.55
(0.37–6.47) | | |
| GC vs. CC | Ethnicity |
| Asian | 16 | 1.09
(1.02–1.17) | 71.6 | <0.001 |
| Caucasian | 5 | 1.06
(0.83–1.36) | 0.0 | 0.446 |
| Digestive cancer
type |
| Biliary
cancer | 2 | 0.61
(0.31–1.17) | | |
| Hepatocellular
cancer | 6 | 1.09
(0.95–1.25) | 0.0 | 0.629 |
| Esophageal
squamous cell carcinoma | 2 | 1.13
(0.85–1.50) | 73 | 0.054 |
| Gastric
cancer | 6 | 1.03
(0.94–1.13) | 77.4 | <0.001 |
| Colorectal
cancer | 4 | 1.28
(1.09–1.50) | 83.5 | <0.001 |
| Pancreatic
cancer | 1 | 1.51
(0.85–2.67) | | |
However, in the subgroup analysis where ethnicity
was analyzed, significantly increased risks were found for Asians
[(for G vs. C: 1.08 (1.04–1.13),
Pheterogeneity<0.001; for GC + GG vs. CC: 1.11
(1.04–1.18), Pheterogeneity<0.001; for GG vs. CC +
GC: 1.12 (1.04–1.19), Pheterogeneity=0.005; for GG vs.
CC: 1.16 (1.06–1.26), Pheterogeneity<0.001; for GC
vs. CC: 1.09 (1.02–1.17), Pheterogeneity<0.001)]. No
significant risk was found to be associated with any of the genetic
models for Caucasians (Table
II).
Statistical sensitivity
Data from one study were omitted and the rest was
analyzed, and the pooled RRs were similar with the overall pooled
RRs (data not shown), supporting the robustness of our results.
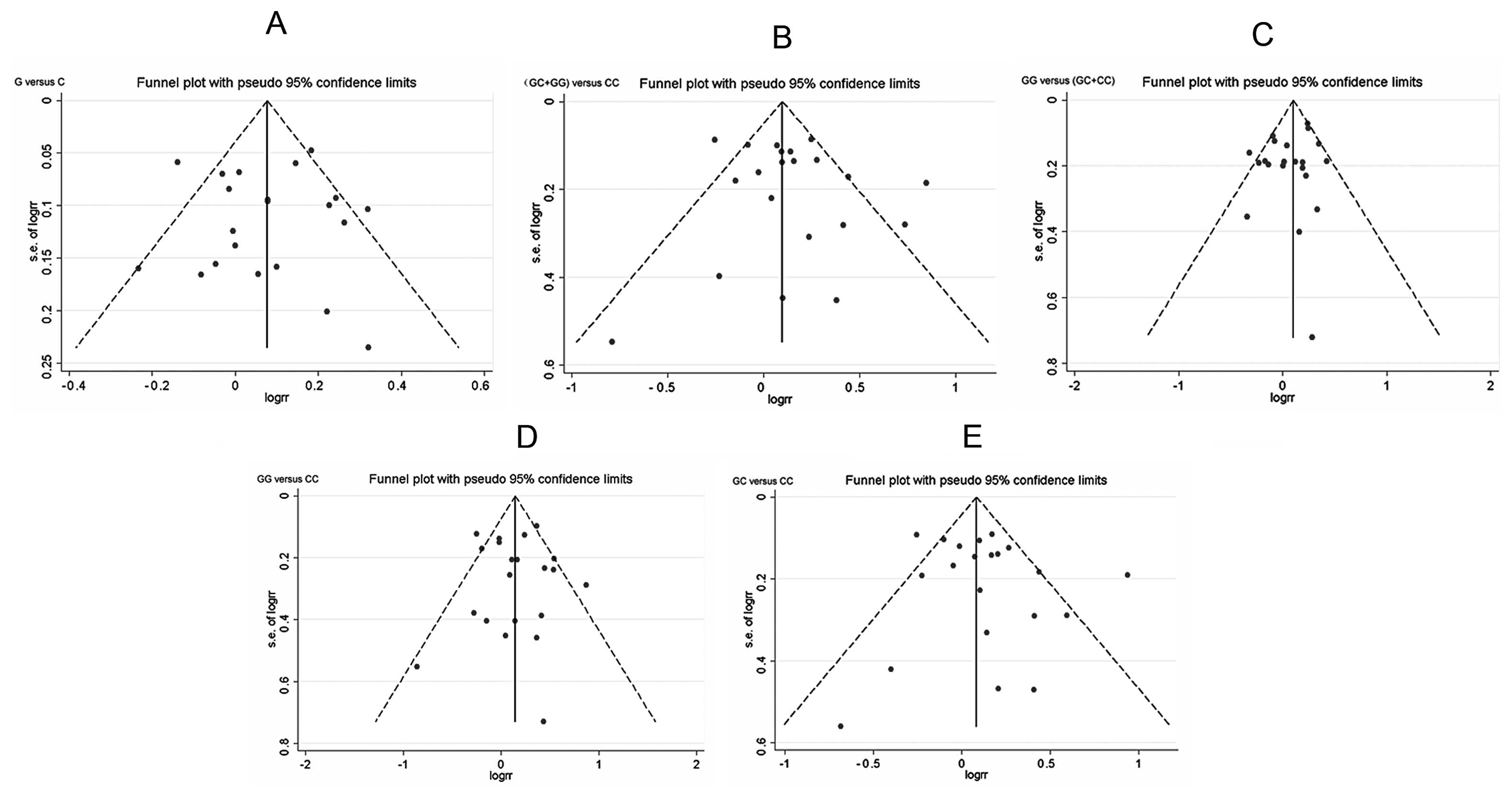
Publication bias
Begg’s funnel plot and Egger’s test were used to
assess the publication bias of the included studies. However, the
results from all the comparisons and from Egger’s test showed no
evidence for a publication bias (Fig.
3): P=0.354 for G vs. C, P=0.912 for GC + GG vs. CC, P=0.352
for GG vs. CC, P=0.795 for GC vs. CC (except P=0.045<0.1 for GG
vs. CC + GC).
Discussion
In the present study, we conducted a comprehensive
statistical analysis of the relationship between the miR-146a
polymorphism rs2910164 and digestive tumors. The results of the
stratified analysis are the following: allele G vs. C (OR=1.08, 95%
CI: 1.04–1.12), dominant genetic model GC + GG vs. CC comparison
(OR=1.11, 95% CI: 1.04–1.18), recessive genetic model GG vs. CC +
GC comparison (OR=1.11, 95% CI: 1.04–1.18), homozygous GG vs. CC
comparison (OR=1.11, 95% CI: 1.04–1.18), heterozygous GC vs. CC
comparison (OR=1.09, 95% CI: 1.02–1.16). We found that the miR-146a
polymorphism rs2910164 might significantly increase the
susceptibility to digestive tumors, especially for esophageal and
colorectal cancer. In addition, the miR-146a polymorphism might
significantly increase the risk for developing digestive tumors in
Asian individuals, while for Caucasians, no obvious correlation
between the polymorphism and the risk for digestive tumors was
found.
miRNAs are small single-stranded regulatory RNAs the
abnormal expression of which has been associated with the
susceptibility to many human diseases, including cancer in the
lung, prostate and bladder, cervical squamous cell carcinoma
(37–40), autoimmune diseases such as systemic
lupus erythematosus, Sjogren’s syndrome and lupus nephritis
(41–43), as well as cardiovascular diseases
such as heart disease, heart failure and myocardial infarction
(44–46).
The miR-146a polymorphism rs2910164 is associated
with the susceptibility to a variety of tumors. Jazdzewski et al
(47) found a significantly
different distribution of genotypes among patients with papillary
thyroid carcinomas as compared to normal subjects, with the GC
genotype being associated with an increased risk of papillary
thyroid carcinoma. Another study indicated that miR-146a might be
involved in the pathogenesis of malignant melanoma, and individuals
with the CG genotype showed an increased risk of developing
malignant melanoma (48). Orsós
et al(49) found that the
pre-miR/146a C allele might contribute to an increased
susceptibility to head and neck cancer. Therefore, the miR-146a
polymorphism rs2910164 appears to be associated with the risk of
developing cancer in a cancer type-specific manner.
The association between miR-146a polymorphisms and
susceptibility to digestive tumors has attracted increased research
in recent years. However, numerous studies on the topic were
characterized by small sample size and thus, might not possess
sufficient statistical power to detect effects of small magnitude
or might have generated a fluctuated risk estimate. Moreover,
conclusions from all these studies have not been uniform, and have
even been contradicting for different types of tumors. Therefore,
it is necessary to collect previously-generated research data and
obtain a large number of samples to get reliable results.
Nevertheless, the present meta-analysis had a number
of limitations. First, our study only concerned the analysis of
unvaried factors. Second, the population characteristics of the
experimental and the control groups were not uniform. Age, gender,
HBV and potentially, other features, might have affected the
reliability of the results. Third, the present study included only
Caucasian and Asian populations; the absence of other ethnicities
in the sample considerably reduces the universal validity of the
results. Furthermore, unconsidered non-neoplastic disease factors
might have impacted on the conclusions. Therefore, a more precise
analysis might need to be performed.
In conclusion, our analysis demonstrated that there
is an apparent association between the miR-146a polymorphism
rs2910164 and digestive cancer. However, the association is
inconsistent with regards to susceptibility to different types of
gastrointestinal cancer. Therefore, it is necessary to collect
large samples of data, perform stratified analyses and gather data
from additional ethnicities to clarify the association between the
miR-146a G/C rs2910164 polymorphism and the susceptibility to
digestive cancer.
Acknowledgements
The present study was partially supported by the
National Natural Science Foundation of China (grant nos. 81172348,
81102078 and 81172597), Health Research Projects in Jiangsu
Province (H201313) and the Priority Academic Program Development of
Jiangsu Higher Education Institutions (PAPD).
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Giovannucci E, Egan KM, Hunter DJ, et al:
Aspirin and the risk of colorectal cancer in women. N Engl J Med.
333:609–614. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Landi D, Moreno V, Guino E, et al:
Polymorphisms affecting micro-RNA regulation and associated with
the risk of dietary-related cancers: a review from the literature
and new evidence for a functional role of rs17281995 (CD86)
and rs1051690 (INSR), previously associated with colorectal
cancer. Mutat Res. 717:109–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu X, Yang X, Xing C, et al: miRNA: the
nemesis of gastric cancer (Review). Oncol Lett. 6:631–641.
2013.PubMed/NCBI
|
|
5
|
Hou Z, Xie L, Yu L, et al: MicroRNA-146a
is down-regulated in gastric cancer and regulates cell
proliferation and apoptosis. Med Oncol. 29:886–892. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vinci S, Gelmini S, Mancini I, et al:
Genetic and epigenetic factors in regulation of microRNA in
colorectal cancers. Methods. 59:138–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He Y, Huang C, Sun X, et al: MicroRNA-146a
modulates TGF-beta1-induced hepatic stellate cell proliferation by
targeting SMAD4. Cell Signal. 24:1923–1930. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomokuni A, Eguchi H, Tomimaru Y, et al:
miR-146a suppresses the sensitivity to interferon-α in
hepatocellular carcinoma cells. Biochem Biophys Res Commun.
414:675–680. 2011.PubMed/NCBI
|
|
9
|
Li Y, Vandenboom TG II, Wang Z, et al:
miR-146a suppresses invasion of pancreatic cancer cells. Cancer
Res. 70:1486–1495. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Tang S, Le SY, et al: Aberrant
expression of oncogenic and tumor-suppressive microRNAs in cervical
cancer is required for cancer cell growth. PLoS One. 3:e25572008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu J, Li A, Hong SM, et al: MicroRNA
alterations of pancreatic intraepithelial neoplasias. Clin Cancer
Res. 18:981–992. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu B, Wang N, Wang X, et al: MiR-146a
suppresses tumor growth and progression by targeting EGFR pathway
and in a p-ERK-dependent manner in castration-resistant prostate
cancer. Prostate. 72:1171–1178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lian H, Wang L and Zhang J: Increased risk
of breast cancer associated with CC genotype of Hsa-miR-146a
Rs2910164 polymorphism in Europeans. PLoS One. 7:e316152012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yue C, Wang M, Ding B, et al: Polymorphism
of the pre-miR-146a is associated with risk of cervical cancer in a
Chinese population. Gynecol Oncol. 122:33–37. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Bi J, Liu X, et al: Hsa-miR-146a
polymorphism (rs2910164) and cancer risk: a meta-analysis of 19
case-control studies. Mol Biol Rep. 39:4571–4579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu T, Zhu Y, Wei QK, et al: A functional
polymorphism in the miR-146a gene is associated with the
risk for hepatocellular carcinoma. Carcinogenesis. 29:2126–2131.
2008.PubMed/NCBI
|
|
17
|
Ye Y, Wang KK, Gu J, et al: Genetic
variations in microRNA-related genes are novel susceptibility loci
for esophageal cancer risk. Cancer Prev Res (Phila). 460–469. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Srivastava K, Srivastava A and Mittal B:
Common genetic variants in pre-microRNAs and risk of gallbladder
cancer in North Indian population. J Hum Genet. 55:495–499. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo H, Wang K, Xiong G, et al: A
functional variant in microRNA-146a is associated with risk of
esophageal squamous cell carcinoma in Chinese Han. Fam Cancer.
9:599–603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zeng Y, Sun QM, Liu NN, et al: Correlation
between pre-miR-146a C/G polymorphism and gastric
cancer risk in Chinese population. World J Gastroenterol.
16:3578–3583. 2010.PubMed/NCBI
|
|
21
|
Min KT, Kim JW, Jeon YJ, et al:
Association of the miR-146aC>G, 149C>T,
196a2C>T, and 499A>G polymorphisms with
colorectal cancer in the Korean population. Mol Carcinog. 51(Suppl
1): E65–E73. 2012.
|
|
22
|
Akkiz H, Bayram S, Bekar A, et al: No
association of pre-microRNA-146a rs2910164 polymorphism and risk of
hepatocellular carcinoma development in Turkish population: a
case-control study. Gene. 486:104–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hishida A, Matsuo K, Goto Y, et al:
Combined effect of miR-146a rs2910164 G/C polymorphism and
Toll-like receptor 4 +3725 G/C polymorphism on the risk of
severe gastric atrophy in Japanese. Dig Dis Sci. 56:1131–1137.
2011.
|
|
24
|
Okubo M, Tahara T, Shibata T, et al:
Association between common genetic variants in pre-microRNAs and
gastric cancer risk in Japanese population. Helicobacter.
15:524–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang XW, Pan SD, Feng YL, et al:
Relationship between genetic polymorphism in microRNAs precursor
and genetic predisposition of hepatocellular carcinoma. Zhonghua Yu
Fang Yi Xue Za Zhi. 45:239–243. 2011.(In Chinese).
|
|
26
|
Xiang Y, Fan S, Cao J, et al: Association
of the microRNA-499 variants with susceptibility to hepatocellular
carcinoma in a Chinese population. Mol Biol Rep. 39:7019–7023.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim WH, Min KT, Jeon YJ, et al:
Association study of microRNA polymorphisms with hepatocellular
carcinoma in Korean population. Gene. 504:92–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou F, Zhu H, Luo D, et al: A functional
polymorphism in Pre-miR-146a is associated with
susceptibility to gastric cancer in a Chinese population. DNA Cell
Biol. 31:1290–1295. 2012.PubMed/NCBI
|
|
29
|
Ahn DH, Rah H, Choi YK, et al: Association
of the miR-146a C>G, miR-149T>C, miR-196a2T>C, and
miR-499A>G polymorphisms with gastric cancer risk and survival
in the Korean population. Mol Carcinog. 52(Suppl 1): 39–51.
2013.
|
|
30
|
Hezova R, Kovarikova A, Bienertova-Vasku
J, et al: Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as
risk factors of colorectal cancer. World J Gastroenterol.
18:2827–2831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mihalache F, Hoblinger A, Acalovschi M, et
al: A common variant in the precursor miR-146a sequence does not
predispose to cholangiocarcinoma in a large European cohort.
Hepatob Pancreat Dis Int. 11:412–417. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou J, Lv R, Song X, et al: Association
between two genetic variants in miRNA and primary liver cancer risk
in the Chinese population. DNA Cell Biol. 31:524–530. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song MY, Su HJ, Zhang L, et al: Genetic
polymorphisms of miR-146a and miR-27a, H.
pylori infection, and risk of gastric lesions in a Chinese
population. PLoS One. 8:e612502013.PubMed/NCBI
|
|
34
|
Ma L, Zhu L, Gu D, et al: A genetic
variant in miR-146a modifies colorectal cancer susceptibility in a
Chinese population. Arch Toxicol. 87:825–833. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lv M, Dong W, Li L, et al: Association
between genetic variants in pre-miRNA and colorectal cancer risk in
a Chinese population. J Cancer Res Clin Oncol. 139:1405–1410. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pavlakis E, Papaconstantinou I, Gazouli M,
et al: MicroRNA gene polymorphisms in pancreatic cancer.
Pancreatology. 13:273–278. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yuan Z, Zeng X, Yang D, et al: Effects of
common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer
risk. PLoS One. 8:e610472013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu J, Liu J, He Y, et al: Genetic
variants in the microRNA machinery gene GEMIN4 are
associated with risk of prostate cancer: a case-control study of
the Chinese Han population. DNA Cell Biol. 31:1296–1302.
2012.PubMed/NCBI
|
|
39
|
Luo J, Cai Q, Wang W, et al: A microRNA-7
binding site polymorphism in HOXB5 leads to differential gene
expression in bladder cancer. PLoS One. 7:e401272012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou B, Wang K, Wang Y, et al: Common
genetic polymorphisms in pre-microRNAs and risk of cervical
squamous cell carcinoma. Mol Carcinog. 50:499–505. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Löfgren SE, Frostegård J, Truedsson L, et
al: Genetic association of miRNA-146a with systemic lupus
erythematosus in Europeans through decreased expression of the
gene. Genes Immun. 13:268–274. 2012.PubMed/NCBI
|
|
42
|
Zilahi E, Tarr T, Papp G, et al: Increased
microRNA-146a/b, TRAF6 gene and decreased IRAK1 gene expressions in
the peripheral mononuclear cells of patients with Sjögren’s
syndrome. Immunol Lett. 141:165–168. 2012.PubMed/NCBI
|
|
43
|
Lu J, Kwan BC, Lai FM, et al: Glomerular
and tubulointerstitial miR-638, miR-198 and miR-146a expression in
lupus nephritis. Nephrology. 17:346–351. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Halkein J, Tabruyn SP, Ricke-Hoch M, et
al: MicroRNA-146a is a therapeutic target and biomarker for
peripartum cardiomyopathy. J Clin Invest. 123:2143–2154. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fan KL, Zhang HF, Shen J, et al:
Circulating microRNAs levels in Chinese heart failure patients
caused by dilated cardiomyopathy. Indian Heart J. 65:12–16. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zidar N, Boštjančič E, Glavač D and Stajer
D: MicroRNAs, innate immunity and ventricular rupture in human
myocardial infarction. Dis Markers. 31:259–265. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jazdzewski K, Murray EL, Franssila K, et
al: Common SNP in pre-miR-146a decreases mature miR
expression and predisposes to papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 105:7269–7274. 2008.
|
|
48
|
Yamashita J, Iwakiri T, Fukushima S, et
al: The rs2910164 G>C polymorphism in microRNA-146a is
associated with the incidence of malignant melanoma. Melanoma Res.
23:13–20. 2013.
|
|
49
|
Orsós Z, Szanyi I, Csejtei A, et al:
Association of pre-miR-146a rs2910164 polymorphism with the risk of
head and neck cancer. Anticancer Res. 33:341–346. 2013.PubMed/NCBI
|