Introduction
DAP1 is one of the members of the death-associated
protein (DAP) family, first isolated as a gene involved in
IFN-γ-induced apoptosis through a technical knockout strategy
(TKO), i.e. random inactivation of genes with antisense cDNA
libraries (1). The DAP family is
comprised of DAP1, DAP2 (DAP kinase), DAP3, DAP4 and DAP5. DAP2 was
found to be involved in apoptosis induced by IFN-γ, TNF-α and Fas
(2–3). Serine/threonine kinase catalytic
activity and the death domain contribute to the pro-apoptotic
function. Based on the loss of DAP2 expression in certain types of
cancers, it has been proposed as a candidate tumour-suppressor
gene. While the clinical significance of other members of the DAP
Family that also exhibit death-promoting functions remain largely
unknown, there have been recent reports on the possible association
between the DAP family and the clinical outcome of patients with
malignant diseases, for example breast cancer (4).
Induction of apoptosis is an important mechanism of
chemotherapeutic agents in the treatment of cancer. Among the DAP
family members, DAP3 was found to be a mitochondrial protein that
has a specific amino acid sequence to recognise and anchor to
mitochondria. Mitochondria play a key role in apoptosis including
release of pro-apoptotic substances such as cytochrome c and
AIF, production of reactive oxygen species (ROS) and reduction in
ATP synthesis (5–8). All of these findings indicate that DAP
may be involved in both the intrinsic and extrinsic apoptosis
pathways. In addition, it was reported that DAP2 methylation is
associated with a reduced response to chemotherapy and poor
prognosis in gastric cancer (9).
In the present study, we investigated the expression
pattern of DAP1 in a cohort of colorectal cancer patients.
Decreased expression of DAP1 transcripts was detected in tumour
tissues. qPCR analysis revealed that DAP1 expression levels were
correlated with clinical and pathological parameters of the
colorectal cancer patients including disease-free survival.
Furthermore, the study reports the role of DAP1 in the growth and
apoptosis of colorectal cancer cells and in the cellular response
of cells to chemotherapeutic agents.
Materials and methods
Materials
HRT18 and HT115 cell lines were obtained from the
European Collection of Cell Cultures (ECACC; Salisbury, UK).
Reagents and kits were obtained from Promega Corporation (Madison,
WI, USA), Bio-Rad Lifescience Company (Hercules, CA, USA), and
Gibco Invitrogen Corporation (Gibco-BRL, Paisley, Scotland, UK).
The secondary and goat polyclonal antibodies raised against a
peptide mapping at the N-terminus of human DAP1 were purchased from
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Colorectal cancer tissues were collected immediately
after surgery. All protocols were reviewed and approved by the
local ethics committee, and consent was obtained from the
patients.
Tissue processing, RNA extraction, cDNA
synthesis and RT-PCR
Frozen sections of the tissues were cut to
thicknesses of 5–10 μm and kept for immunohistochemistry and
routine histology (10). RNA was
isolated using Total RNA reagent (Promega Corporation). cDNA
synthesis and RT-PCR were performed using standard methods.
Quantitative analysis of DAP1
DAP1 mRNA expression in the colon cancer tissues was
determined by quantitative real-time PCR, based on the Amplifluor™
technology as previously described (11). DAP1 qPCR primers as follows were
designed using Beacon Designer software (Premier Biosoft, Palo
Alto, CA, USA): sense, 5′-ATGGACAAGCATCCCTTCC-3′ and antisense,
5′-ACTGAACCTGACCGTACACTCTGTCAGG
GAAATACCAA-3′, exon 10–12). The underlined sequence is
complementary to the universal Z probe (TCS Biologicals Ltd.,
Oxford, UK).
Immunohistochemical staining of the DAP1
protein
Frozen sections of tissues were cut to thicknesses
of 5–10 μm, and IHC was performed using the anti-DAP1 antibody and
Vectastain ABC kits (Vector Laboratories, Burlingame, CA, USA).
Ribozyme transgene targeting human
DAP1
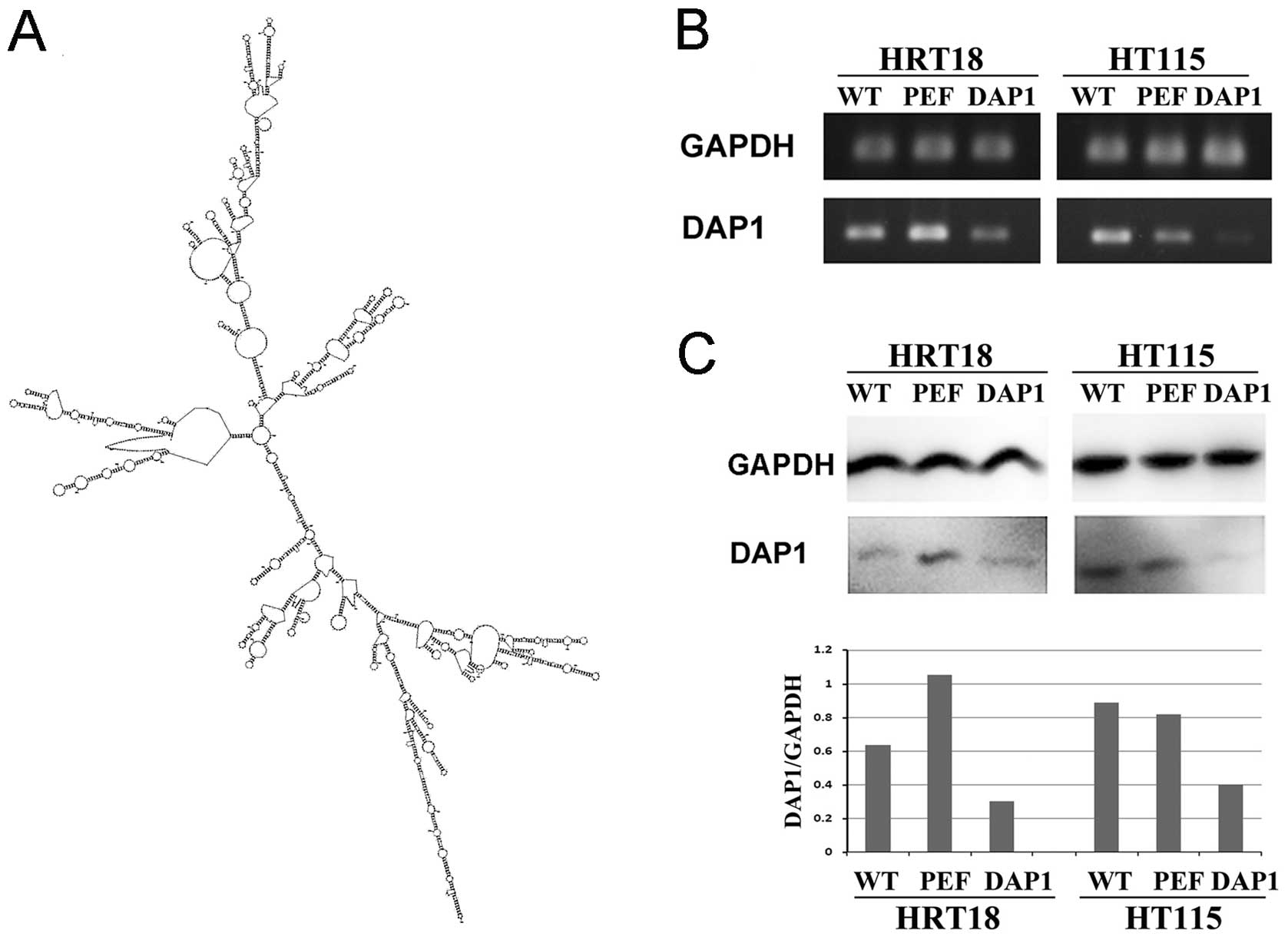
Anti-human DAP1 hammerhead ribozymes were designed
according to the secondary structure of the gene (Fig. 2A) using the Zuker RNA mFold program
(sense, 5′-CTGCAGTTCAACCACTT CTCCCAGAGCTGATGAGTCCGTGAGGA-3′ and
antisense, 5′-ACTAGTAGAGAAAGCACTGAGAAAGGGAGT TTCGTCCTCACGGACT-3′)
(12). The ribozymes were
accordingly synthesised using touchdown PCR and cloned into the
pEF6/V5-His TOPO TA Expression kit (Invitrogen) following the
manufacturer’s protocol. Transfection was performed using an
Easyject Plus electroporator (EquiBio, Kent, UK). After up to 5
days of selection with blasticidin, the transfectants from the DAP1
knockdown and control cells were verified using RT-PCR and then
used in the subsequent experiments.
Western blot analysis
The protein concentrations in the cell lysates were
determined using the DC protein assay kit (Bio-Rad) and an ELx800
spectrophotometer (BioTek). Proteins were probed with the anti-DAP1
(1:200) and anti-GAPDH-antibodies (1:500) (both from Santa Cruz
Biotechnology, Inc.) as an internal control, followed by a
peroxidase-conjugated secondary antibody (1:1,000). Protein bands
were visualised and photographed using an UVITech imager (UVITech,
Inc.).
In vitro cell growth assay under normal
culture condition, or following exposure to gradient concentrations
of 5FU and oxaliplatin
Equal amounts of cells were plated into 96-well
plates at 2,500 cells/well with a gradient concentration of a
combination of 5FU and oxaliplatin at a 1:5 dilution. The dilution
started from 10 times that of the threshold concentration
(2×10−6). Cell growth was assessed after 1, 3 and 5
days. Crystal violet was used to stain the cells, and the
absorbance was determined at a wavelength of 540 nm using a
spectrophotometer ELx800.
The blood and tissue fluid drug concentrations of
5FU and oxaplatin were determined to be 5×10−5 M and
1×10−4 M, respectively, according to the drug dosage
used in clinical chemotherapy regimens. The wild-type HRT18 and
HT115 cells were plated with a gradient concentration with a 1:5
dilution of 5 times that of the blood concentration. The
concentration causing obviously increased cell growth was set as a
threshold dosage.
Cell matrix adhesion assay
The cell matrix adhesion assay was conducted as
previously described (13). Cells
were added to a 96-well plate precoated with Matrigel
(Collaborative Research Products, Bedford, MA, USA) (5 μg/well).
After 40 min of incubation, the non-adherent cells were washed off
using BSS buffer. The remaining cells were fixed, stained and
counted.
Wound healing assay
The wound healing assay was performed as previously
described (14). The monolayer of
cells was scraped with a fine gauge needle. The movement of cells
to close the wound was recorded on a time lapse video recorder and
analysed using Optimas 6.0 motion analysis (Meyer Instruments,
Inc., Houston, TX, USA).
In vitro invasion assay
The in vitro invasion assay was performed as
previously described (14).
Transwell inserts with an 8-μm pore size were coated with 50 μg of
Matrigel and air-dried. Following rehydration, 40,000 cells were
added to each well. After 3 days of incubation, cells that had
migrated through the matrix to the other side of the insert were
fixed, stained and counted.
Flow cytometric analysis of apoptosis
following treatment with a combination of 5FU and oxaliplatin
DAP1-RIB and empty vector PEF-transfected HRT18 and
HT115 cells were plated into a 6-well plate at a density of
3×105 cells/well. Each cell line was plated into 3-wells
at the same time. The first well was used as the control with no
treatment, and the second and third wells received treatment with a
combination of 5FU and oxaliplatin at concentrations of
2×10−6 M and 1×10−5 M, respectively. All
cells, including those floating in the culture medium were
harvested after 6 h of incubation. The apoptotic population was
determined using a Vybrant® Apoptosis Assay kit and flow
cytometry and FlowMax software package as previously described
(15).
Statistical analysis
Statistical analysis was performed using SPSS
software (SPSS standard version 13.0; SPSS Inc.). The relationship
between DAP1 expression and tumor grade, TNM stage and nodal status
were assessed using Mann-Whitney U test and Kruskal-Wallis test.
(The error bar shown in the graph represents SEM). Survival curves
were analyzed using Kaplan-Meier survival analysis. Differences
were considered statistically significant at p<0.05.
Results
DAP1 mRNA expression in the colorectal
adenocarcinoma tissues
DAP1 transcript expression was examined in specimens
from the 94 colorectal adenocarcinoma patients using real-time
quantitative PCR (expressed as mean DAP1 transcript copies/μl of
RNA from 50 ng total RNA and standardized with GAPDH). The cohort
comprised 61 men (71.8%) and 24 women (28.2%). The average age of
all patients was 56.9 years. A significantly lower mRNA expression
level of DAP1 was observed in the tumour tissues when compared with
the normal background tissues (p=0.027).
DAP1 expression correlates with tumour
invasiveness and clinical stage
The relationship between DAP1 expression and
pathological status was also assessed in the present study through
quantitative analysis of DAP1 transcript. DAP1 levels were
initially assessed in relation to tumour invasiveness. Invasive
tumours appeared to have reduced levels of DAP1 when compared with
non-invasive tumours, but the difference did not reach statistical
significance.
The relationship between DAP1 expression and
clinical TNM stage is shown in Table
I. The DAP1 expression level was relatively higher in lymph
node-negative patients in comparison with the lymph node-positive
group. Although the difference was not significant (p=0.21), a
trend was observed that DAP1 expression decreased along with the
upgrading of T stage (p=0.218). Expression levels of DAP1 were also
decreased in the advanced disease cases. Statistical analysis
revealed a significant difference (p=0.039). This is in line with
the reduced expression of DAP1 in tumours of Dukes’ B and C groups,
which was significantly lower than its level in tumours of the
Dukes’ A group (p=0.036).
 | Table IDAP1 transcript levels in colorectal
tumour tissues. |
Table I
DAP1 transcript levels in colorectal
tumour tissues.
| DAP1 transcripts
(copies/50 ng RNA) means ± SD | P-value |
|---|
| Location | | 0.744 |
| Left colon | 1,803±2,749 | |
| Right colon | 845±2,046 | |
| Transcolon | 0.426±0.531 | |
| Rectum | 82±1,063 | |
| Differentiation | | 0.007 |
|
Well-differentiated | 5,099±7,207 | |
| Moderately
differentiated | 709±1,614 | |
| Poorly
differentiated | 1,859±2,836 | |
| Dukes’ stage | | 0.036 |
| A | 3,203±3,975 | |
| B | 995±2,192 | |
| C | 689±1,602 | |
| T stage | | 0.218 |
| T1 | 3,589±2,404 | |
| T2 | 1,944±3,587 | |
| T3 | 872±2,008 | |
| T4 | 801±1,764 | |
| N stage | | 0.21 |
| Negative | 1,379±1,443 | |
| Positive | 711±1,623 | |
| TNM stage | | 0.039 |
| I | 3,071±3,646 | |
| II | 913±2,142 | |
| III | 537±1,532 | |
| IV | 1,347±1,877 | |
| Invasiveness | | 0.43 |
| Non-invasive | 72.1±188.7 | |
| Invasive | 46.7±110.4 | |
| Incidents | | 0.037 |
| Disease-free | 1,441±2,695 | |
| Incidents
(recurrence and metastases) | 335±1,013 | |
| Survival | | 0.14 |
| Alive | 1,384±2,672 | |
| Death | 570±1,404 | |
| Distant
metastasis | | 0.19 |
| Yes | 586±1,495 | |
| No | 1,229±2,398 | |
| Recurrence | | 0.016 |
| No recurrence | 1,111±2,307 | |
| Local
recurrence | 206±495 | |
DAP1 expression in relation to recurrence
and metastasis
The correlations between DAP1 expression levels and
occurrence of incidents (local recurrence and distant metastasis)
were also investigated. We found a significant correlation between
DAP1 expression levels and the incidents (p=0.037) (Table I).
Quantitative studies showed that DAP1 expression
levels in patients with either recurrence or metastasis were
significantly lower than those without any of these
complications.
Correlation between DAP1 expression and
survival of the colorectal adenocarcinoma patients
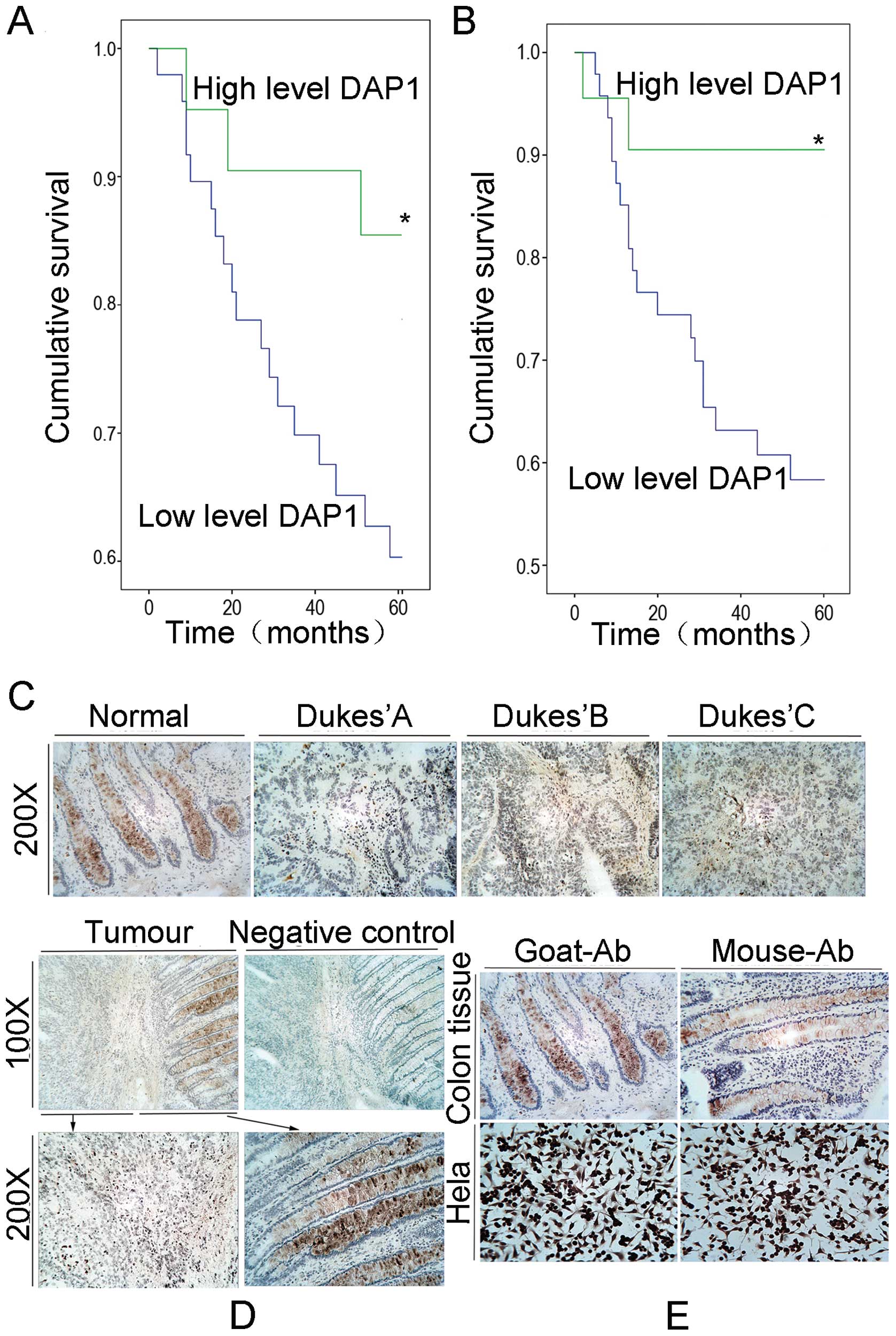
The median follow-up period was 21.7 months (0.7–88
months) for the current cohort. According to a threshold level
(average DAP1 level of Dukes’ B), Kaplan-Meier analysis
demonstrated that patients with higher levels of DAP1 expression in
tumours had a prolonged overall survival (Fig. 1A) and longer disease-free survival
time (Fig. 1B) when compared with
the lower expression level group (p<0.01).
In the stratified survival analysis according to
node status, node-negative patients with high DAP1 levels had a
significantly prolonged survival in comparison with the low level
group (p=0.011). In the node-positive patients, no significant
association was found between DAP1 expression and survival.
Finally, multivariate analysis using gender, age, grade, TNM, nodal
status and DAP1 expression levels as variants showed that nodal
status (p=0.015), TNM (p=0.006), grade (p=0.026), age (p=0.020) and
DAP1 (p=0.037) are all independent factors for overall
survival.
Immunohistochemical staining of human
colorectal adenocarcinoma specimens
To assess the expression pattern of DAP1 at the
protein level, we performed immunohistochemical analysis of DAP1 in
the human colorectal adenocarcinoma tissue sections (n=20 pairs).
Using a specific anti-DAP1 polyclonal antibody, DAP1 was detected
both in the cytoplasm of the tumours and in the non-tumour cells.
The staining of DAP1 was significantly reduced or absent in the
tumour tissues when compared with the non-tumour tissues. No
obvious staining of DAP1 was observed in the stromal cells in
either normal or tumour tissues (Fig.
1C–E).
Stable knockdown of DAP1
To investigate the role of DAP1 in colorectal
adenocarcinoma, knockdown of DAP1 expression was performed in the
HRT18 and HT115 cell lines which expressed DAP1. Reduced transcript
levels of DAP1 were verified in the HRT18 and HT115 cells which
were transfected with ribozyme transgenes using RT-PCR (Fig. 2B). A reduction in the protein
expression of DAP1 was further confirmed using western blotting
(Fig. 2C). These DAP1-modified cell
lines were used for the following in vitro studies.
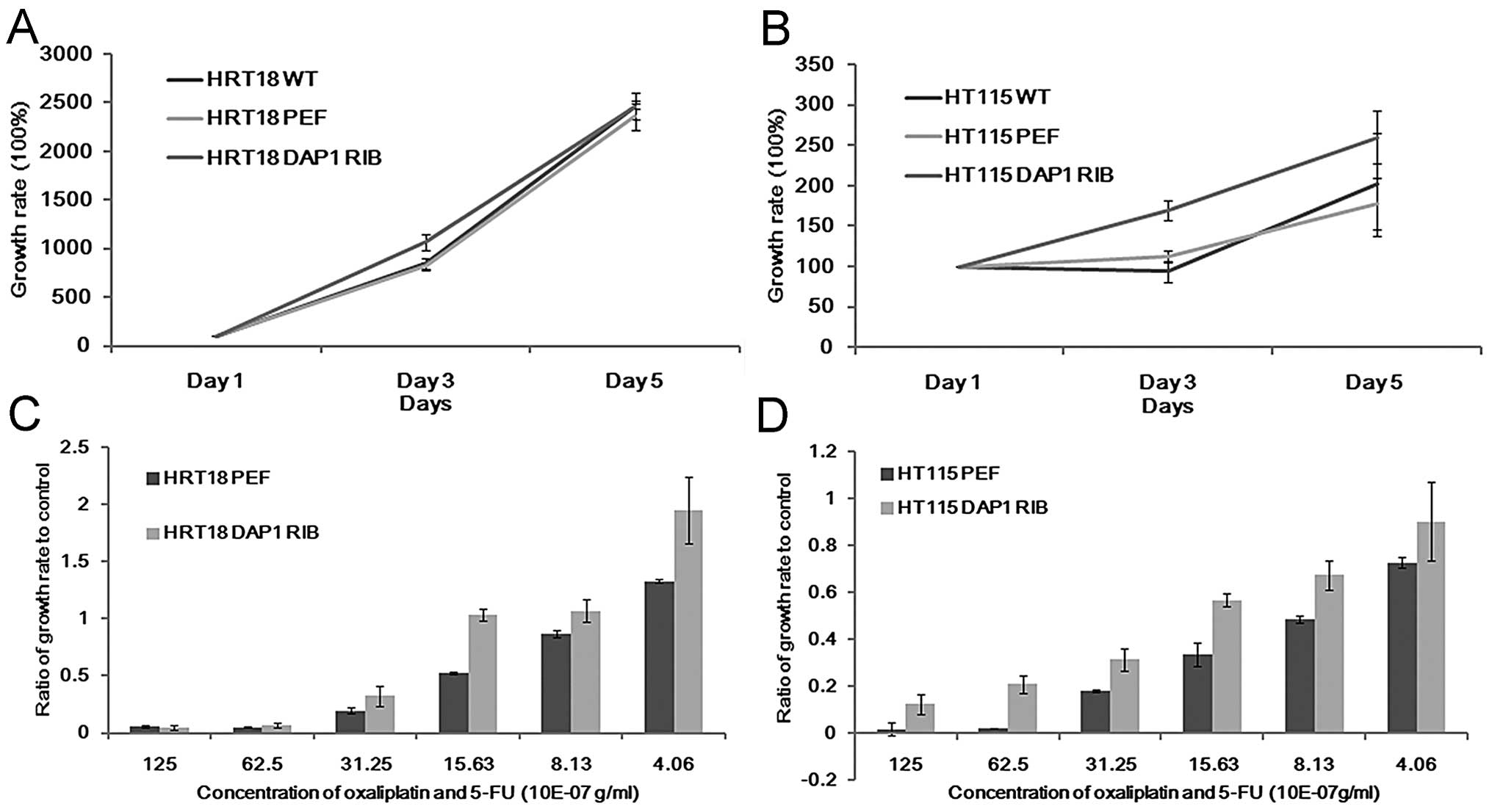
Impact of DAP1 knockdown on in vitro cell
growth following treatment with gradient concentrations of 5FU and
oxaliplatin
Although there was no significant difference noted
in the DAP1-knockdown cells in comparison with the controls in
regards to cell growth (Fig. 3A and
B), these cells appeared to be less responsive to treatment
with 5FU alone or in combination with oxaliplatin. The growth rate
of DAP1-RIB cells significantly exceeded that of the control cells.
This effect was noted in both the HRT18 and HT115 cell lines
(Fig. 3C and D).
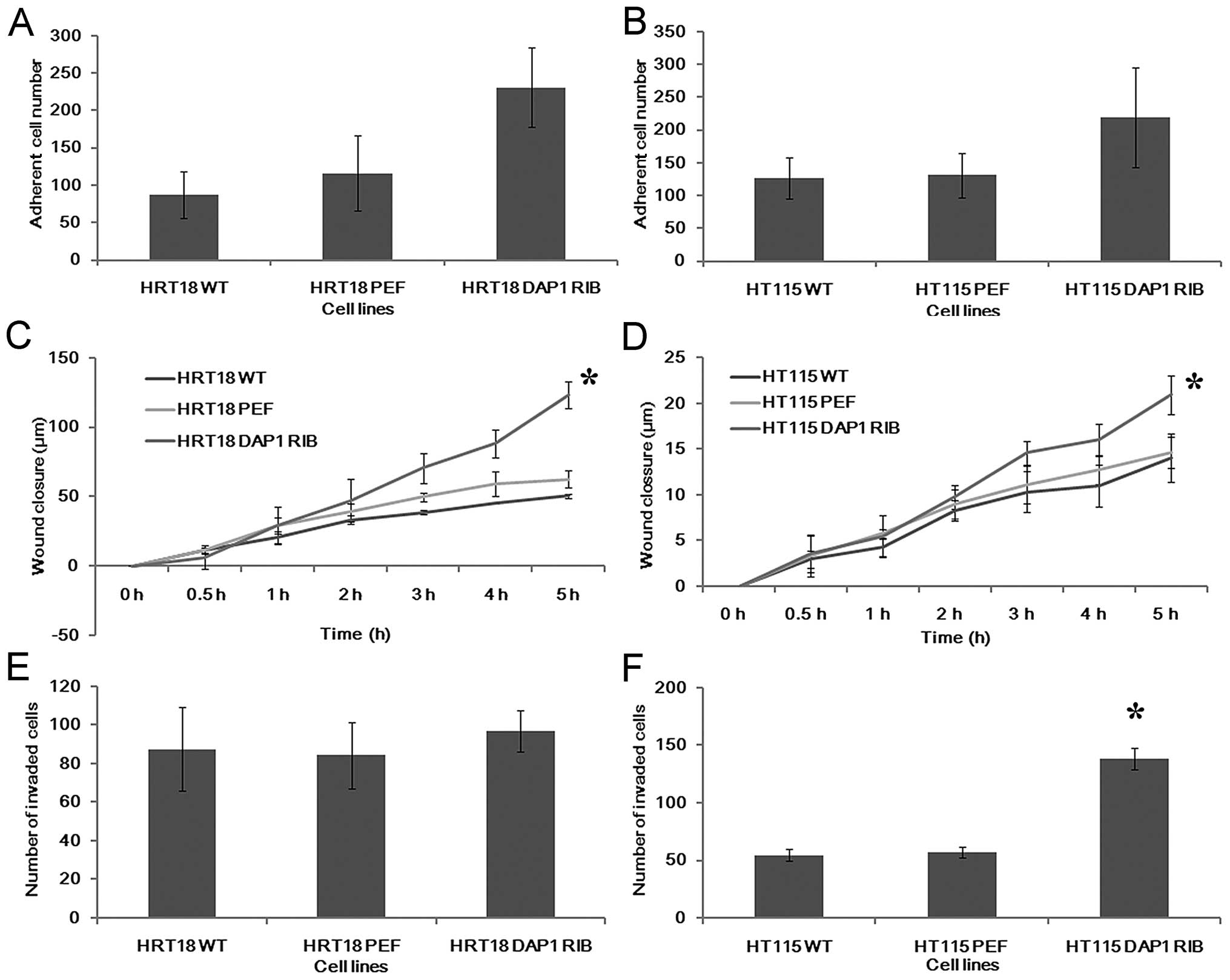
Effects of DAP1 knockdown on adhesion,
migration and invasion of colorectal cancer cells
We further examined the influence of DAP1 on the
adhesive nature of the colorectal cancer cells (Fig. 4A and B). Knockdown of DAP1
expression resulted in an increase in adhesive ability of both
HRT18 and HT115 cells.
A wound healing assay was employed to examine the
influence of DAP1 knockdown on migration. Downregulation of DAP1
significantly promoted cell migration to close the wound when
compared with the control in the HRT18 cell line (p<0.05). The
migration was also promoted by DAP1 knockdown to a lesser degree in
the HT115 cell line (Fig. 4C and
D).
Finally, the presence of DAP1 has also been shown to
affect colorectal cancer cell invasion. Knockdown of DAP1 in the
HT115 cell line resulted in a marked increase in cell invasiveness
(p<0.0001 vs. controls). However, no similar effect was observed
in the HRT18 cell line (Fig. 4E and
F).
Effects of DAP1 knockdown on apoptosis in
response to chemotherapeutic agents
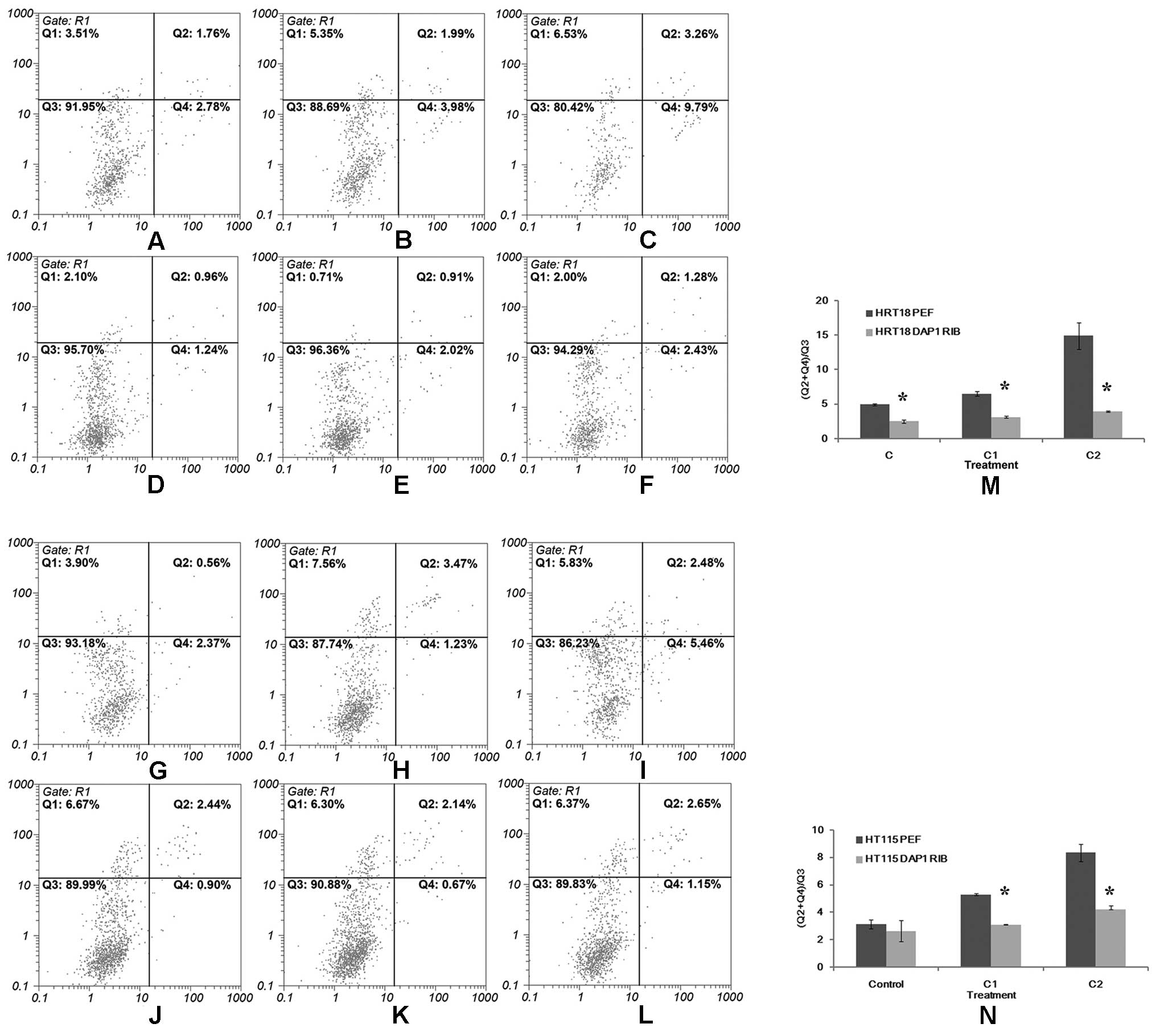
As shown in Fig. 5,
when compared to the control group, the percentage of cells
undergoing apoptosis was significantly higher in both the HRT18 and
HT115 cells treated with the chemotherapeutic agents. The apoptotic
population was increased in a concentration-dependent manner. In
comparison with the cells transfected with empty vectors, both
HRT18DAP1RIB and HT115DAP1RIB cells consisted of a significantly
lower percentage of apoptotic cells when exposed to different
concentrations of the chemotherapeutic agents (Fig. 5).
Discussion
DAP1 belongs to the DAP family which was initially
isolated through technical knockout (TKO) strategy, and was
characterized as an apoptosis-associated protein because of its
involvement in IFN-γ-induced apoptosis. The DAP family consists of
DAP1, DAP2 (DAP kinase), DAP3, DAP4 and DAP5. These members share
some common feature domains which confer a pro-apoptotic function.
However, the expression pattern of the DAP family differs among
different types of human tumours. DAP2 has been reported to be
below the limit of detection in 80% of B cell leukemia cell lines,
while the frequency of loss of DAP2 expression in breast, bladder
and renal carcinoma cell lines ranges from 30 to 40% (16) Promoter hypermethylation of DAP2 was
also reported in various types of human tumours, including B cell
lymphoma, non-small cell lung, head and neck and colon cancer
(17,18). DAP3 was found to be overexpressed in
human thyroid oncocytic tumours and correlated with human thymoma
stage (19,20). In breast cancer, however, DAP3 was
found to be lost in aggressive tumours, and low levels of DAP-3
were found to be linked to a shorter survival of breast cancer
patients (20). In the present
study, the DAP1 expression pattern in colorectal adenocarcinoma was
similar to that of DAP2. Immunohistochemistry revealed that DAP1
expression in tumour cells was significantly lower than that of the
background normal tissues. qPCR analysis of DAP1 in our clinical
colorectal cancer cohort revealed that lower expression of DAP1 was
correlated with lymph node involvement, higher T stage, tumour
invasiveness, local recurrence, metastases, higher TNM stage and
higher Dukes’ stage. A higher level of DAP1 expression indicated a
longer disease-free survival time. From the above data and reports
of similar research, it can be argued that DAP1 acts as a
tumour-suppressor gene in colorectal cancer.
In order to clarify the role of DAP1 in colorectal
cancer, we transfected anti-human DAP1 hammerhead ribozymes into
two colorectal tumour cell lines, HRT18 and HT115, and investigated
the role of DAP1 through an in vitro cell function test.
There was no significant difference in the growth rate between
DAP1-knockdown cell lines and the corresponding controls. However,
the in vitro cell-matrix adhesion assay and wound healing
assay both indicated that DAP1-knockdown HRT18 and HT115 cancer
cell lines presented more aggressive behaviour than the
corresponding control cell lines. Faster wound closure speed and
stronger adherent ability were observed in the DAP1-RIB-transfected
cell lines, which was consistent with the clinical cohort qPCR
result that lower expression of DAP1 is correlated with tumour
invasiveness. However only the HT115 DAP1RIB cell line exhibited
invasive behaviour, which is only partially consistent with the
clinical data indicating that patients with higher T stage or
invasive tumours have a lower DAP1 expression level.
In the TKO strategy, IFN-γ was initially utilized as
a killing agent to induce cell apoptosis, and later various
apoptosis-inducing stimuli such as TNF-α and FAS were all found to
be able to be adopted as killing agents. Taken together, with the
DAP1 expression patterns in colorectal cancer tissue and paired
background normal tissue and the results of the cell function
tests, it can be concluded that downregulation of DAP1 is part of
the complicated development of tumour progression. Resistance to
apoptotic stimuli by virtue of DAP1 downregulation may confer
tumour cells a better chance to elude attacks from the immune
system. This may be correlated with the results from the clinical
cohort.
In the present study, we also investigated the
growth rate of different cell lines exposed to 5-FU and
oxaliplatin. The DAP1-knockdown cells exhibited higher tolerance to
the genotoxic agents and higher growth rates, when compared with
the control or wild-type cells. Thus, we can conclude that
downregulation of DAP1 expression prevents colorectal tumour cell
lines from undergoing apoptosis induced by chemotherapeutic agents
and contributes to resistance to chemotherapy. When we consider the
clinical cohort data, we found that, although chemotherapy
information was not complete, a trend was observed that among the
patients who received adjuvant chemotherapy or chemoradiotherapy,
those with higher DAP1 expression had prolonged survival when
compared with those with lower DAP1 expression. At present, DAP1
has only been reported to be involved in the extrinsic apoptosis
pathway, but there are a number of studies indicating that DAP3 is
a mitochondrial protein and is involved in the regulation of
mitochondria morphology during the process of apoptosis (21–25).
As a member of the DAP family, DAP1 may share many common molecular
structures with DAP3 and may also be involved in the endogenous
apoptosis pathway. However, the detailed mechanism of DAP1-related
apoptosis remains to be elucidated.
In conclusion, DAP1 expression is downregulated in
colorectal cancer and is correlated with advanced TNM/Dukes’ stage
and disease-free survival; knockdown of DAP1 expression enhanced
the ability of HRT18 and HT115 cell lines to adhere and migrate.
The resistance to chemotherapeutic agents may also be promoted by
the downregulation of DAP1 expression. DAP1 may act as a
tumour-suppressor gene in colorectal adenocarcinoma.
Acknowledgements
The authors wish to thank the Cancer Research Wales,
and the Albert Hung Foundation for supporting the present study. Dr
Y.J. is a recipient of the Cardiff University’s China Medical
Scholarship.
References
|
1
|
Inbal B, Shani G, Cohen O, Kissil JL and
Kimchi A: Death-associated protein kinase-related protein 1, a
novel serine/threonine kinase involved in apoptosis. Mol Cell Biol.
20:1044–1054. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cohen O, Inbal B, Kissil JL, et al:
DAP-kinase participates in TNF-α- and Fas-induced apoptosis and its
function requires the death domain. J Cell Biol. 146:141–148.
1999.
|
|
3
|
Inbal B, Cohen O, Polak-Charcon S, et al:
DAP kinase links the control of apoptosis to metastasis. Nature.
390:180–184. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lehmann U, Celikkaya G, Hasemeier B,
Länger F and Kreipe H: Promoter hypermethylation of the
death-associated protein kinase gene in breast cancer is
associated with the invasive lobular subtype. Cancer Res.
62:6634–6638. 2002.
|
|
5
|
Green DR and Reed JC: Mitochondria and
apoptosis. Science. 281:1309–1312. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Korsmeyer SJ, Wei MC, Saito M, Weiler S,
Oh KJ and Schlesinger PH: Pro-apoptotic cascade activates BID,
which oligomerizes BAK or BAX into pores that result in the release
of cytochrome c. Cell Death Differ. 7:1166–1173. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kroemer G and Reed JC: Mitochondrial
control of cell death. Nat Med. 6:513–519. 2000. View Article : Google Scholar
|
|
8
|
Martinou JC and Green DR: Breaking the
mitochondrial barrier. Nat Rev Mol Cell Biol. 2:63–67. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sugita H, Iida S, Inokuchi M, et al:
Methylation of BNIP3 and DAPK indicates lower
response to chemotherapy and poor prognosis in gastric cancer.
Oncol Rep. 25:513–518. 2011.
|
|
10
|
Jiang WG, Watkins G, Lane J, et al:
Prognostic value of rho GTPases and rho guanine nucleotide
dissociation inhibitors in human breast cancers. Clin Cancer Res.
9:6432–6440. 2003.PubMed/NCBI
|
|
11
|
Nazarenko IA, Bhatnagar SK and Hohman RJ:
A closed tube format for amplification and detection of DNA based
on energy transfer. Nucleic Acids Res. 25:2516–2521. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zuker M: Mfold web server for nucleic acid
folding and hybridization prediction. Nucleic Acids Res.
31:3406–3415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang WG, Hiscox S, Hallett MB, Scott C,
Horrobin DF and Puntis MC: Inhibition of hepatocyte growth
factor-induced motility and in vitro invasion of human colon cancer
cells by gamma-linolenic acid. Br J Cancer. 71:744–752. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang WG, Hiscox SE, Parr C, et al:
Antagonistic effect of NK4, a novel hepatocyte growth factor
variant, on in vitro angiogenesis of human vascular endothelial
cells. Clin Cancer Res. 5:3695–3703. 1999.PubMed/NCBI
|
|
15
|
Ye L, Kynaston H and Jiang WG: Bone
morphogenetic protein-9 induces apoptosis in prostate cancer cells,
the role of prostate apoptosis response-4. Mol Cancer Res.
6:1594–1606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kissil JL, Feinstein E, Cohen O, et al:
DAP-kinase loss of expression in various carcinoma and B-cell
lymphoma cell lines: possible implications for role as tumor
suppressor gene. Oncogene. 15:403–407. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Esteller M, Sanchez-Cespedes M, Rosell R,
Sidransky D, Baylin SB and Herman JG: Detection of aberrant
promoter hypermethylation of tumor suppressor genes in serum DNA
from non-small cell lung cancer patients. Cancer Res. 59:67–70.
1999.PubMed/NCBI
|
|
18
|
Sanchez-Cespedes M, Esteller M, Wu L, et
al: Gene promoter hypermethylation in tumors and serum of head and
neck cancer patients. Cancer Res. 60:892–895. 2000.PubMed/NCBI
|
|
19
|
Sasaki H, Ide N, Yukiue H, et al: Arg and
DAP3 expression was correlated with human thymoma stage. Clin Exp
Metastasis. 21:507–513. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jacques C, Fontaine JF, Franc B, et al:
Death-associated protein 3 is overexpressed in human thyroid
oncocytic tumours. Br J Cancer. 101:132–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Suzuki T, Terasaki M, Takemoto-Hori C, et
al: Proteomic analysis of the mammalian mitochondrial ribosome.
Identification of protein components in the 28 S small subunit. J
Biol Chem. 276:33181–33195. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cavdar Koc E, Burkhart W, Blackburn K,
Moseley A and Spremulli LL: The small subunit of the mammalian
mitochondrial ribosome. Identification of the full complement of
ribosomal proteins present. J Biol Chem. 276:19363–19374.
2001.PubMed/NCBI
|
|
23
|
Zhang Z and Gerstein M: Identification and
characterization of over 100 mitochondrial ribosomal protein
pseudogenes in the human genome. Genomics. 81:468–480. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mukamel Z and Kimchi A: Death-associated
protein 3 localizes to the mitochondria and is involved in the
process of mitochondrial fragmentation during cell death. J Biol
Chem. 279:36732–36738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cavdar Koc E, Ranasinghe A, Burkhart W, et
al: A new face on apoptosis: death-associated protein 3 and PDCD9
are mitochondrial ribosomal proteins. FEBS Lett. 492:166–170.
2001.PubMed/NCBI
|