|
1
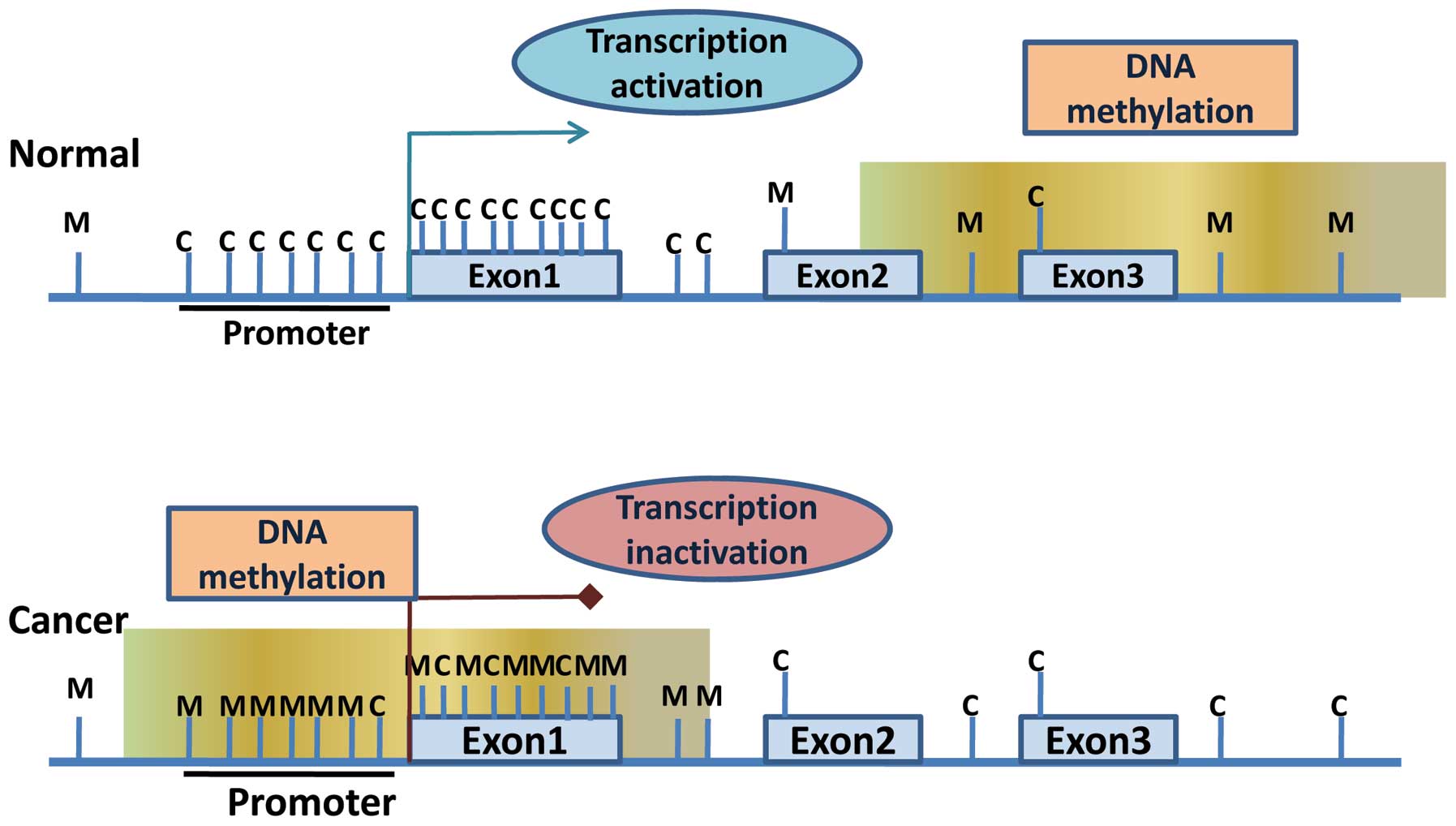
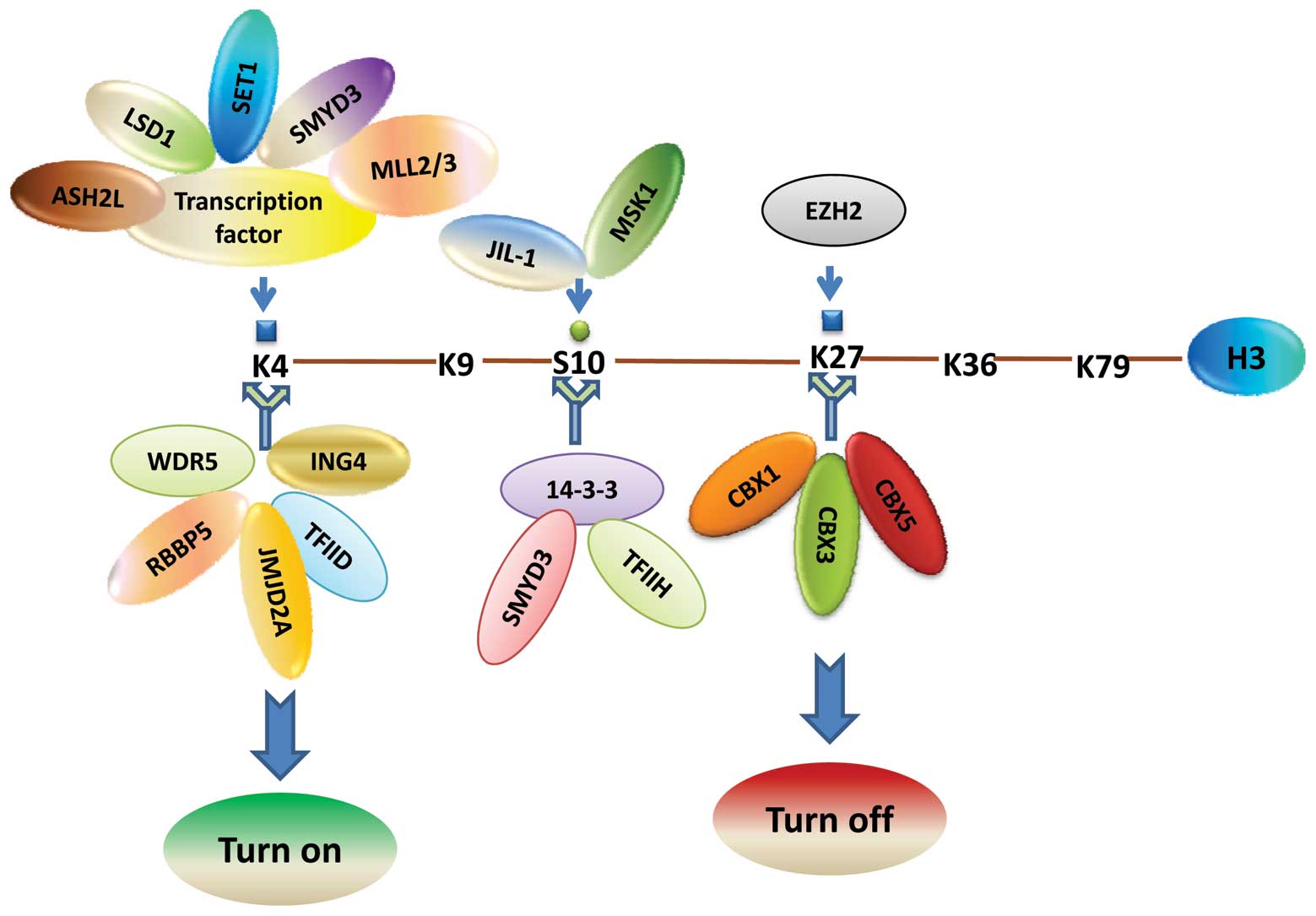
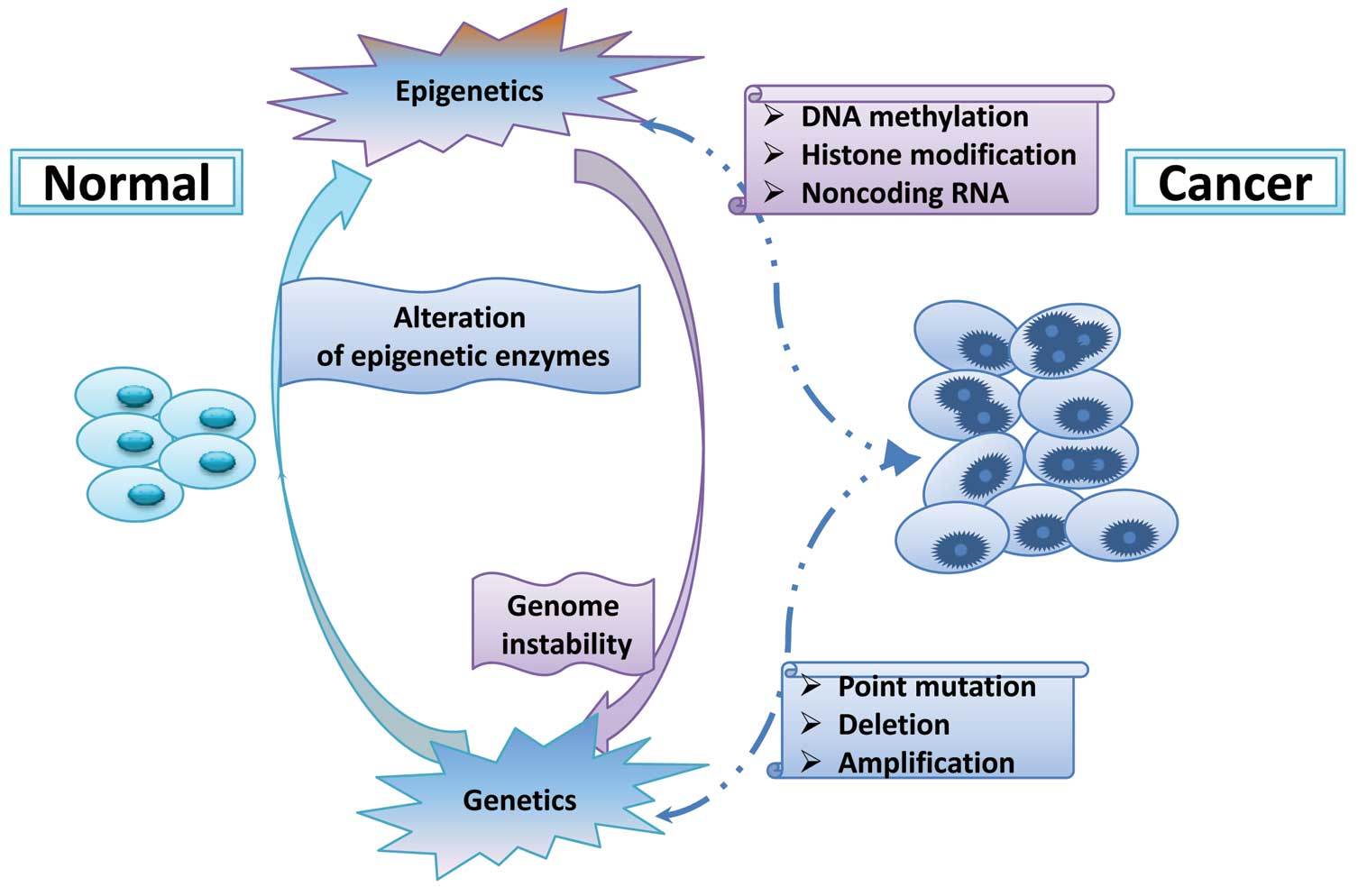
|
Luger K, Mader AW, Richmond RK, Sargent DF
and Richmond TJ: Crystal structure of the nucleosome core particle
at 2.8 A resolution. Nature. 389:251–260. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar
|
|
3
|
Suzuki MM and Bird A: DNA methylation
landscapes: provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rijnkels M, Kabotyanski E,
Montazer-Torbati MB, Beauvais CH, Vassetzky Y, Rosen JM and Devinoy
E: The epigenetic landscape of mammary gland development and
functional differentiation. J Mammary Gland Biol Neoplasia.
15:85–100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bernstein BE, Meissner A and Lander ES:
The mammalian epigenome. Cell. 128:669–681. 2007. View Article : Google Scholar
|
|
6
|
Watt F and Molloy PL: Cytosine methylation
prevents binding to DNA of a HeLa cell transcription factor
required for optimal expression of the adenovirus major late
promoter. Genes Dev. 2:1136–1143. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nan X, Ng HH, Johnson CA, Laherty CD,
Turner BM, Eisenman RN and Bird A: Transcriptional repression by
the methyl-CpG-binding protein MeCP2 involves a histone deacetylase
complex. Nature. 393:386–389. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weber M, Hellmann I, Stadler MB, Ramos L,
Paabo S, Rebhan M and Schubeler D: Distribution, silencing
potential and evolutionary impact of promoter DNA methylation in
the human genome. Nat Genet. 39:457–466. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamada Y, Shirakawa T, Taylor TD, et al: A
comprehensive analysis of allelic methylation status of CpG islands
on human chromosome 11q: comparison with chromosome 21q. DNA Seq.
17:300–306. 2006.PubMed/NCBI
|
|
10
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheung P, Allis CD and Sassone-Corsi P:
Signaling to chromatin through histone modifications. Cell.
103:263–271. 2000. View Article : Google Scholar
|
|
12
|
Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau
LA, Whetstine JR and Price BD: Histone H3 methylation links DNA
damage detection to activation of the tumour suppressor Tip60. Nat
Cell Biol. 11:1376–1382. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li B, Carey M and Workman JL: The role of
chromatin during transcription. Cell. 128:707–719. 2007. View Article : Google Scholar
|
|
14
|
Li Y, Sun L, Zhang Y, et al: The histone
modifications governing TFF1 transcription mediated by estrogen
receptor. J Biol Chem. 286:13925–13936. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rouhi A, Mager DL, Humphries RK and
Kuchenbauer F: MiRNAs, epigenetics, and cancer. Mamm Genome.
19:517–525. 2008. View Article : Google Scholar
|
|
17
|
Lim LP, Glasner ME, Yekta S, Burge CB and
Bartel DP: Vertebrate microRNA genes. Science. 299:15402003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lai EC, Tomancak P, Williams RW and Rubin
GM: Computational identification of Drosophila microRNA
genes. Genome Biol. 4:R422003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Griffiths-Jones S: The microRNA Registry.
Nucleic Acids Res. 32:D109–D111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iorio MV, Piovan C and Croce CM: Interplay
between microRNAs and the epigenetic machinery: an intricate
network. Biochim Biophys Acta. 1799:694–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Friedman JM, Liang G, Liu CC, et al: The
putative tumor suppressor microRNA-101 modulates the cancer
epigenome by repressing the polycomb group protein EZH2. Cancer
Res. 69:2623–2629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan W, Zhu S, Yuan M, et al: MicroRNA-21
and microRNA-148a contribute to DNA hypomethylation in lupus
CD4+ T cells by directly and indirectly targeting DNA
methyltransferase 1. J Immunol. 184:6773–6781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barber BA and Rastegar M: Epigenetic
control of Hox genes during neurogenesis, development, and disease.
Ann Anat. 192:261–274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martin GS: The road to Src. Oncogene.
23:7910–7917. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dietrich D, Lesche R, Tetzner R, et al:
Analysis of DNA methylation of multiple genes in microdissected
cells from formalin-fixed and paraffin-embedded tissues. J
Histochem Cytochem. 57:477–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yballe CM, Vu TH and Hoffman AR:
Imprinting and expression of insulin-like growth factor-II and H19
in normal breast tissue and breast tumor. J Clin Endocrinol Metab.
81:1607–1612. 1996.PubMed/NCBI
|
|
28
|
Li S, Rong M and Iacopetta B: DNA
hypermethylation in breast cancer and its association with
clinicopathological features. Cancer Lett. 237:272–280. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Parrella P, Poeta ML, Gallo AP, et al:
Nonrandom distribution of aberrant promoter methylation of
cancer-related genes in sporadic breast tumors. Clin Cancer Res.
10:5349–5354. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Swift-Scanlan T, Vang R, Blackford A,
Fackler MJ and Sukumar S: Methylated genes in breast cancer:
associations with clinical and histopathological features in a
familial breast cancer cohort. Cancer Biol Ther. 11:853–865. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fackler MJ, McVeigh M, Evron E, et al: DNA
methylation of RASSF1A, HIN-1, RAR-β,
Cyclin D2 and Twist in in situ and invasive
lobular breast carcinoma. Int J Cancer. 107:970–975. 2003.
|
|
32
|
Yan L, Yang X and Davidson NE: Role of DNA
methylation and histone acetylation in steroid receptor expression
in breast cancer. J Mammary Gland Biol Neoplasia. 6:183–192. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lehmann U, Hasemeier B, Romermann D,
Muller M, Langer F and Kreipe H: Epigenetic inactivation of
microRNA genes in mammary carcinoma. Verh Dtsch Ges Pathol.
91:214–220. 2007.(In German).
|
|
34
|
Kapoor-Vazirani P, Kagey JD, Powell DR and
Vertino PM: Role of hMOF-dependent histone H4 lysine 16 acetylation
in the maintenance of TMS1/ASC gene activity. Cancer Res.
68:6810–6821. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang X, Karuturi RK, Sun F, et al: CDKN1C
(p57) is a direct target of EZH2 and suppressed by multiple
epigenetic mechanisms in breast cancer cells. PLoS One.
4:e50112009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Luo Q, Yuan Y, et al: Pygo2
associates with MLL2 histone methyltransferase and GCN5 histone
acetyltransferase complexes to augment Wnt target gene expression
and breast cancer stem-like cell expansion. Mol Cell Biol.
30:5621–5635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shi L, Sun L, Li Q, et al: Histone
demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes
hormonally responsive breast carcinogenesis. Proc Natl Acad Sci
USA. 108:7541–7546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Zhang H, Chen Y, et al: LSD1 is a
subunit of the NuRD complex and targets the metastasis programs in
breast cancer. Cell. 138:660–672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sieuwerts AM, Mostert B, Bolt-de Vries J,
et al: mRNA and microRNA expression profiles in circulating tumor
cells and primary tumors of metastatic breast cancer patients. Clin
Cancer Res. 17:3600–3618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
O’Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010.
|
|
41
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Walter BA, Gomez-Macias G, Valera VA,
Sobel M and Merino MJ: miR-21 expression in pregnancy-associated
breast cancer: a possible marker of poor prognosis. J Cancer.
2:67–75. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim SJ, Kelly WK, Fu A, Haines K, Hoffman
A, Zheng T and Zhu Y: Genome-wide methylation analysis identifies
involvement of TNF-α mediated cancer pathways in prostate cancer.
Cancer Lett. 302:47–53. 2011.
|
|
46
|
Bedford MT and van Helden PD:
Hypomethylation of DNA in pathological conditions of the human
prostate. Cancer Res. 47:5274–5276. 1987.PubMed/NCBI
|
|
47
|
Pakneshan P, Szyf M and Rabbani SA:
Hypomethylation of urokinase (uPA) promoter in breast and prostate
cancer: prognostic and therapeutic implications. Curr Cancer Drug
Targets. 5:471–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cho B, Lee H, Jeong S, et al: Promoter
hypomethylation of a novel cancer/testis antigen gene CAGE is
correlated with its aberrant expression and is seen in premalignant
stage of gastric carcinoma. Biochem Biophys Res Commun. 307:52–63.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tokizane T, Shiina H, Igawa M, et al:
Cytochrome P450 1B1 is overexpressed and regulated by
hypomethylation in prostate cancer. Clin Cancer Res. 11:5793–5801.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Laner T, Schulz WA, Engers R, Muller M and
Florl AR: Hypomethylation of the XIST gene promoter in prostate
cancer. Oncol Res. 15:257–264. 2005.PubMed/NCBI
|
|
51
|
Reibenwein J, Pils D, Horak P, et al:
Promoter hypermethylation of GSTP1, AR, and 14-3-3σ in serum of
prostate cancer patients and its clinical relevance. Prostate.
67:427–432. 2007.PubMed/NCBI
|
|
52
|
Dammann R, Schagdarsurengin U, Liu L, et
al: Frequent RASSF1A promoter hypermethylation and K-ras
mutations in pancreatic carcinoma. Oncogene. 22:3806–3812.
2003.PubMed/NCBI
|
|
53
|
Woodson K, Hayes R, Wideroff L, Villaruz L
and Tangrea J: Hypermethylation of GSTP1, CD44, and
E-cadherin genes in prostate cancer among US Blacks and Whites.
Prostate. 55:199–205. 2003.
|
|
54
|
Phe V, Cussenot O and Roupret M: Interest
of methylated genes as biomarkers in urothelial cell carcinomas of
the urinary tract. BJU Int. 104:896–901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pulukuri SM, Patibandla S, Patel J, Estes
N and Rao JS: Epigenetic inactivation of the tissue inhibitor of
metalloproteinase-2 (TIMP-2) gene in human prostate tumors.
Oncogene. 26:5229–5237. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Henrique R, Costa VL, Cerveira N, et al:
Hypermethylation of Cyclin D2 is associated with loss of mRNA
expression and tumor development in prostate cancer. J Mol Med.
84:911–918. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Seligson DB, Horvath S, Shi T, Yu H, Tze
S, Grunstein M and Kurdistani SK: Global histone modification
patterns predict risk of prostate cancer recurrence. Nature.
435:1262–1266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ellinger J, Kahl P, von der Gathen J, et
al: Global levels of histone modifications predict prostate cancer
recurrence. Prostate. 70:61–69. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Muller I, Wischnewski F, Pantel K and
Schwarzenbach H: Promoter- and cell-specific epigenetic regulation
of CD44, Cyclin D2, GLIPR1 and PTEN by methyl-CpG binding proteins
and histone modifications. BMC Cancer. 10:2972010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yu J, Cao Q, Mehra R, et al: Integrative
genomics analysis reveals silencing of β-adrenergic signaling by
polycomb in prostate cancer. Cancer Cell. 12:419–431. 2007.
|
|
61
|
Sikand K, Slaibi JE, Singh R, Slane SD and
Shukla GC: miR 488* inhibits androgen receptor expression in
prostate carcinoma cells. Int J Cancer. 129:810–819. 2011.
|
|
62
|
Majid S, Dar AA, Saini S, et al:
MicroRNA-205-directed transcriptional activation of tumor
suppressor genes in prostate cancer. Cancer. 116:5637–5649. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mercatelli N, Coppola V, Bonci D, et al:
The inhibition of the highly expressed miR-221 and miR-222 impairs
the growth of prostate carcinoma xenografts in mice. PLoS One.
3:e40292008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dong Q, Meng P, Wang T, et al: MicroRNA
let-7a inhibits proliferation of human prostate cancer cells in
vitro and in vivo by targeting E2F2 and CCND2. PLoS One.
5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Peng X, Guo W, Liu T, et al:
Identification of miRs-143 and -145 that is associated with bone
metastasis of prostate cancer and involved in the regulation of
EMT. PLoS One. 6:e203412011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fujita Y, Kojima K, Hamada N, et al:
Effects of miR-34a on cell growth and chemoresistance in prostate
cancer PC3 cells. Biochem Biophys Res Commun. 377:114–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lodygin D, Tarasov V, Epanchintsev A, et
al: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lin SL, Chiang A, Chang D and Ying SY:
Loss of mir-146a function in hormone-refractory prostate cancer.
RNA. 14:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rauch TA, Zhong X, Wu X, et al:
High-resolution mapping of DNA hypermethylation and hypomethylation
in lung cancer. Proc Natl Acad Sci USA. 105:252–257. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Glazer CA, Smith IM, Ochs MF, et al:
Integrative discovery of epigenetically derepressed cancer testis
antigens in NSCLC. PLoS One. 4:e81892009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Otterson GA, Khleif SN, Chen W, Coxon AB
and Kaye FJ: CDKN2 gene silencing in lung cancer by DNA
hypermethylation and kinetics of p16INK4 protein induction by 5-aza
2′deoxycytidine. Oncogene. 11:1211–1216. 1995.PubMed/NCBI
|
|
72
|
Paz MF, Avila S, Fraga MF, et al:
Germ-line variants in methyl-group metabolism genes and
susceptibility to DNA methylation in normal tissues and human
primary tumors. Cancer Res. 62:4519–4524. 2002.PubMed/NCBI
|
|
73
|
Pereira MA, Tao L, Liu Y, Li L, Steele VE
and Lubet RA: Modulation by budesonide of DNA methylation and mRNA
expression in mouse lung tumors. Int J Cancer. 120:1150–1153. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Katayama H, Hiraki A, Fujiwara K, et al:
Aberrant promoter methylation profile in pleural fluid DNA and
clinicopathological factors in patients with non-small cell lung
cancer. Asian Pac J Cancer Prev. 8:221–224. 2007.PubMed/NCBI
|
|
75
|
Licchesi JD, Westra WH, Hooker CM and
Herman JG: Promoter hypermethylation of hallmark cancer genes in
atypical adenomatous hyperplasia of the lung. Clin Cancer Res.
14:2570–2578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Barski A, Cuddapah S, Cui K, et al:
High-resolution profiling of histone methylations in the human
genome. Cell. 129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Seligson DB, Horvath S, McBrian MA, et al:
Global levels of histone modifications predict prognosis in
different cancers. Am J Pathol. 174:1619–1628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Feng Y, Wang X, Xu L, et al: The
transcription factor ZBP-89 suppresses p16 expression through a
histone modification mechanism to affect cell senescence. FEBS J.
276:4197–4206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Nonaka D, Fabbri A, Roz L, et al: Reduced
FEZ1/LZTS1 expression and outcome prediction in lung cancer. Cancer
Res. 65:1207–1212. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nishioka M, Kohno T, Tani M, et al:
MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1,
deleted, mutated, and methylated in human lung cancer. Proc Natl
Acad Sci USA. 99:12269–12274. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen MW, Hua KT, Kao HJ, et al: H3K9
histone methyltransferase G9a promotes lung cancer invasion and
metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer
Res. 70:7830–7840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Grosshans H, Johnson T, Reinert KL,
Gerstein M and Slack FJ: The temporal patterning microRNA let-7
regulates several transcription factors at the larval to adult
transition in C. elegans. Dev Cell. 8:321–330. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lujambio A and Esteller M: CpG island
hypermethylation of tumor suppressor microRNAs in human cancer.
Cell Cycle. 6:1455–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Saito M, Schetter AJ, Mollerup S, et al:
The association of microRNA expression with prognosis and
progression in early-stage, non-small cell lung adenocarcinoma: a
retrospective analysis of three cohorts. Clin Cancer Res.
17:1875–1882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Fang JY, Lu R, Mikovits JA, Cheng ZH, Zhu
HY and Chen YX: Regulation of hMSH2 and hMLH1
expression in the human colon cancer cell line SW1116 by DNA
methyltransferase 1. Cancer Lett. 233:124–130. 2006.
|
|
88
|
Goel A, Arnold CN, Niedzwiecki D, et al:
Frequent inactivation of PTEN by promoter hypermethylation in
microsatellite instability-high sporadic colorectal cancers. Cancer
Res. 64:3014–3021. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yuan BZ, Durkin ME and Popescu NC:
Promoter hypermethylation of DLC-1, a candidate tumor suppressor
gene, in several common human cancers. Cancer Genet Cytogenet.
140:113–117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thangaraju M, Carswell KN, Prasad PD and
Ganapathy V: Colon cancer cells maintain low levels of pyruvate to
avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J.
417:379–389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Spurling CC, Godman CA, Noonan EJ,
Rasmussen TP, Rosenberg DW and Giardina C: HDAC3 overexpression and
colon cancer cell proliferation and differentiation. Mol Carcinog.
47:137–147. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Godman CA, Joshi R, Tierney BR, et al:
HDAC3 impacts multiple oncogenic pathways in colon cancer cells
with effects on Wnt and vitamin D signaling. Cancer Biol Ther.
7:1570–1580. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tuttle R, Simon M, Hitch DC, et al:
Senescence-associated gene YPEL3 is downregulated in human colon
tumors. Ann Surg Oncol. 18:1791–1796. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li Q and Chen H: Transcriptional silencing
of N-Myc downstream-regulated gene 1 (NDRG1) in metastatic colon
cancer cell line SW620. Clin Exp Metastasis. 28:127–135. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Menigatti M, Cattaneo E, Sabates-Bellver
J, et al: The protein tyrosine phosphatase receptor type R gene is
an early and frequent target of silencing in human colorectal
tumorigenesis. Mol Cancer. 8:1242009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yadav S, Singhal J, Singhal SS and Awasthi
S: hSET1: a novel approach for colon cancer therapy. Biochem
Pharmacol. 77:1635–1641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Krieg AJ, Rankin EB, Chan D, Razorenova O,
Fernandez S and Giaccia AJ: Regulation of the histone demethylase
JMJD1A by hypoxia-inducible factor 1α enhances hypoxic gene
expression and tumor growth. Mol Cell Biol. 30:344–353. 2010.
|
|
98
|
Vlaicu SI, Tegla CA, Cudrici CD, et al:
Epigenetic modifications induced by RGC-32 in colon cancer. Exp Mol
Pathol. 88:67–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yin Q, Wang X, Fewell C, et al: MicroRNA
miR-155 inhibits bone morphogenetic protein (BMP) signaling and
BMP-mediated Epstein-Barr virus reactivation. J Virol.
84:6318–6327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu C, Kelnar K, Liu B, et al: The
microRNA miR-34a inhibits prostate cancer stem cells and metastasis
by directly repressing CD44. Nat Med. 17:211–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li T, Li D, Sha J, Sun P and Huang Y:
MicroRNA-21 directly targets MARCKS and promotes apoptosis
resistance and invasion in prostate cancer cells. Biochem Biophys
Res Commun. 383:280–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tang JT, Wang JL, Du W, et al:
MicroRNA-345, a methylation-sensitive microRNA is involved in cell
proliferation and invasion in human colorectal cancer.
Carcinogenesis. 32:1207–1215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Schneider AC, Heukamp LC, Rogenhofer S, et
al: Global histone H4K20 trimethylation predicts cancer-specific
survival in patients with muscle-invasive bladder cancer. BJU Int.
1082:E290–E296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Elsheikh SE, Green AR, Rakha EA, et al:
Global histone modifications in breast cancer correlate with tumor
phenotypes, prognostic factors, and patient outcome. Cancer Res.
69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Van Den Broeck A, Brambilla E,
Moro-Sibilot D, et al: Loss of histone H4K20 trimethylation occurs
in preneoplasia and influences prognosis of non-small cell lung
cancer. Clin Cancer Res. 14:7237–7245. 2008.PubMed/NCBI
|
|
107
|
Ellinger J, Kahl P, Mertens C, et al:
Prognostic relevance of global histone H3 lysine 4 (H3K4)
methylation in renal cell carcinoma. Int J Cancer. 127:2360–2366.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ke XS, Qu Y, Rostad K, et al: Genome-wide
profiling of histone h3 lysine 4 and lysine 27 trimethylation
reveals an epigenetic signature in prostate carcinogenesis. PLoS
One. 4:e46872009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Li Q, Wang X, Lu Z, et al: Polycomb CBX7
directly controls trimethylation of histone H3 at lysine 9 at the
p16 locus. PLoS One. 5:e137322010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
McGarvey KM, Van Neste L, Cope L, et al:
Defining a chromatin pattern that characterizes DNA-hypermethylated
genes in colon cancer cells. Cancer Res. 68:5753–5759. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wei Y, Xia W, Zhang Z, et al: Loss of
trimethylation at lysine 27 of histone H3 is a predictor of poor
outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog.
47:701–706. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Pogribny IP, Tryndyak VP, Muskhelishvili
L, Rusyn I and Ross SA: Methyl deficiency, alterations in global
histone modifications, and carcinogenesis. J Nutr. 137:216S–222S.
2007.PubMed/NCBI
|
|
113
|
Canaani E, Nakamura T, Rozovskaia T, Smith
ST, Mori T, Croce CM and Mazo A: ALL-1/MLL1, a homologue of
Drosophila TRITHORAX, modifies chromatin and is directly
involved in infant acute leukaemia. Br J Cancer. 90:756–760.
2004.
|
|
114
|
Liu H, Takeda S, Kumar R, et al:
Phosphorylation of MLL by ATR is required for execution of
mammalian S-phase checkpoint. Nature. 467:343–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Scacheri PC, Davis S, Odom DT, et al:
Genome-wide analysis of menin binding provides insights into MEN1
tumorigenesis. PLoS Genet. 2:e512006. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Seigne C, Fontaniere S, Carreira C, et al:
Characterisation of prostate cancer lesions in heterozygous Men1
mutant mice. BMC Cancer. 10:3952010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Feng ZJ, Gao SB, Wu Y, Xu XF, Hua X and
Jin GH: Lung cancer cell migration is regulated via repressing
growth factor PTN/RPTP β/ζ signaling by menin. Oncogene.
29:5416–5426. 2010.PubMed/NCBI
|
|
118
|
Wang J, Zhou Y, Yin B, et al: ASH2L:
alternative splicing and downregulation during induced
megakaryocytic differentiation of multipotential leukemia cell
lines. J Mol Med. 79:399–405. 2001. View Article : Google Scholar
|
|
119
|
Magerl C, Ellinger J, Braunschweig T, et
al: H3K4 dimethylation in hepatocellular carcinoma is rare compared
with other hepatobiliary and gastrointestinal carcinomas and
correlates with expression of the methylase Ash2 and the
demethylase LSD1. Hum Pathol. 41:181–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kobayashi Y, Absher DM, Gulzar ZG, et al:
DNA methylation profiling reveals novel biomarkers and important
roles for DNA methyltransferases in prostate cancer. Genome Res.
21:1017–1027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Velichutina I, Shaknovich R, Geng H,
Johnson NA, Gascoyne RD, Melnick AM and Elemento O: EZH2-mediated
epigenetic silencing in germinal center B cells contributes to
proliferation and lymphomagenesis. Blood. 116:5247–5255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Tell R, Rivera CA, Eskra J, Taglia LN,
Blunier A, Wang QT and Benya RV: Gastrin-releasing peptide
signaling alters colon cancer invasiveness via heterochromatin
protein 1Hsβ. Am J Pathol. 178:672–678. 2011.PubMed/NCBI
|
|
124
|
Xi Y, Formentini A, Nakajima G, Kornmann M
and Ju J: Validation of biomarkers associated with 5-fluorouracil
and thymidylate synthase in colorectal cancer. Oncol Rep.
19:257–262. 2008.PubMed/NCBI
|
|
125
|
Wang XQ, Miao X, Cai Q, Garcia-Barcelo MM
and Fan ST: SMYD3 tandem repeats polymorphism is not associated
with the occurrence and metastasis of hepatocellular carcinoma in a
Chinese population. Exp Oncol. 29:71–73. 2007.PubMed/NCBI
|
|
126
|
Oue N, Mitani Y, Motoshita J, et al:
Accumulation of DNA methylation is associated with tumor stage in
gastric cancer. Cancer. 106:1250–1259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Fang W, Piao Z, Buyse IM, Simon D, Sheu
JC, Perucho M and Huang S: Preferential loss of a polymorphic RIZ
allele in human hepatocellular carcinoma. Br J Cancer. 84:743–747.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhao Q, Caballero OL, Levy S, et al:
Transcriptome-guided characterization of genomic rearrangements in
a breast cancer cell line. Proc Natl Acad Sci USA. 106:1886–1891.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lucio-Eterovic AK, Singh MM, Gardner JE,
Veerappan CS, Rice JC and Carpenter PB: Role for the nuclear
receptor-binding SET domain protein 1 (NSD1) methyltransferase in
coordinating lysine 36 methylation at histone 3 with RNA polymerase
II function. Proc Natl Acad Sci USA. 107:16952–16957. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Berdasco M, Ropero S, Setien F, et al:
Epigenetic inactivation of the Sotos overgrowth syndrome gene
histone methyltransferase NSD1 in human neuroblastoma and glioma.
Proc Natl Acad Sci USA. 106:21830–21835. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Nimura K, Ura K, Shiratori H, Ikawa M,
Okabe M, Schwartz RJ and Kaneda Y: A histone H3 lysine 36
trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome.
Nature. 460:287–291. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Taketani T, Taki T, Nakamura H, Taniwaki
M, Masuda J and Hayashi Y: NUP98-NSD3 fusion gene in
radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15)
and expression pattern of NSD family genes. Cancer Genet Cytogenet.
190:108–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Morishita M and di Luccio E: Cancers and
the NSD family of histone lysine methyltransferases. Biochim
Biophys Acta. 1816:158–163. 2011.PubMed/NCBI
|
|
134
|
Watanabe H, Soejima K, Yasuda H, et al:
Deregulation of histone lysine methyltransferases contributes to
oncogenic transformation of human bronchoepithelial cells. Cancer
Cell Int. 8:152008. View Article : Google Scholar
|
|
135
|
Visakorpi T, Suikki HE, Kujala PM, Tammela
TLJ, van Weerden WM and Vessella RL: Genetic alterations and
changes in expression of histone demethylases in prostate cancer.
Prostate. 70:889–898. 2010.PubMed/NCBI
|
|
136
|
Fukuda T, Tokunaga A, Sakamoto R and
Yoshida N: Fbxl10/Kdm2b deficiency accelerates neural progenitor
cell death and leads to exencephaly. Mol Cell Neurosci. 46:614–624.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Vinatzer U, Gollinger M, Mullauer L,
Raderer M, Chott A and Streubel B: Mucosa-associated lymphoid
tissue lymphoma: novel translocations including rearrangements of
ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 14:6426–6431. 2008.
View Article : Google Scholar
|
|
138
|
Yang ZQ, Imoto I, Fukuda Y, et al:
Identification of a novel gene, GASC1, within an amplicon at
9p23–24 frequently detected in esophageal cancer cell lines. Cancer
Res. 60:4735–4739. 2000.PubMed/NCBI
|
|
139
|
Zeng J, Ge Z, Wang L, et al: The histone
demethylase RBP2 is overexpressed in gastric cancer and its
inhibition triggers senescence of cancer cells. Gastroenterology.
138:981–992. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Rao M, Chinnasamy N, Hong JA, et al:
Inhibition of histone lysine methylation enhances cancer-testis
antigen expression in lung cancer cells: implications for adoptive
immunotherapy of cancer. Cancer Res. 71:4192–4204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liggins AP, Lim SH, Soilleux EJ, Pulford K
and Banham AH: A panel of cancer-testis genes exhibiting
broad-spectrum expression in haematological malignancies. Cancer
Immun. 10:82010.PubMed/NCBI
|
|
142
|
Jankowska A, Makishima H, Tiu RV, et al:
Mutational spectrum analysis of chronic myelomonocytic leukemia
includes genes associated with epigenetic regulation: UTX, EZH2,
and DNMT3A. Blood. 116:268–269. 2010.PubMed/NCBI
|
|
143
|
Xiang Y, Zhu Z, Han G, Lin H, Xu L and
Chen CD: JMJD3 is a histone H3K27 demethylase. Cell Res.
17:850–857. 2007. View Article : Google Scholar : PubMed/NCBI
|