Introduction
Ovarian cancer is the leading cause of death among
malignancies of the female reproductive system, with a high rate of
mortality worldwide. Early-stage ovarian cancer is frequently
asymptomatic and difficult to detect; thus, most patients are in
advanced stages [International Federation of Gynecology and
Obstetrics (FIGO) stage III and IV] at the time of initial
diagnosis (1), and 5-year survival
rates are less than 40% with only modestly improved survival noted
over the past 40 years (2). The
current therapy for ovarian cancer is debulking surgery followed by
cisplatin-centered chemotherapy (3). Although cisplatin-centered
chemotherapy achieves a complete response rate of 40 to 60% in
advanced ovarian cancer patients (4), long-term survival remains poor as a
result of recurrence and emergence of drug resistance that finally
leads to fatal disease (5).
Drug resistance, both intrinsic and acquired,
results from a variety of factors including individual variations
in patients and somatic cell genetic differences in tumors
(6). A number of factors such as
decreased cell-associated drugs, altered drug inactivation,
increased DNA damage tolerance/repair, increased anti-apoptotic
regulator activity and growth factor receptor deregulation are
considered to be responsible for drug resistance in ovarian cancer
(7,8). However, regardless of the mechanisms,
abnormal expression of drug resistance-related genes often plays an
important role in drug resistance. And among all of these drug
resistance-related genes, oncogenes are clearly the key
players.
Oncogenes refer to genes whose activation can
contribute to the development of cancer (9) and many are involved in drug resistance
in varied types of cancers. For example, the overexpression and
activation of the c-myc oncogene is associated with drug
resistance in small cell lung carcinoma (10); the LRF oncogene is a survival
factor in chondrosarcoma and contributes to tumor malignancy and
drug resistance (11) and the
STAT3 oncogene is a predictive marker of drug resistance in
cancer (12). In ovarian cancer, a
total of 13 oncogenes including CCNE1, PIK3CA,
RAB25, MYC, PRKC1, FGF1, NOTCH3,
PIK3R1, AKT2, EGFR, ERBB2, KRAS
and AURKA are associated with cancer development (13), and several genes such as KRAS
(14), ERBB2 (15), PIK3CA (16,17)
and PIK3R1 (18) are
involved in drug resistance.
NEK2 [NIMA (never in mitosis gene A)-related
expressed kinase 2], a serine/threonine centrosomal kinase, is
highly expressed and activated during the S and G2 phases of the
cell cycle (19). NEK2 has emerged
as an important oncogene due to its regulatory role in mitosis
(20). NEK2 has been proven to play
pivotal roles in the development of several types of cancers, while
its relationship with ovarian cancer is rare. More recently, NEK2
has become important since two studies indicate that high
expression of NEK2 induces drug resistance in multiple myeloma
(21,22). However, despite these study, the
research on NEK2 in regards to drug resistance in cancers is still
limited, and its relationship with drug resistance in ovarian
cancer has never been reported. In the present study, on the basis
of a comprehensive bioinformatic analysis, for the first time, we
report that NEK2 may contribute to drug resistance in ovarian
cancer.
Materials and methods
The target gene, NEK2, which is closely associated
with drug resistance in multiple myeloma (21,22)
was selected for bioinformatic analysis.
Databases
The microarray data of NEK2 in ovarian cancer
tissues was retrieved from the Oncomine online database (https://www.oncomine.org/resource/main.html); the
microarray data of NEK2 in ovarian cancer cells was retrieved from
the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geoprofiles/) (23). The protein/gene-protein/gene
interaction network was generated using the GeneMANIA online tool
(http://www.genemania.org/) (24); annotation of biological processes
was performed using the COREMINE online tool (http://www.coremine.com/medical/) (25). The microRNAs (miRNAs) targeted to
the gene were predicted by miRWalk online tool which included 7
prediction tools (DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid,
PICTAR5, Targetscan) (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)
(26).
NEK2 expression in ovarian cancer cells retrieved
from the GEO database was normalized. Unpaired, two-tailed t-test
assuming homogeneity of the variances was performed with Excel
software.
Results
Function of NEK2 in cancers
Genetic screening for cell division cycle mutants in
the filamentous fungus Aspergillus nidulans resulted in the
discovery of never in mitosis A (NIMA), a serine/threonine kinase
that is essential for mitotic entry. Since then, NIMA-related
kinases (NEKs) have been identified in most eukaryotes, including
humans in which 11 genetically distinct proteins named NEK1 to
NEK11 have been discovered (27).
The NEKs play crucial roles in several aspects of mitotic
progression, such as chromatin condensation, nuclear envelope
breakdown, spindle assembly checkpoint signaling and cytokinesis.
Of the human NEKs, NEK2 is the most closely related to
Aspergillus NIMA and is the first NEK which has been
relatively well studied (27).
Similar to NIMA, NEK2 is a cell cycle regulated kinase, with a peak
of expression and activity in the S and G2 phase of the cell cycle,
and it is a core component of the human centrosome (28,29).
NEK2 also contributes to the establishment of the microtubule-based
mitotic spindle (27).
It has been proven that the NEK2 protein is elevated
2- to 5-fold in cell lines derived from a wide range of human
tumors including those of cervical, ovarian, breast, prostate and
leukemic origin (30), suggesting
its potential roles in cancer development. The role of NEK2 in
breast cancer has been extensively studied. In various human breast
cancer cell lines, suppression of NEK2 was found to induce
aneuploidy and cell cycle arrest resulting in cell death.
Significantly, the breast cancer cell line which was most sensitive
to NEK2 depletion was of the triple negative breast cancer subtype.
These results indicate that NEK2 plays crucial roles in breast
cancer growth at primary and secondary sites (31). Similarly, Wang et al
(32) indicated that the abnormal
expression of NEK2 and β-catenin may be one of the mechanisms for
tumorigenesis, particularly for abnormal tumor proliferation, and
the cytoplasmic expression of NEK2 is associated with both tumor
grade and tumor size. These results suggest that NEK2 and β-catenin
may represent new potential targets for therapeutic intervention.
Moreover, a study indicated that the genetic polymorphisms of NEK2
are related to breast cancer susceptibility in Chinese Han women
(33). In addition, the NEK2C,
which is a splice variant of NEK2, which was found to have
significantly upregulated expression in breast cancer cell lines as
well as in human primary breast cancer tissue, plays a crucial role
in breast cancer development and NEK2C inhibition may be a useful
therapeutic target for human breast tumors (34). In addition to the important roles in
breast cancer, NEK2 also plays a role in other types of cancer. For
instance, NEK2 is shown to be involved in the development of lung
cancer induced by smoking and affects patient survival (35). In cervical cancer, NEK2 was induced
in 102 cancer biopsies when compared with 24 normal controls,
indicating its potential roles in cervical cancer (36).
The role of NEK2 in ovarian cancer is less
understood. Limited studies indicate that NEK2 is overexpressed in
ovarian cancer cells (30) and its
expression is regulated by NR2F2 which is associated with a
significantly shorter disease-free interval (37), yet no further research has been
reported. Likewise, the relationship between NEK2 and drug
resistance is unclear, with only several studies indicating its
drug resistance-related functions. The combined use of both NEK2
siRNA and cisplatin inhibited cell proliferation and induced
apoptotic cell death in vitro, suggesting that NEK2 may be
associated with drug resistance in colorectal cancer (38). Similarly, the combination of NEK2
siRNA with paclitaxel and doxorubicin was found to improve the
sensitivity of breast cancer cells during chemotherapy treatment
(20). More recently, two studies
strongly support the idea that NEK2 contributes to drug resistance
in cancers. Zhou et al (21)
and Harrison (22) revealed that
high expression of NEK2 induced drug resistance mainly through
activation of efflux pumps and it may be a strong predictor of drug
resistance and poor prognosis in multiple myeloma and other types
of cancers.
Taken together, NEK2, an oncogene, plays crucial
roles in the development of various types of cancers, but related
studies in ovarian cancer are rare. In addition, although several
studies have revealed the drug resistance-related functions of NEK2
in several cancers, studies in this area are limited and its
association with drug resistance in ovarian cancer has never been
reported.
Expression of NEK2 in ovarian cancer and
drug-resistant cells
NEK2 is considered as an oncogene and is
overexpressed in various tumors. Thus, as an oncogene, NEK2 should
be overexpressed in ovarian cancer when compared to the normal
control and should be upregulated in drug-resistant cells when
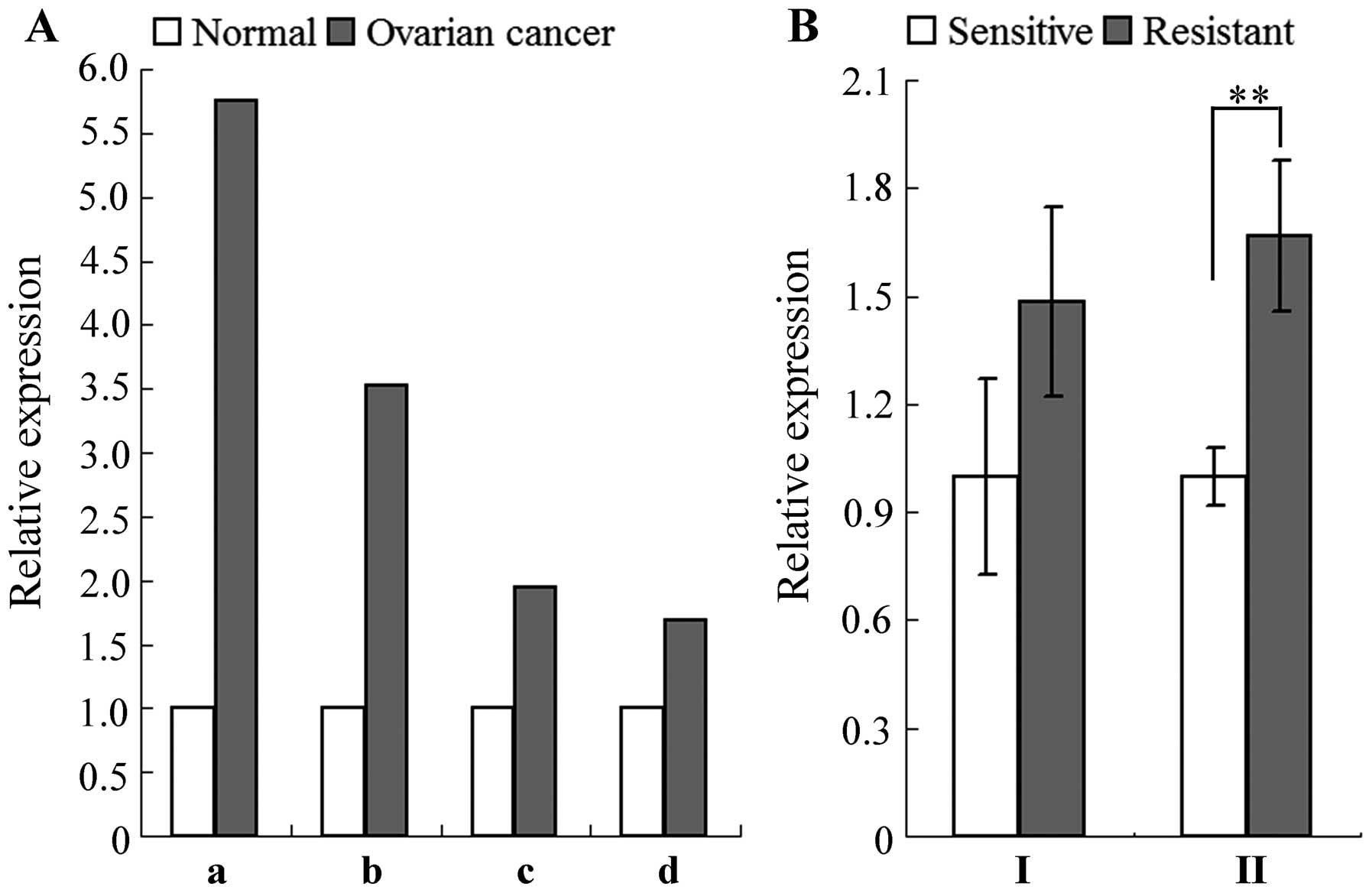
compared to sensitive cells. Based on microarray data retrieved
from the Oncomine online database, we revealed that among 4
independent microarray analyses which covered a total of 759
ovarian cancer tissues and 26 normal controls, NEK2 was
overexpressed 1.687- to 5.754-fold in ovarian cancer tissues when
compared with the normal controls (Fig.
1A). Regarding two microarray data sets from the GEO Profiles
database, NEK2 was found to be elevated 1.5- to 1.7-fold in
cisplatin-resistant A2780 ovarian cancer cells when compared with
the sensitive counterparts (Fig.
1B). These results suggest that NEK2 may be involved in the
development of ovarian cancer, including the regulation of drug
resistance.
Functional prediction and analysis based
on protein/gene-protein/gene interaction
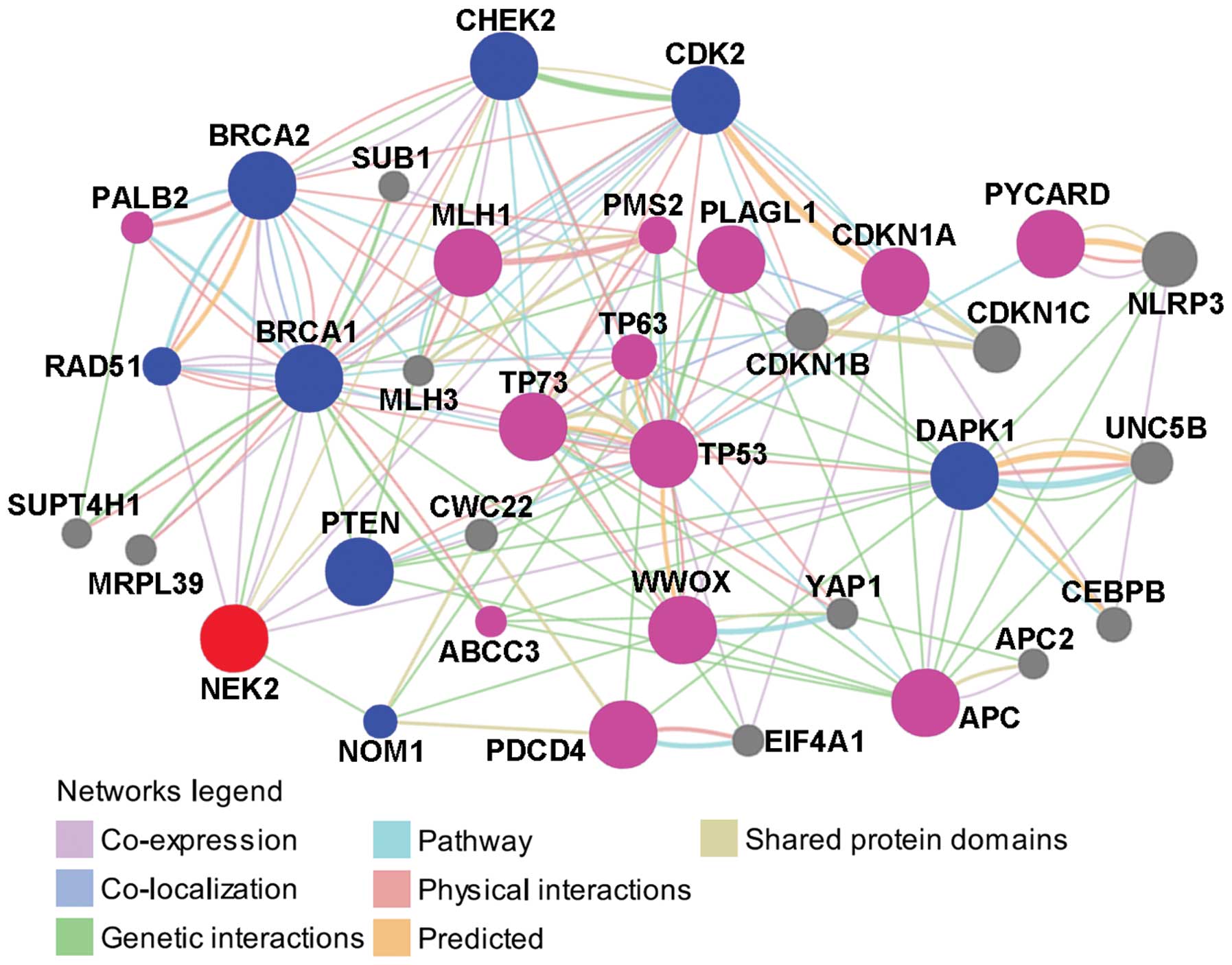
The protein/gene interaction of NEK2 with other
proteins/genes was analyzed using GeneMANIA online tool. As shown
in Fig. 2, NEK2 has direct
interactions with 8 proteins/genes. Among these, NEK2 was
co-expressed with RAD51, BRCA2, PTEN and DAPK1, was co-expressed
and had genetic interaction with BRCA1, was co-expressed and shared
protein domains with CDK2, shared protein domains with CHEK2 and
had genetic interactions with NOM1. With the exception of NOM1, the
other 7 proteins/genes are all associated with drug resistance in
ovarian cancer.
BRCA1 and BRCA2 proteins are critically important
for the repair of double-strand breaks (DSB) by homologous
recombination (HR) (39). They
cooperate in DNA damage responses in a PALB2-dependent manner and
have important implications for the genesis of ovarian cancer and
for chemotherapy with DNA interstrand cross-linking agents
(40). The mRNA expression of BRCA1
has a negative correlation with the clinical sensitivity of
platinum-based chemotherapy (41).
In addition, BRCA1 is positively correlated with MDR1 which is
significantly involved in drug resistance and disease progression
(42). In regards to BRCA2,
previous research indicates that the functional restoration of
BRCA2 due to a secondary BRCA2 mutation is involved in acquired
drug resistance of BRCA2-mutated ovarian carcinomas (43). These results implicate the important
role of BRCA1 and BRCA2 in drug resistance in ovarian cancer. Rad51
functionally interacts with BRCA1 in the meiotic and mitotic cell
cycles (44), and, on the basis of
an interaction network (Fig. 2),
Rad51 co-expressed, physically interacted and shared pathways with
BRCA1, and co-expressed, and shared pathways with BRCA2. These
results indicate that Rad51 which is closely associated with BRCA1
and BRCA2 may also have drug resistance-related functions in
ovarian cancer. The PTEN tumor-suppressor is a central negative
regulator of the PI3K/AKT signaling cascade and suppresses cell
survival as well as cell proliferation. It is found to be either
inactivated or mutated in various human malignancies. Previously
studies suggest that PTEN may be involved in drug resistance via
the PI3K/AKT pathway and the p53-mediated apoptotic cascade.
Reduction in PTEN expression was found to result in the development
of drug resistance in OVCAR-3 cells and the alterations conferred
resistance to cisplatin through the activation of PI3K/Akt and the
inhibition of Bax translocation (17). Further research indicates that
overexpression of PTEN reverses chemoresistance to cisplatin in
human ovarian cancer cells through inactivation of the PI3K/AKT
cell survival pathway and may serve as a potential molecular target
for the treatment of chemoresistant ovarian cancer (45). Moreover, overexpression of PTEN
upregulates p53 content and increases the sensitivity of
chemoresistant cells to cisplatin-induced apoptosis without
detectable changes in the levels of phosphorylated Akt, suggesting
that PTEN may be involved in drug resistance through a p53-mediated
apoptotic cascade independent of the PI3K/Akt pathway (46). CHEK2 is one of the critical kinases
governing cell apoptosis, cell cycle checkpoint and DNA damage
repair. In ovarian cancer cells, CHEK2 is degraded at the protein
level in response to cisplatin through the ubiquitin-proteasome
pathway, suggesting that degradation or decreased expression of
CHEK2 is partially responsible for chemoresistance (47). The expression of DAPK1 and DAPK2 is
altered in multi-drug-resistant gastric cancer cell lines and thus
these genes may be an integral part of the mechanisms responsible
for chemoresistance (48). In
ovarian cancer, DAPK1 has direct interactions with FBXO32, PDCD4,
PTEN, TP53 and TP73, and has indirect interactions with BRCA1,
BRCA2, CDKN1A, IL24, MLH1 and SULF1, which are all involved in drug
resistance in ovarian cancer, suggesting its potential role in drug
resistance in ovarian cancer (49).
As for CDK2, a previous study revealed that cisplatin consistently
induces transient S-phase arrest by inhibiting the CDK2/cyclin A
complex in the S-phase at 12 h after treatment with cisplatin or
DAP in combination with the mitotic inhibitor nocodazole and also
potently inhibits G1-phase CDK2/cyclin E activities at 18 h
(50), indicating that CDK2 can
directly respond to cisplatin in ovarian cancer cells.
In addition to those with direct interactions with
NEK2, there were other proteins/genes in the network which
indirectly interacted with NEK2 (Fig.
2). Among those, MLH1 (51,52),
PDCD4 (53), TP53 (54), TP73 (55), WWOX (56), CDKN1A (57), PLAGL1, PYCARD, APC and TP63
(49), PALB2 (40), ABCC3 (58) and PMS2 (59) are associated with drug resistance in
ovarian cancer.
Collectively, based on the protein/gene interaction
network analysis, 8 proteins/genes were identified which directly
interact with NEK2, and 7 of them were found to contribute to drug
resistance in ovarian cancer; 24 proteins/genes were found to
indirectly interact with NEK2 and 13 of them were found to be
associated with drug resistance in ovarian cancer. These results
indicate a potential role of NEK2 in drug resistance in ovarian
cancer.
Functional annotation of NEK2 with its interacting
proteins/genes was performed to further reveal the relationship of
NEK2 with drug resistance. As shown in Table I, the main functions of NEK2 with
its interacting proteins/genes are cell cycle-related and
microtubule-related functions, which were both proven to be
associated with drug resistance in ovarian and other cancer types.
Cell cycle-mediated drug resistance is best described as a relative
insensitivity to a chemotherapeutic agent because of the position
of the cells in the cell cycle. The cell cycle is closely involved
in the chemosensitivity to combination chemotherapy, and the
chemotherapeutic agents correlated with cell cycle events include
taxanes, platinum, camptothecin and fluorouracil (60). It has been proven that the cell
cycle is closely associated with drug resistance in ovarian cancer.
Integration of DNA methylation and gene expression reveals specific
platinum resistance-related signaling pathways in ovarian cancer,
which include cell growth-promoting pathways, PI3K/Akt and cell
cycle progression (61). Moreover,
numerous genes participate in drug resistance through regulation of
the cell cycle. For example, Phb1 induces G0/G1 phase cell cycle
arrest and promotes cancer cell survival, and silencing of Phb1
expression is a valuable therapeutic approach for chemoresistant
ovarian cancer by increasing the sensitivity of cancer cells to
apoptosis (62). Likewise,
comprehensive bioinformatic analysis revealed that 15
tumor-suppressor genes (TSGs) associated with drug resistance in
ovarian cancer perform their drug resistance-related functions
through 5 pathways including the cell cycle (49). In addition, the cell cycle is also
involved in drug transportation in cancer. Multidrug resistance
driven by ABC membrane transporters is one of the major reasons for
treatment failure in human malignancies, and modulation of MDR by
cell cycle-related factors has been observed in MCF-7 breast cancer
cells (63).
 | Table IAnnotated functions of NEK2 with its
interacting proteins/genes based on the protein/gene-protein/gene
interaction network. |
Table I
Annotated functions of NEK2 with its
interacting proteins/genes based on the protein/gene-protein/gene
interaction network.
| Annotated
function | FDRa | NEK2 and other
proteins/genes in the network |
|---|
| Cell
cycle-related |
| Regulation of
mitotic cell cycle | 2.08e-7 | NEK2, BRCA2, TP53,
TP63, TP73, CDK2, APC, PTEN, CDKN1A, CDKN1B |
| Centrosome
cycle | 2.02e-4 | NEK2, BRCA1, BRCA2,
CDK2 |
| G2/M transition of
mitotic cell cycle | 1.99e-2 | NEK2, CHEK2, CDK2,
CDKN1A |
| Regulation of
mitosis | 5.76e-2 | NEK2, PTEN,
APC |
| Centrosome
organization | 8.97e-4 | NEK2, BRCA1, BRCA2,
CDK2 |
|
Microtubule-related |
| Microtubule
cytoskeleton organization | 3.66e-4 | NEK2, BRCA1, BRCA2,
CDK2, CHEK2, APC |
| Microtubule
organizing center organization | 1.18e-3 | NEK2, BRCA1, BRCA2,
CDK2 |
| Microtubule-based
process | 1.47e-3 | NEK2, BRCA1, BRCA2,
CDK2, CHEK2, APC |
| Regulation of
microtubule cytoskeleton organization | 3.07e-2 | NEK2, BRCA1,
APC |
| Regulation of
microtubule-based process | 4.43e-2 | NEK2, BRCA1,
APC |
Microtubules are intracellular tubular structures
found in all eukaryotic cells. Microtubules have various functions
including organization of intracellular structure, cell division
and intracellular transport (64).
However, abnormal changes in microtubules lead to drug resistance
in cancers. For instance, RASSF1A, tumor-suppressor protein, forms
an endogenous complex with tubulin and promotes the stabilization
of microtubules. Previous research revealed a strong correlation
between the reduced relative expression of RASSF1A and taxol
resistance in primary ovarian cancer. The reason is that the loss
of RASSF1A expression sensitizes cells to microtubule destabilizing
stimuli finally resulting in the development of drug resistance
(65). In ovarian cancer treatment,
anticancer drugs including paclitaxel, epothilone B (EpoB) and
discodermolide all specifically interfere with microtubules and
arrest cells in the G2/M phase of the cell cycle (66).
Collectively, on the basis of functional annotation,
the two ovarian cancer drug resistance-related functions, including
cell cycle- and microtubule-related functions, were annotated to be
the major functions of NEK2 with its interacting proteins/genes of
which most are associated with drug resistance in ovarian cancer
(Fig. 2). These results suggest
that NEK2 may contribute to drug resistance in a cell cycle- and/or
microtubule-mediated manner in ovarian cancer.
Functional prediction and analysis based
on annotation of biological processes
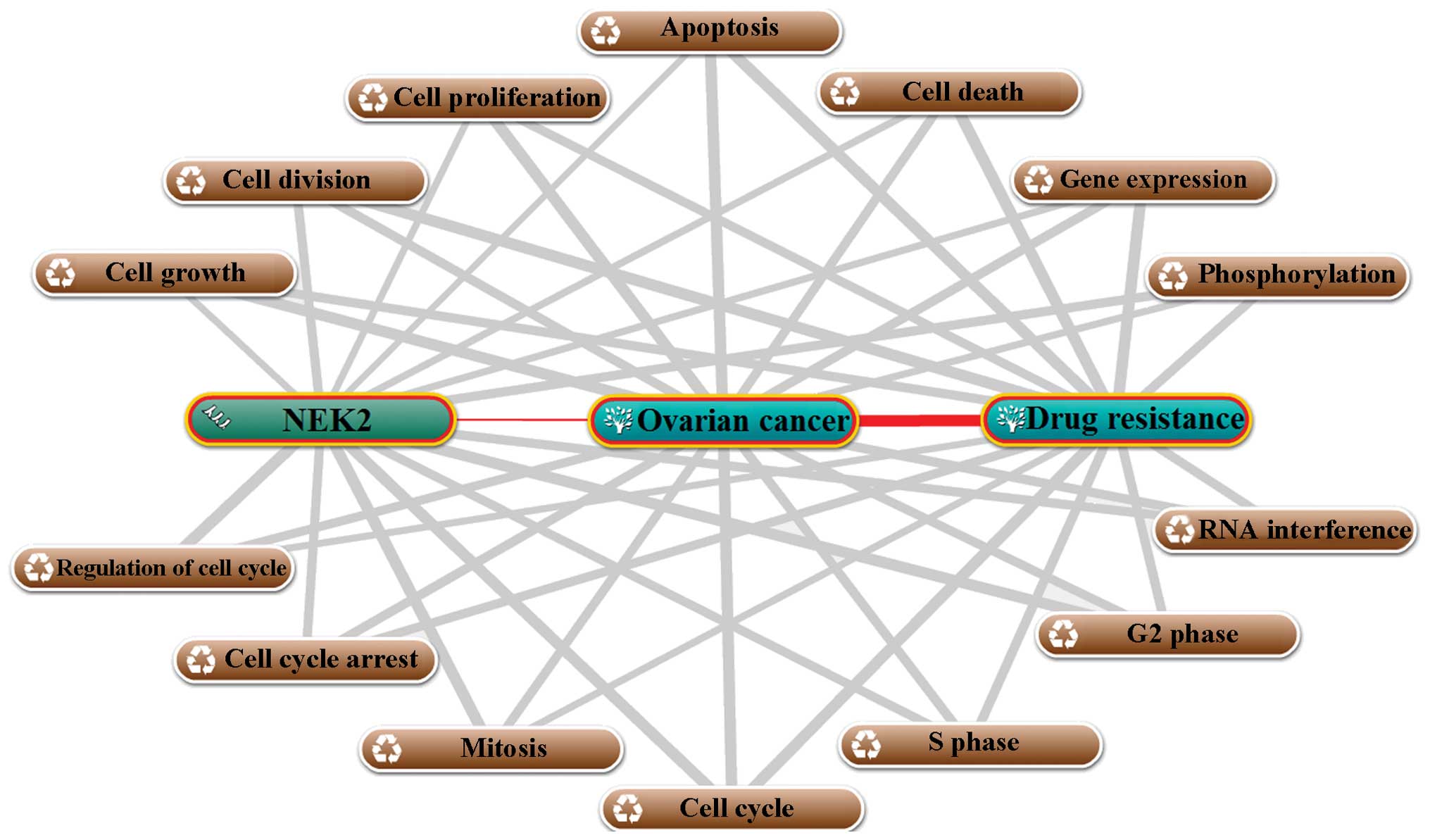
The biological process annotation was performed
using COREMINE online database/tool. As shown in Fig. 3, a total of 14 biological processes
were annotated with NEK2, ovarian cancer and drug resistance
(p<0.00265), which suggested that, on the one hand, those
biological processes contribute to the development of ovarian
cancer and drug resistance, and on the other hand, NEK2 was
associated with these biological processes and it may be a
regulator for those processes. Taken together, the annotation of
biological processes suggested that NEK2 may contribute to drug
resistance via 14 biological processes in ovarian cancer. The 14
biological processes may be mainly divided into two groups
including growth/death-related processes which include cell growth,
cell division, cell proliferation, apoptosis and cell death and
cell cycle-related processes including cell cycle arrest,
regulation of cell cycle, mitosis, the cell cycle, S and G2 phase.
These results indicate that NEK2 may participate in the regulation
of ovarian cancer drug resistance mainly through the cell cycle,
cell growth and death, in particular, through the cell cycle. This
conclusion was consistent with the result of our functional
annotation (Table I).
Functional prediction and analysis based
on miRNA-mRNA interaction
As shown in Table
II, the top 10 miRNAs targeted to NEK2 were predicted by 7
prediction tools, and the drug resistance-related functions of
these miRNAs were reviewed and integrated. Among the top 10 miRNAs,
7 are associated with drug resistance in ovarian and other cancers.
For instance, the expression levels of miR-27a were found to be
increased in paclitaxel-resistant ovarian cancer cell line
A2780/taxol when compared with its parental line A2780.
Transfection of A2780/taxol cells with inhibitors of miR-27a
enhanced the sensitivity of A2780/taxol cells to paclitaxel and
increased paclitaxel-induced apoptosis, which suggest that the
deregulation of miR-27a may be involved in the development of drug
resistance in ovarian cancer (67).
miRNA expression profiles revealed that miR-27b is downregulated in
most drug-resistant Ehrlich ascites tumor cells, suggesting this
miRNA may be involved in drug resistance (68), even though no further results have
been reported. miR-150 was found to have low expression in most
primary ovarian tumors with significantly increased expression in
omental lesions, and miR-150 increased the number of residual
surviving cells by 2- to 4-fold when challenged with lethal
cisplatin concentrations, indicating that the upregulation of
miR-150 may stimulate survival and increase drug tolerance. Thus,
miR-150 may be a critical regulator for the emergence of
drug-resistant disease (69).
miR-374a was found to be downregulated in head and neck squamous
cell carcinoma cells exposed to cisplatin, resulting in the
subsequent modulation of mRNA expression of several targets
including DICER1, DDIT1 and DDIT4 which are involved in the
apoptotic process. This indicates that miR-374a may mediate key
pathways implicated in the response of cancer cells to
chemotherapeutic drugs (70).
miR-630 was found to arrest non-small cell lung cancer A549 cells
in the G0–G1 phase of the cell cycle, correlating with increased
levels of the cell cycle inhibitor p27 as well as with reduced
proliferation rates resulting in greatly diminished sensitivity of
A549 cells to the late S-G2-M cell cycle arrest mediated by
cisplatin. These results suggest that miR-630 may be a novel
modulator of the cisplatin response in non-small cell lung cancer
(71). In addition, miR-24-2 is
capable of inducing apoptosis by modulating different apoptotic
pathways and targeting BCL-2. Furthermore, cells overexpressing
miR-24-2 are hypersensitive to DNA-damaging drugs such as cisplatin
and undergo apoptotic cell death, suggesting that miR-24-2 may be
involved in drug resistance in human cancer cells (MCF-7 and HeLa)
(72). miR-128 is obviously
expressed at a higher level in drug-resistant breast cancer samples
than its expression in drug-sensitive samples. Following
transfection with a precursor of miR-128 or antisense-miR-128
oligonucleotides, the chemosensitivity of MDA-MB-231 cell was
upregulated (73), suggesting a
direct association of miR-128 with drug resistance in cancer.
 | Table IIThe top 10 miRNAs targeted by NEK2 as
predicted by miRNA-mRNA interactions and their drug
resistance-related functions in cancer. |
Table II
The top 10 miRNAs targeted by NEK2 as
predicted by miRNA-mRNA interactions and their drug
resistance-related functions in cancer.
| microRNA | microRNA-mRNA
prediction tool | Drug
resistance-related functions of the microRNAs in cancers
(ref.) |
|---|
|
|---|
| a | b | c | d | e | f | g |
|---|
| hsa-miR-27a | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Drug resistance in
ovarian cancer (67) |
| hsa-miR-27b | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Drug resistance in
Ehrlich ascites tumor cells (68) |
| hsa-miR-150 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | Drug resistance and
metastasis in ovarian cancer (69) |
| hsa-miR-374a | 1 | 1 | 1 | 1 | 0 | 1 | 1 | Drug resistance in
head and neck squamous cell carcinoma cells (70) |
| hsa-miR-374b | 1 | 1 | 1 | 1 | 0 | 1 | 1 | No reported
functions in cancer |
| hsa-miR-630 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | Drug resistance in
non-small cell lung cancer (71) |
| hsa-miR-24-2 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | Drug resistance in
human cancer cells (MCF-7 and HeLa) (72) |
| hsa-miR-216b | 1 | 1 | 0 | 1 | 0 | 1 | 1 | Cell proliferation,
invasion and tumor growth and cellular senescence in nasopharyngeal
carcinoma and colorectal cancer (74,75) |
| hsa-miR-128 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | Drug resistance in
breast cancer (73) |
| hsa-miR-486-5p | 1 | 1 | 0 | 1 | 0 | 1 | 1 | Cell migration and
invasion in non-small cell lung cancer (76) |
In regards to miR-216b and miR-486-5p, no study has
indicated their associations with drug resistance in cancer, but
they contribute to several biological processes during cancer
development. It has been proven that downregulated expression of
miR-216b is directly related to advanced clinical stage and lymph
node metastasis. Both in vitro and in vivo assays
revealed that miR-216b attenuates cell proliferation, invasion and
tumor growth in nude mice (74). In
addition, miR-216b, miR-186, miR-337-3p and miR-760 were found to
cooperatively promote cellular senescence through the p53-p21
(Cip1/WAF1) pathway in human colorectal cancer cells (75). Given that the overexpression of
miR-486-5p is closely correlated with the downregulated expression
of ARHGAP5 in lung tumor tissues, which considerably inhibits lung
cancer cell migration and invasion, miR-486-5p may act as a tumor
suppressor contributing to the progression and metastasis of
non-small cell lung cancer (76).
Actually, the biological processes including cell proliferation,
invasion, growth and metastasis in which miR-216b and miR-486-5p
are involved are closely related to the development of drug
resistance in cancers (49,77), and the annotation of biological
processes also suggests that cell proliferation and growth are
associated with drug resistance. All these results indicate that
miR-216b and miR-486-5p may be associated with drug resistance in
an indirectly way.
Discussion
Inferring the functional role of proteins/genes is a
primary task in biology, for purposes ranging from general
knowledge to drug discovery and diagnostic development (78). Protein/gene function prediction
based on various genome-wide data is a potential, feasible and
valuable strategy for gene function mining, and many large-scale
networks of molecular interactions within the cell have made it
possible to go beyond the one dimensional approach to study protein
function in the context of a network (79). For example, on the basis of
comprehensive bioinformatic analysis, Yin et al (77) reported that SPARCL1 and CCL21 are
associated with drug resistance in ovarian cancer. Similarly, 15
TSGs associated with drug resistance in ovarian cancer were
analyzed by comprehensive bioinformatics to overview the
relationship of the 15 TSGs with drug resistance and to discover
potential TSGs related with drug resistance (49).
Oncomine is a cancer microarray database and
web-based data-mining platform aimed at facilitating the discovery
from genome-wide expression analyses. Differential expression
analyses comparing most major types of cancer with respective
normal tissues are available for exploration. Data can be queried
and visualized for a selected gene across all analyses (80). The Gene Expression Omnibus (GEO) at
the National Center for Biotechnology Information (NCBI) has
emerged as the leading fully public repository for gene expression
data, predominantly gene expression data generated by DNA
microarray technology (23). By
2006, GEO stored approximately a billion individual gene expression
measurements, derived from over 100 organisms, submitted by over
1,500 laboratories, addressing a wide range of biological phenomena
(81). Thus, the expression data of
NEK2 in ovarian cancer tissues and drug-resistant cells retrieved
from the two online databases are reliable.
GeneMANIA is a web-based database and a tool for the
prediction of gene functions on the basis of multiple networks
derived from different genomic or proteomic data/sources (24). Six organisms are currently supported
(Arabidopsis thaliana, Caenorhabditis elegans,
Drosophila melanogaster, Mus musculus, Homo
sapiens and Saccharomyces cerevisiae), and hundreds of data
sets have been collected from GEO, BioGRID, Pathway Commons and
I2D, as well as organism-specific functional genomics data sets
(82). Thus, it is fast enough to
predict gene functions with great accuracy using this software.
COREMINE Medical is a product of the PubGene Company designed to be
used for searching information on health, medicine and biology
(25). COREMINE Medical grew out of
Pubgene online tool which is a gene/protein database and a
web-based tool for literature mining. Pubgene carries out automated
extraction of experimental and theoretical biomedical knowledge
from publicly available gene and text databases to create a
gene-to-gene co-citation network for millions of named human genes
by automated analysis of titles and abstracts in over 10 million
MEDLINE records (83). Therefore,
the functions of NEK2 predicted by these two software programs were
potentially accurate.
miRNAs are a class of small (22 bp) endogenous
non-coding RNAs which regulate gene expression at the
post-transcriptional and translational levels (84,85).
miRNA-mediated post-transcriptional gene regulation is considered
as a significant regulator of many cellular processes including
cell proliferation, apoptosis, differentiation, angiogenesis,
invasion, metastasis and drug resistance, both physiological and
pathological (86–88). miRNAs perform their functions
through the regulation of their target genes, and it has been well
established that miRNAs represent a class of genes with a great
potential for use in diagnostics, prognosis and therapy (89). Therefore, we can predict the
function of genes through the functions of the miRNAs targeting the
gene. miRWalk is a comprehensive database on miRNAs, which collects
predicted and validated miRNA binding sites on all known genes of
the human, mouse and rat. More importantly, the miRWalk is a
potential real-time database, in which the ‘Predicted Target
module’ is updated every 6 months and the ‘Validated Target module’
is updated every month (26).
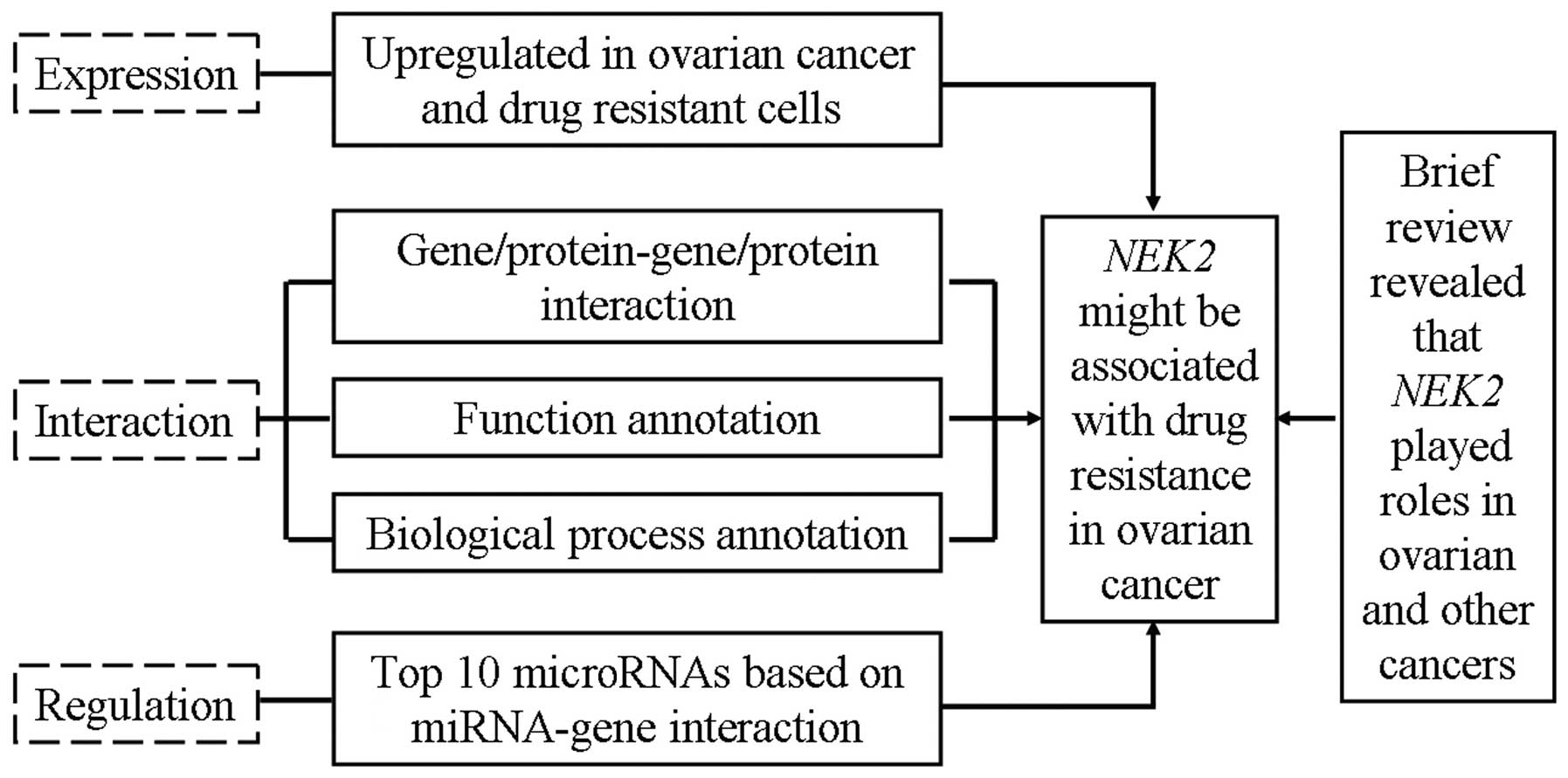
Taken together, based on the comprehensive
bioinformatic analysis including microarray data interpretation,
protein/gene-protein/gene interaction, annotation of biological
processes and miRNA-mRNA interaction (Fig. 4), we revealed that the expression of
NEK2 in drug-resistant ovarian cancer cells was elevated and that
NEK2 directly/indirectly interacts with 20 proteins/genes which all
contribute to drug resistance in ovarian and other cancers
(Fig. 2). Further analysis based on
the annotation of biological processes suggested that NEK2 was
closely related to 14 biological processes which are involved in
ovarian cancer and drug resistance (Fig. 3). Moreover, 7 of 10 miRNAs targeting
NEK2 are involved in drug resistance in ovarian and other cancers
(Table II). Given that NEK2
interacts with numerous proteins/genes, miRNAs and biological
processes which were all found to contribute to drug resistance in
ovarian and other cancers, upregulation of NEK2 in ovarian cancer
and drug-resistant cancer cells may also contribute to
drug-resistance.
Annotation of biological processes revealed that
among the 14 biological processes annotated for NEK2, 6 were cell
cycle-related (Fig. 3), and the
molecular function annotation-based protein/gene-protein/gene
interaction obtained similar results. These results suggest that
NEK2 may participate in the regulation of drug resistance mainly
through regulation of the cell cycle. In addition,
microtubule-related function was another major function of NEK2
with its interacting proteins in accordance with
protein/gene-protein/gene interaction (Table I), which implicated that NEK2 may
also exert its drug resistant functions via microtubules. Actually,
NEK2 has been reported to play important roles in both mitosis and
meiosis (90) and contributes to
the establishment of the microtubule-based mitotic spindle
(27).
Collectively, for the first time, we report that the
overexpression of NEK2 in ovarian cancers/drug-resistant cells may
contribute to drug resistance through regulation of the cell cycle
and microtubules. The present study provides important information
for further investigation of the drug resistant-related functions
of NEK2 in ovarian cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81302283) and the
Youth Science Foundation of Guangxi Medical University (no.
GXMUYSF201205).
References
|
1
|
Landis SH, Murray T, Bolden S and Wingo
PA: Cancer statistics, 1999. CA Cancer J Clin. 49:8–31. 1999.
View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
3
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, Friedlander M, Gabra H, Kaye SB, Lord CJ, Lengyel
E, Levine DA, McNeish IA, Menon U, Mills GB, Nephew KP, Oza AM,
Sood AK, Stronach EA, Walczak H, Bowtell DD and Balkwill FR:
Rethinking ovarian cancer: recommendations for improving outcomes.
Nat Rev Cancer. 11:719–725. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
5
|
Cannistra SA: Cancer of the ovary. N Engl
J Med. 351:2519–2529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson SW, Ozols RF and Hamilton TC:
Mechanisms of drug resistance in ovarian cancer. Cancer. 71(Suppl
2): S644–S649. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osborne C, Wilson P and Tripathy D:
Oncogenes and tumor suppressor genes in breast cancer: potential
diagnostic and therapeutic applications. Oncologist. 9:361–377.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Van Waardenburg RC, Prins J, Meijer C,
Uges DR, De Vries EG and Mulder NH: Effects of c-myc oncogene
modulation on drug resistance in human small cell lung carcinoma
cell lines. Anticancer Res. 16:1963–1970. 1996.
|
|
11
|
Kumari R, Li H, Haudenschild DR, Fierro F,
Carlson CS, Overn P, Gupta L, Gupta K, Nolta J, Yik JH and Di
Cesare PE: The oncogene LRF is a survival factor in chondrosarcoma
and contributes to tumor malignancy and drug resistance.
Carcinogenesis. 33:2076–2083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barre B, Vigneron A, Perkins N, Roninson
IB, Gamelin E and Coqueret O: The STAT3 oncogene as a
predictive marker of drug resistance. Trends Mol Med. 13:4–11.
2007.
|
|
13
|
Zhao M, Sun J and Zhao Z: Distinct and
competitive regulatory patterns of tumor suppressor genes and
oncogenes in ovarian cancer. PLoS One. 7:e441752012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ratner ES, Keane FK, Lindner R, Tassi RA,
Paranjape T, Glasgow M, Nallur S, Deng Y, Lu L, Steele L, Sand S,
Muller RU, Bignotti E, Bellone S, Boeke M, Yao X, Pecorelli S,
Ravaggi A, Katsaros D, Zelterman D, Cristea MC, Yu H, Rutherford
TJ, Weitzel JN, Neuhausen SL, Schwartz PE, Slack FJ, Santin AD and
Weidhaas JB: A KRAS variant is a biomarker of poor outcome,
platinum chemotherapy resistance and a potential target for therapy
in ovarian cancer. Oncogene. 31:4559–4566. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu L, Wu A and Jiang K: Effect of
antisense c-erbB2 on biologic behaviour and chemotherapeutic drug
sensitivity in human ovarian cancer cells. Zhonghua Fu Chan Ke Za
Zhi. 31:169–172. 1996.(In Chinese).
|
|
16
|
Fu S, Hennessy BT, Ng CS, Ju Z, Coombes
KR, Wolf JK, Sood AK, Levenback CF, Coleman RL, Kavanagh JJ,
Gershenson DM, Markman M, Dice K, Howard A, Li J, Li Y, Stemke-Hale
K, Dyer M, Atkinson E, Jackson E, Kundra V, Kurzrock R, Bast RC Jr
and Mills GB: Perifosine plus docetaxel in patients with platinum
and taxane resistant or refractory high-grade epithelial ovarian
cancer. Gynecol Oncol. 126:47–53. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee S, Choi EJ, Jin C and Kim DH:
Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA
amplification contributes to cisplatin resistance in an ovarian
cancer cell line. Gynecol Oncol. 97:26–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stronach EA, Alfraidi A, Rama N, Datler C,
Studd JB, Agarwal R, Guney TG, Gourley C, Hennessy BT, Mills GB,
Mai A, Brown R, Dina R and Gabra H: HDAC4-regulated STAT1
activation mediates platinum resistance in ovarian cancer. Cancer
Res. 71:4412–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fry AM, Schultz SJ, Bartek J and Nigg EA:
Substrate specificity and cell cycle regulation of the Nek2 protein
kinase, a potential human homolog of the mitotic regulator NIMA of
Aspergillus nidulans. J Biol Chem. 270:12899–12905. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee J and Gollahon L: Nek2-targeted ASO or
siRNA pretreatment enhances anticancer drug sensitivity in
triplenegative breast cancer cells. Int J Oncol. 42:839–847.
2013.PubMed/NCBI
|
|
21
|
Zhou W, Yang Y, Xia J, Wang H, Salama ME,
Xiong W, Xu H, Shetty S, Chen T, Zeng Z, Shi L, Zangari M, Miles R,
Bearss D, Tricot G and Zhan F: NEK2 induces drug resistance
mainly through activation of efflux drug pumps and is associated
with poor prognosis in myeloma and other cancers. Cancer Cell.
23:48–62. 2013. View Article : Google Scholar
|
|
22
|
Harrison C: Cancer: a target for drug
resistance. Nat Rev Drug Discov. 12:1902013. View Article : Google Scholar
|
|
23
|
Barrett T and Edgar R: Mining microarray
data at NCBI’s Gene Expression Omnibus (GEO)*. Methods
Mol Biol. 338:175–190. 2006.
|
|
24
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: a real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9(Suppl 1): S42008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
de Leeuw N, Dijkhuizen T, Hehir-Kwa JY,
Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van
Ravenswaaij-Arts CM, Scherer SW, Shams S, Van Vooren S, Sijmons R,
Swertz M and Hastings R: Diagnostic interpretation of array data
using public databases and internet sources. Hum Mutat. Feb
14–2012.(Epub ahead of print). View Article : Google Scholar
|
|
26
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011.
|
|
27
|
Fry AM, O’Regan L, Sabir SR and Bayliss R:
Cell cycle regulation by the NEK family of protein kinases. J Cell
Sci. 125:4423–4433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Andersen JS, Wilkinson CJ, Mayor T,
Mortensen P, Nigg EA and Mann M: Proteomic characterization of the
human centrosome by protein correlation profiling. Nature.
426:570–574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fry AM, Meraldi P and Nigg EA: A
centrosomal function for the human Nek2 protein kinase, a member of
the NIMA family of cell cycle regulators. EMBO J. 17:470–481. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hayward DG, Clarke RB, Faragher AJ, Pillai
MR, Hagan IM and Fry AM: The centrosomal kinase Nek2 displays
elevated levels of protein expression in human breast cancer.
Cancer Res. 64:7370–7376. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cappello P, Blaser H, Gorrini C, Lin DC,
Elia AJ, Wakeham A, Haider S, Boutros PC, Mason JM, Miller NA,
Youngson B, Done SJ and Mak TW: Role of Nek2 on centrosome
duplication and aneuploidy in breast cancer cells. Oncogene. May
27–2013.(Epub ahead of print). View Article : Google Scholar
|
|
32
|
Wang S, Li W, Lv S, Wang Y, Liu Z, Zhang
J, Liu T and Niu Y: Abnormal expression of Nek2 and β-catenin in
breast carcinoma: clinicopathological correlations. Histopathology.
59:631–642. 2011.
|
|
33
|
Wang H, Xie YT, Han JY, Ruan Y, Song AP,
Zheng LY, Zhang WZ, Sajdik C, Li Y, Tian XX and Fang WG: Genetic
polymorphisms in centrobin and Nek2 are associated
with breast cancer susceptibility in a Chinese Han population.
Breast Cancer Res Treat. 136:241–251. 2012.
|
|
34
|
Liu Z, Wang Y, Wang S, Zhang J, Zhang F
and Niu Y: Nek2C functions as a tumor promoter in human breast
tumorigenesis. Int J Mol Med. 30:775–782. 2012.PubMed/NCBI
|
|
35
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW,
Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder
S, Shih JH, Caporaso NE and Jen J: Gene expression signature of
cigarette smoking and its role in lung adenocarcinoma development
and survival. PLoS One. 3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Koch M and Wiese M: Gene expression
signatures of angiocidin and darapladib treatment connect to
therapy options in cervical cancer. J Cancer Res Clin Oncol.
139:259–267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hawkins SM, Loomans HA, Wan YW,
Ghosh-Choudhury T, Coffey D, Xiao W, Liu Z, Sangi-Haghpeykar H and
Anderson ML: Expression and functional pathway analysis of nuclear
receptor NR2F2 in ovarian cancer. J Clin Endocrinol Metab.
98:E1152–E1162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Suzuki K, Kokuryo T, Senga T, Yokoyama Y,
Nagino M and Hamaguchi M: Novel combination treatment for
colorectal cancer using Nek2 siRNA and cisplatin. Cancer Sci.
101:1163–1169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gudmundsdottir K and Ashworth A: The roles
of BRCA1 and BRCA2 and associated proteins in the maintenance of
genomic stability. Oncogene. 25:5864–5874. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang F, Fan Q, Ren K and Andreassen PR:
PALB2 functionally connects the breast cancer susceptibility
proteins BRCA1 and BRCA2. Mol Cancer Res. 7:1110–1118. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao D, Zhang W, Li XG, Wang XB, Li M, Li
YF, Tian HM, Song PP, Liu J, Chang QY and Wu LY: The mRNA
expression of BRCA1, ERCC1, TUBB3, PRR13 genes and their
relationship with clinical chemosensitivity in primary epithelial
ovarian cancer. Zhonghua Zhong Liu Za Zhi. 34:196–200. 2012.(In
Chinese).
|
|
42
|
Lu L, Katsaros D, Wiley A, Rigault de la
Longrais IA, Puopolo M and Yu H: Expression of MDR1 in epithelial
ovarian cancer and its association with disease progression. Oncol
Res. 16:395–403. 2007.PubMed/NCBI
|
|
43
|
Sakai W, Swisher EM, Jacquemont C,
Chandramohan KV, Couch FJ, Langdon SP, Wurz K, Higgins J, Villegas
E and Taniguchi T: Functional restoration of BRCA2 protein
by secondary BRCA2 mutations in BRCA2-mutated ovarian
carcinoma. Cancer Res. 69:6381–6386. 2009.
|
|
44
|
Scully R, Chen J, Plug A, Xiao Y, Weaver
D, Feunteun J, Ashley T and Livingston DM: Association of BRCA1
with Rad51 in mitotic and meiotic cells. Cell. 88:265–275. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu H, Cao Y, Weng D, Xing H, Song X, Zhou
J, Xu G, Lu Y, Wang S and Ma D: Effect of tumor suppressor gene
PTEN on the resistance to cisplatin in human ovarian cancer cell
lines and related mechanisms. Cancer Lett. 271:260–271. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yan X, Fraser M, Qiu Q and Tsang BK:
Over-expression of PTEN sensitizes human ovarian cancer cells to
cisplatin-induced apoptosis in a p53-dependent manner. Gynecol
Oncol. 102:348–355. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang P, Gao W, Li H, Reed E and Chen F:
Inducible degradation of checkpoint kinase 2 links to
cisplatin-induced resistance in ovarian cancer cells. Biochem
Biophys Res Commun. 328:567–572. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang X, Yashiro M, Qiu H, Nishii T,
Matsuzaki T and Hirakawa K: Establishment and characterization of
multidrug-resistant gastric cancer cell lines. Anticancer Res.
30:915–921. 2010.PubMed/NCBI
|
|
49
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Tumor suppressor genes associated with drug resistance in
ovarian cancer (Review). Oncol Rep. 30:3–10. 2013.PubMed/NCBI
|
|
50
|
He G, Kuang J, Khokhar AR and Siddik ZH:
The impact of S- and G2-checkpoint response on the fidelity of
G1-arrest by cisplatin and its comparison to a non-cross-resistant
platinum(IV) analog. Gynecol Oncol. 122:402–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Plumb JA, Strathdee G, Sludden J, Kaye SB
and Brown R: Reversal of drug resistance in human tumor xenografts
by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1
gene promoter. Cancer Res. 60:6039–6044. 2000.
|
|
52
|
Strathdee G, MacKean MJ, Illand M and
Brown R: A role for methylation of the hMLH1 promoter in
loss of hMLH1 expression and drug resistance in ovarian cancer.
Oncogene. 18:2335–2341. 1999.PubMed/NCBI
|
|
53
|
Zhang X, Wang X, Song X, Liu C, Shi Y,
Wang Y, Afonja O, Ma C, Chen YH and Zhang L: Programmed cell death
4 enhances chemosensitivity of ovarian cancer cells by activating
death receptor pathway in vitro and in vivo. Cancer Sci.
101:2163–2170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Reles A, Wen WH, Schmider A, Gee C,
Runnebaum IB, Kilian U, Jones LA, El-Naggar A, Minguillon C,
Schönborn I, Reich O, Kreienberg R, Lichtenegger W and Press MF:
Correlation of p53 mutations with resistance to
platinum-based chemotherapy and shortened survival in ovarian
cancer. Clin Cancer Res. 7:2984–2997. 2001.
|
|
55
|
Al-Bahlani S, Fraser M, Wong AY, Sayan BS,
Bergeron R, Melino G and Tsang BK: P73 regulates cisplatin-induced
apoptosis in ovarian cancer cells via a calcium/calpain-dependent
mechanism. Oncogene. 30:4219–4230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu YY, Li L, Li DR, Zhang W and Wang Q:
Suppression of WWOX gene by RNA interference reverses platinum
resistance acquired in SKOV3/SB cells. Zhonghua Fu Chan Ke Za Zhi.
43:854–858. 2008.(In Chinese).
|
|
57
|
Takai N and Narahara H: Histone
deacetylase inhibitor therapy in epithelial ovarian cancer. J
Oncol. 2010:4584312010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Auner V, Sehouli J, Oskay-Oezcelik G,
Horvat R, Speiser P and Zeillinger R: ABC transporter gene
expression in benign and malignant ovarian tissue. Gynecol Oncol.
117:198–201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fink D, Nebel S, Aebi S, Nehme A and
Howell S: Loss of DNA mismatch repair due to knockout of MSH2 or
PMS2 results in resistance to cisplatin and carboplatin. Int J
Oncol. 11:539–542. 1997.PubMed/NCBI
|
|
60
|
Shah MA and Schwartz GK: Cell
cycle-mediated drug resistance: an emerging concept in cancer
therapy. Clin Cancer Res. 7:2168–2181. 2001.PubMed/NCBI
|
|
61
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 2:342009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gregory-Bass RC, Olatinwo M, Xu W,
Matthews R, Stiles JK, Thomas K, Liu D, Tsang B and Thompson WE:
Prohibitin silencing reverses stabilization of mitochondrial
integrity and chemoresistance in ovarian cancer cells by increasing
their sensitivity to apoptosis. Int J Cancer. 122:1923–1930. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Koshkin V and Krylov SN: Correlation
between multi-drug resistance-associated membrane transport in
clonal cancer cells and the cell cycle phase. PLoS One.
7:e413682012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fojo T and Menefee M: Mechanisms of
multidrug resistance: the potential role of microtubule-stabilizing
agents. Ann Oncol. 18(Suppl 5): v3–v8. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kassler S, Donninger H, Birrer MJ and
Clark GJ: RASSF1A and the taxol response in ovarian cancer. Mol
Biol Int. 2012:2632672012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pellicciotta I, Yang CP, Venditti CA,
Goldberg GL and Shahabi S: Response to microtubule-interacting
agents in primary epithelial ovarian cancer cells. Cancer Cell Int.
13:332013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L
and Wang Z: MiR-27a modulates MDR1/P-glycoprotein expression by
targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol.
119:125–130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Husted S, Søkilde R, Rask L, Cirera S,
Busk PK, Eriksen J and Litman T: MicroRNA expression profiles
associated with development of drug resistance in Ehrlich ascites
tumor cells. Mol Pharm. 8:2055–2062. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Vang S, Wu HT, Fischer A, Miller DH,
MacLaughlan S, Douglass E, Steinhoff M, Collins C, Smith PJ, Brard
L and Brodsky AS: Identification of ovarian cancer metastatic
miRNAs. PLoS One. 8:e582262013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Huang Y, Chuang A, Hao H, Talbot C, Sen T,
Trink B, Sidransky D and Ratovitski E: Phospho-ΔNp63α is a key
regulator of the cisplatin-induced microRNAome in cancer cells.
Cell Death Differ. 18:1220–1230. 2011.
|
|
71
|
Galluzzi L, Morselli E, Vitale I, Kepp O,
Senovilla L, Criollo A, Servant N, Paccard C, Hupé P, Robert T,
Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E and
Kroemer G: miR-181a and miR-630 regulate cisplatin-induced cancer
cell death. Cancer Res. 70:1793–1803. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Srivastava N, Manvati S, Srivastava A, Pal
R, Kalaiarasan P, Chattopadhyay S, Gochhait S, Dua R and Bamezai
RN: miR-24–2 controls H2AFX expression regardless of gene
copy number alteration and induces apoptosis by targeting
antiapoptotic gene BCL-2: a potential for therapeutic
intervention. Breast Cancer Res. 13:R392011.
|
|
73
|
Ji S, Shao G, Lv X, Liu Y, Fan Y, Wu A and
Hu H: Downregulation of miRNA-128 sensitises breast cancer cell to
chemodrugs by targeting Bax. Cell Biol Int. 37:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, Li X, Ma J and Li G: miR-216b
suppresses tumor growth and invasion by targeting KRAS in
nasopharyngeal carcinoma. J Cell Sci. 124:2997–3005. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kim SY, Lee YH and Bae YS: MiR-186,
miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular
senescence by targeting α subunit of protein kinase CKII in human
colorectal cancer cells. Biochem Biophys Res Commun. 429:173–179.
2012.PubMed/NCBI
|
|
76
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, Mannoor K, Deepak J, Donahue
JM, Stass SA, Xing L and Jiang F: Downregulation of miR-486-5p
contributes to tumor progression and metastasis by targeting
protumorigenic ARHGAP5 in lung cancer. Oncogene. Mar 11–2013.(Epub
ahead of print). View Article : Google Scholar
|
|
77
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Bioinformatic analysis of chemokine (C-C motif) ligand 21 and
SPARC-like protein 1 revealing their associations with drug
resistance in ovarian cancer. Int J Oncol. 42:1305–1316.
2013.PubMed/NCBI
|
|
78
|
Jiang X, Gold D and Kolaczyk ED:
Network-based auto-probit modeling for protein function prediction.
Biometrics. 67:958–966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: a cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Barrett T and Edgar R: Gene expression
Omnibus: microarray data storage, submission, retrieval, and
analysis. Methods Enzymol. 411:352–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD and
Morris Q: The GeneMANIA prediction server: biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Jenssen TK, Laegreid A, Komorowski J and
Hovig E: A literature network of human genes for high-throughput
analysis of gene expression. Nat Genet. 28:21–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Behm-Ansmant I, Rehwinkel J and Izaurralde
E: MicroRNAs silence gene expression by repressing protein
expression and/or by promoting mRNA decay. Cold Spring Harb Symp
Quant Biol. 71:523–530. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yi B, Piazza GA, Su X and Xi Y: MicroRNA
and cancer chemoprevention. Cancer Prev Res. 6:401–409. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tili E, Michaille JJ, Gandhi V, Plunkett
W, Sampath D and Calin GA: miRNAs and their potential for use
against cancer and other diseases. Future Oncol. 3:521–537. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tanaka K, Parvinen M and Nigg EA: The in
vivo expression pattern of mouse Nek2, a NIMA-related kinase,
indicates a role in both mitosis and meiosis. Exp Cell Res.
237:264–274. 1997. View Article : Google Scholar : PubMed/NCBI
|