In the eukaryotic nucleus, DNA is compacted into a
chromatin structure with the nucleosome as the basic unit, in which
histone octamer is surrounded by the 147 bases of DNA for 1.7 laps.
The histone octamer includes two elements of the core histone (H3,
H4, H2A and H2B) (1). The packaging
of DNA into chromatin presents a potential barrier to factors that
require DNA as their template. There are mainly three modifications
regulating chromatin structure and epigenetic mechanisms of gene
expression, including DNA methylation, histone covalent
modification and microRNAs (miRNAs) (2). These modifications jointly constitute
the ‘Epigenetic code’ to modulate the expression of the mammalian
genome in different cell types, through developmental stages and in
diverse disease states including cancer (2–4).
DNA methylation is a widespread modification in
bacteria, plants and mammals, and this covalent molecular
transformation is a natural modification of DNA. DNA methylation
which is produced during DNA replication is considered as a stable
gene-silencing mechanism. In eukaryotic cells, DNA methylation is
the covalent modification taking place at the 5′ end of the CpG
dinucleotide of the cytosine ring and with S-adenosyl-methionine as
its methyl donor. This reaction is catalyzed by the DNMT family,
including DNMT1, DNMT3A and DNMT3B. During the process of embryo
formation, DNMT3A and DNMT3B are required for DNA methylation from
scratch, while DNMT1 is considered to be the methyltransferase
maintaining the methylation status (5).
This covalent modification can inhibit the activity
of gene transcription; either by blocking the combination of a
transcription factor and its binding sites (6), or through recruitment of methylated
binding domain proteins that mediate inhibition of gene expression
(7). In mammalian cells, DNA
methylation occurs mainly in CpG dinucleotides (8). However, CpG sites are not randomly
distributed in the genome, but are concentrated in short CpG-rich
DNA fragments or DNA fragments in the long repeat so-called ‘CpG
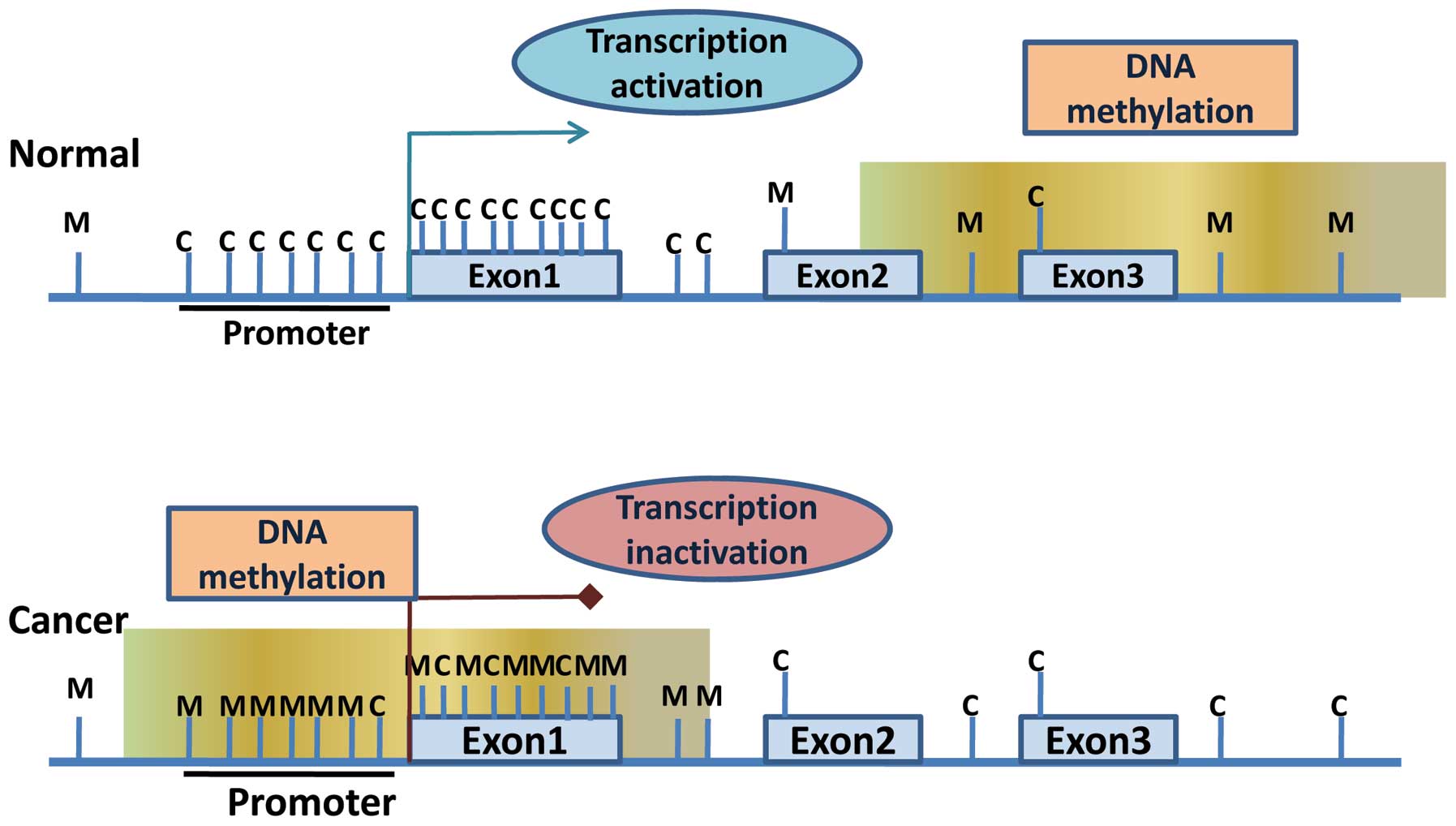
islands’ (8,9). Although for normal cells, the majority
of CpG sites of the genome are methylated, usually the cytosine in
CpG islands is not methylated in the development and
differentiation of tissues. However, in normal cells, certain
subsets of CpG islands at the promoter can be methylated leading to
long-term silencing of transcription. The DNA methylation pattern
is formed during cell differentiation, but it also causes cells to
partially or completely lose the ability to divide. DNA methylation
profiles are tissue-specific, and the functions of methylation
profiles in different cells are not the same. CpG island-containing
gene promoters are usually unmethylated in normal cells to maintain
euchromatic structure, which is the transcriptional active
conformation allowing gene expression. However, during cancer
development, many of these genes are hypermethylated at their CpG
island-containing promoters to inactivate their expression by
changing open euchromatic structure to compact heterochromatic
structure (Fig. 1).
Histones including H2A, H2B, H3 and H4, together
form the histone octamer that is the basic structure of nucleosome
components (1). N-terminals of
histones protrude out of the nucleosome core, and amino acids of
N-terminals easily undergo a series of covalent modifications, such
as methylation, acetylation, phosphorylation, ubiquitination and
sumolation (10,11). Acting individually or in
combination, these modifications are believed to encipher
inheritable epigenetic programs that encode distinct nucleosome
functions such as gene transcription, X-chromosome inactivation,
heterochromatin formation, mitosis, and DNA repair and replication
(2–4,10). For
example, a previous study showed that direct interaction between
the chromodomain of Tip60 and histone H3 trimethylated on lysine 9
(H3K9me3) at DSBs activates the acetyltransferase activity of
Tip60. Depletion of intracellular H3K9me3 blocks activation of the
acetyltransferase activity of Tip60, resulting in defective ATM
activation and widespread defects in DSB repair (12). Mechanistically, these functions are
mediated either directly by altering nucleosome interactions with
chromatin or indirectly by recruiting effector proteins that
possess characteristic modules that recognize specific histone
modifications in a sequence-dependent manner. The underlying basis
of these epigenetic codes resides in the substrate specificity of
the enzymes that catalyze the numerous covalent modifications as
well as the enzymes that remove these marks to alter the
modifications.
Given that chromatin is the physiological template
for all DNA-mediated processes, it is not surprising that histone
modifications represent an essential component in controlling the
structure and/or function of the chromatin, with different
modifications yielding distinct functional consequences. Indeed,
previous research has shown that site-specific histone
modifications correlate well with particular biological functions
such as gene transcription (13).
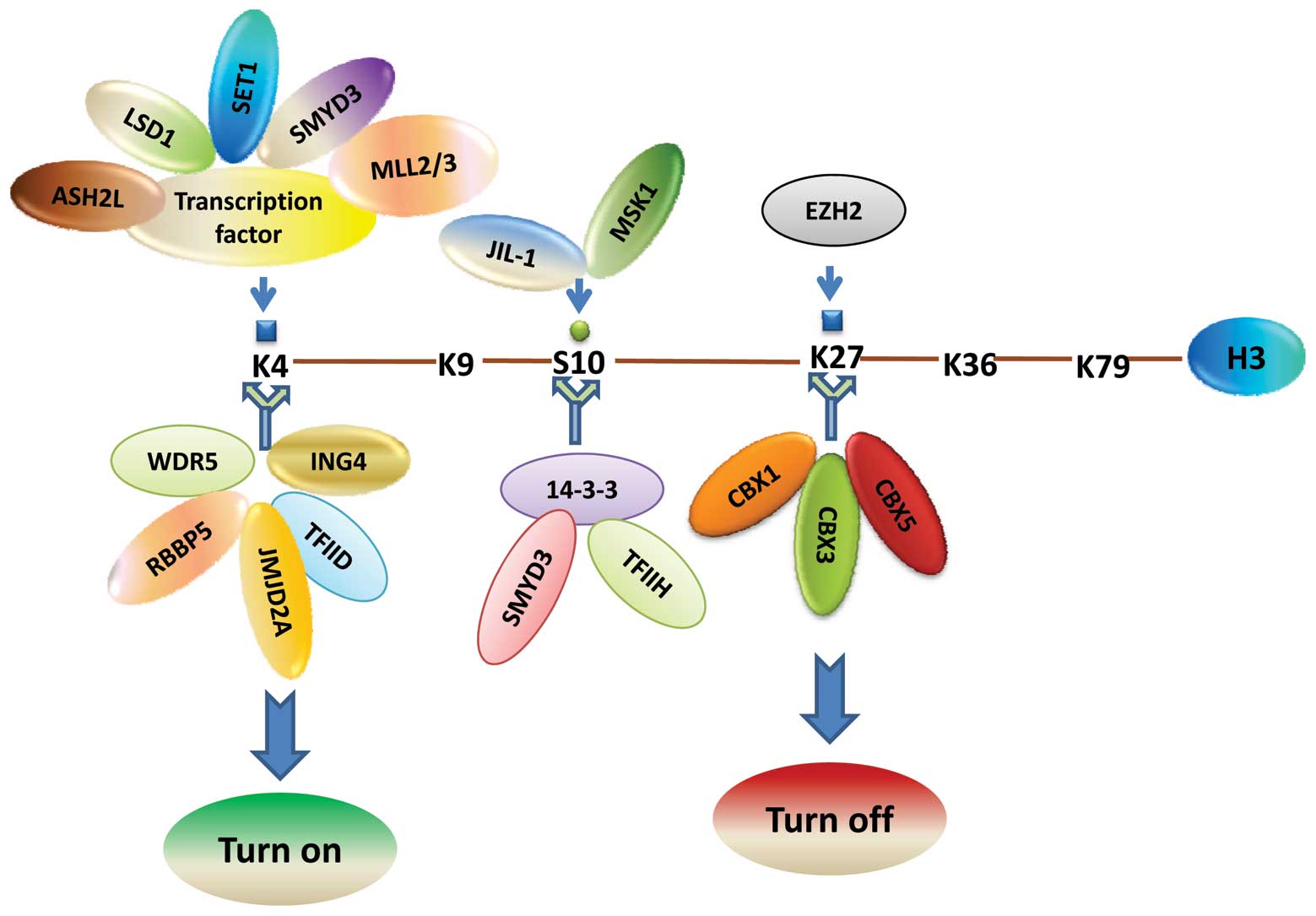
For instance, histone H3 lysine 9 acetylation (H3K9ac), H3 serine
10 phosphorylation (H3S10ph), and H3 lysine 4 trimethylation
(H3K4me3) are reported to be associated with transcriptional
activation (14). Conversely,
H3K27me3 and hypoacetylation of H3 and H4 have been shown to be
correlated with transcriptional repression. As stated above,
ultimately, the functions of histone modifications are uncovered by
the recognition of histone code or histone language by particular
cellular machinery such as the transcription apparatus (15). Our previous study found that the
histone H3S10 phosphorylation mark is catalyzed by mitogen and
stress-activated protein kinase 1 (MSK1) and is recognized by a
14-3-3ɛ/14-3-3γ heterodimer through its interaction with H3K4
trimethyltransferase SMYD3 and the p52 subunit of TFIIH (14) (Fig.
2).
microRNAs (miRNAs) are endogenous, short, 19–25
nucleotide long, evolutionarily conserved non-coding RNAs, which
partially or perfectly match the 3′ untranslated regions (3′UTR) of
target mRNAs to regulate gene expression by post-transcriptional
silencing and/or by the degradation of target mRNAs (16). Bioinformatics and experimental
studies have shown that more than 30% of human genes are direct
miRNA targets, which implies that miRNAs function in almost all
biological processes including cell cycle regulation, cell growth,
apoptosis, cell differentiation and stress reactions. In various
species, including the human, a growing number of miRNAs have been
determined in the past few years. Genome-wide studies estimate that
miRNA genes represent ~1% of the entire genome in different
species; this percentage is similar to other large gene families
with regulatory functions such as the home-domain transcription
factor family (17,18). The number of genes demonstrated to
be the targets of miRNAs is growing rapidly. The latest release of
the Sanger miRNA Registry currently annotates more than 800 human
miRNAs (http://microrna.sanger.ac.uk; release
13.0), yet many more miRNAs are expected to be identified in the
future (19). It is not surprising
that miRNAs, just like protein-coding genes, have to be tightly
regulated in order to contribute to a distinct transcriptome of a
normal cell. In cancer, however, miRNAs have been found to be
massively deregulated.
The direct interaction between miRNAs and epigenetic
mechanisms is believed to be a quite complicated regulatory network
(20). On the one hand, expression
of miRNAs is tissue-specific, and is subject to fine and strict
regulation by epigenetic mechanisms such as DNA methylation and
histone modifications (21); on the
other hand, in turn, miRNAs can also affect epigenetic mechanisms
and regulate gene transcription; the ability to target
post-transcriptional gene-silencing (22).
Epigenetic mechanisms are required to maintain
normal growth and development and gene expression in different
organs (23). Abnormal epigenetic
regulation may alter gene expression and function which may lead to
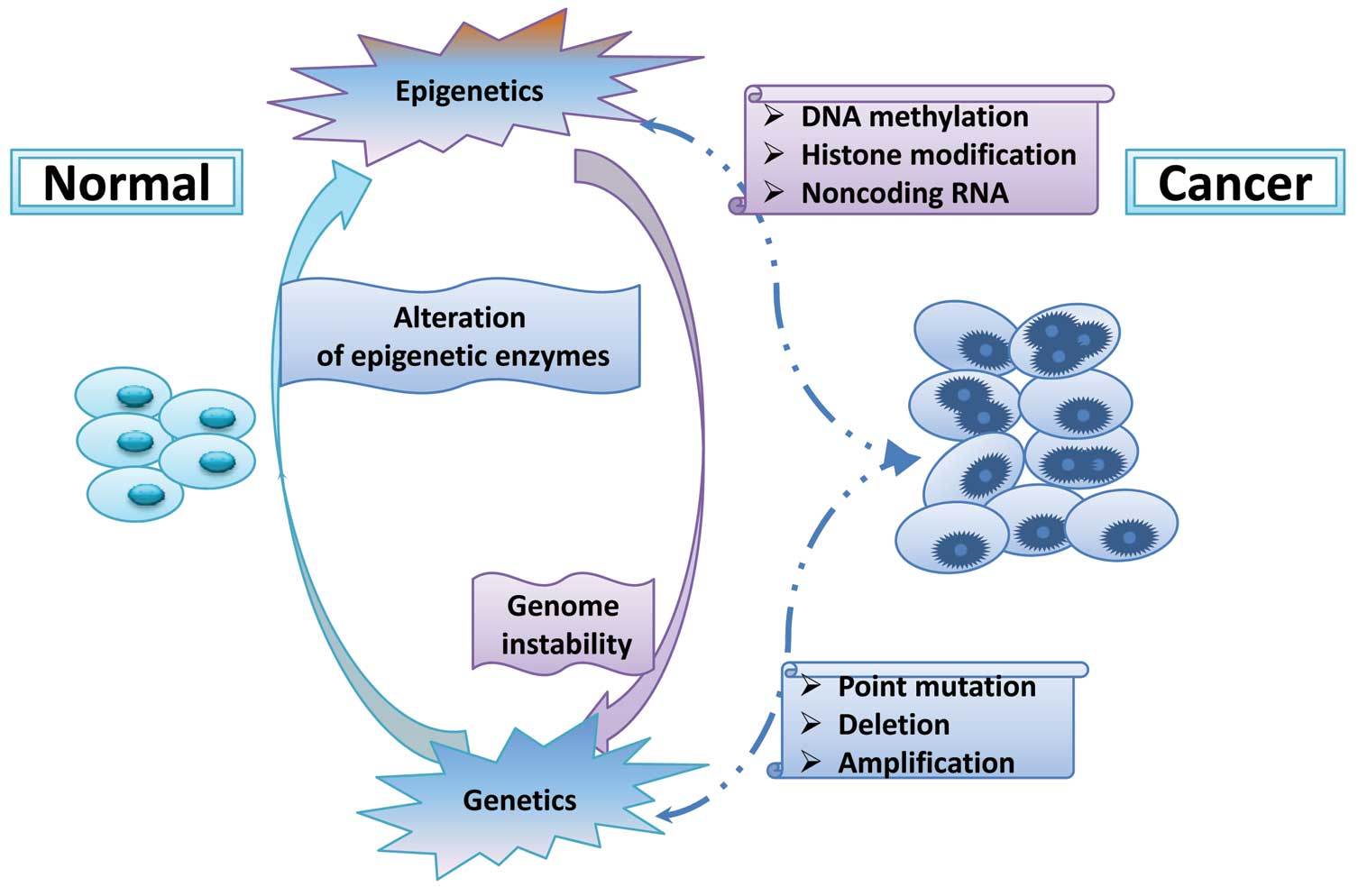
diseases such as cancer. Human tumors, in essence, are a genetic
disease, since during cancer formation, a large number of genes are
mutated or abnormally activated (24,25).
However, recent studies indicate that carcinogenesis cannot be
accounted for by genetic alterations alone, but also involve
epigenetic changes such as DNA methylation, histone modifications
and microRNAs (Fig. 3). Global
levels of lysine methylations are quite different between cell
types and these molecular changes have been considered to be
correlated with various types of cancers (Table I). In addition, the lysine
methyltransferases and demethylases are reported to be de-regulated
in a variety of cancers (Tables II
and III). These molecular
alterations lead to permanent changes in the patterns of gene
expression that regulate the neoplastic phenotype, such as cellular
growth and invasiveness. In this part of the present review, we
focus on recent discoveries of epigenetic alterations in several
types of tumors including breast, prostate, lung and colon
cancer.
Global DNA hypomethylation is frequently reported in
breast tumors, but the number of hypomethylated genes is relatively
small. DNA hypomethylation of FEN1, BCSG1,
PLAU, IGF2 and CDH3 has been detected in
breast cancer cells (26,27). However, more than 100 genes have
been considered to be hypermethyated in breast cancer, and these
aberrantly methylated genes play critical roles in all types of
cell processes including cell-cycle regulation, apoptosis, tissue
invasion and metastasis, angiogenesis and hormone signaling
(28). For instance, CCND2
and p16ink4A/CDKN2A which function as crucial regulators of
the cell cycle are commonly found to be methylated in breast cancer
(29); APC, TWIST and
HOXA5 which play key roles in apoptosis are silenced due to
DNA hypermethylation (30,31); ERα and PR which are critical in
hormone regulation are also frequently methylated (32). In addition to protein-coding genes,
recent research shows that microRNAs with tumor-suppressor function
could be silenced in breast cancer cells through DNA methylation
(33). These findings strongly
indicate that DNA hypermethylation plays a crucial role in breast
carcinogenesis, which cooperatively and synergistically interact
with other genetic alterations to promote the development of breast
cancer.
A growing number of histone modifications and
histone modification enzymes have been found to be deregulated in
breast cancer. H4K16ac and its responsible enzyme hMOF were found
to be markedly reduced in primary breast carcinomas and
medulloblastomas (34). EZH2, which
is a subunit of the polycomb-repressive complex 2 (PRC2) and
catalyzes the trimethylation of histone H3 on Lys 27 (H3K27), is
amplified and overexpressed in breast cancer (35). Furthermore, histone demethylases are
shown to function during breast tumorigenesis. Pygo2 associates
with histone-modifying enzymatic complexes, specifically the MLL2
histone methyltransferase (HMT) and STAGA histone acetyltransferase
(HAT) complexes, to facilitate their interaction with β-catenin and
to augment Wnt1-induced, TCF/LEF-dependent transcriptional
activation in breast cancer cells (36). Depletion of H3K9 trimethyl
demethylase JMJD2B, which is shown to be an integral component of
the H3K4-specific methyltransferase, the mixed-lineage leukemia
(MLL) 2 complex, impairs the estrogen-induced G (1)/S transition of the cell cycle in
vitro and inhibits breast tumorigenesis in vivo
(37). Previous results demonstrate
that LSD1 is downregulated in breast carcinomas and that its
expression level is negatively correlated with that of TGFβ1 which
inhibits the invasion of breast cancer cells in vitro and
suppresses breast cancer metastatic potential in vivo
(38).
Recent genome-wide approaches have revealed that
miRNAs are globally downregulated in breast cancer. They identified
29 differentially expressed candidates, of which 15 predictive
miRNAs were able to distinguish between breast cancer and normal
breast tissue (39). Depletion of
the let-7 family (containing at least 11 homologous miRNAs) in
breast cancer causes enhanced tumorigenicity and is associated with
clinical features, such as PgR status (let-7c), a positive lymph
node status (let-7f-1, let-7a-3 and let-7a-2), or a high
proliferation index (let-7c and let-7d) (40,41).
In addition, miR-15/16 is shown to be downregulated in breast
cancer which leads to aberrant expression of BCL2 (42). AIB1 which plays an important role in
the ERα signaling pathway is overexpressed in breast cancer due to
downregulation of miR-17-5p (43).
However, certain miRNAs are found to be frequently amplified in
breast cancer; for example, miR-21, whose overexpression in breast
cancer confers increased invasive capacities and promotes tumor
metastasis to the lung (44).
Decreased Dicer expression was recently observed in breast cancer,
where loss of expression represented an independent prognostic
factor for metastatic disease, and reduced expression of Dicer was
associated with the highly aggressive mesenchymal phenotype.
Prostate cancer is the most common cancer in men in
Western countries and its incidence is increasing steadily
worldwide. Genome-wide DNA hypomethylation has been observed in
prostate cancer cells, which may lead to structural and functional
changes of the genome. It has been reported that global
hypomethylation is considerately lower in patients with metastatic
prostate cancer in contrast to non-metastatic prostate cancer
(45,46). Gene-specific hypomethylation has
also been found in prostate cancers and functions during a variety
of cellular processes, such as tumor invasion and metastasis
(urokinase plasminogen activator, cellular proliferation gene
heparanase) (47), cell cycle
control (cancer/testis antigen)(48), hydroxylation of estrogens and
activation of carcinogens (cytochrome P450 1B1) (49), X-chromosome inactivation [X
(inactivate)-specific transcript] (50). DNA hypermethylation has been the
most common and best-characterized epigenetic event in cancer,
including prostate cancer. In prostate cancer, a large number of
genes have been found to be hypermethylated. These genes are
involved in a variety of biological processes including DNA damage
repair (Glutathione S transferase P1) (51), signal transduction (RASSF1A)
(52), adhesion (E-cadherin, CD44
and galectins) (53), hormonal
responses (retinoic acid receptor, androgen receptor and estrogen
receptor), apoptosis (death-associated protein kinase) (54), invasion and metastasis (tissue
inhibitors of metalloproteinases and galectins) (55) and cell cycle control (cyclins,
cyclin-dependent kinases) (56).
Research indicates that alterations of histone
modifications play crucial roles during prostate tumorigenesis
(57). The increased active histone
modifications in prostate cancer facilitate activation of
proto-oncogenes and other genes involved in cell growth and
survival, while increased repression of histone modifications leads
to tumor-suppressor gene silencing. For instance, H3K4me1 and
H3K4me2 are found to be increased at the AR enhancers of cell cycle
genes (e.g. CDK1), which facilitates upregulation of these cell
cycle genes to promote cellular growth (58). H3K4me3 is shown to be enriched in
prostate cancer cells, and is correlated with activation of genes
involved in cell growth and survival (e.g. BCL2) (59). H3K9me1, H3K9me2 and H3K9me3 have
been involved in repression of AR target genes in LNCaP cells
(58). In addition, H3K27me3
enrichment at the promoters of genes (e.g., tumor-suppressor genes
GAS2, PIK3CG and ADRB2) in metastatic prostate cancer represses the
expression of these genes, leading to prostate cancer cell growth,
survival and invasion (60).
More than 50 miRNAs have been found to be aberrantly
regulated in prostate cancer, including upregulation of several
oncogenic miRNAs (miR-488, miR-15a/16, miR-221/-222, miR-21,
miR-125b, miR-32, miR-26a, miR-196a, miR-181a, miR-25, miR-93,
miR-92 and let-7i) (61) and
downregulation of various tumor-suppressor miRNAs (miR-101,
miR-126, miR-205, miR-31, miR-146a, miR-330, miR-34 cluster,
miR-218, miR-128, miR-203 and miR-200 family) (62). In prostate cancer cells and primary
tumor cells, the cell cycle inhibitor p27Kip1 was found to be
extensively downregulated by extra introduction of miR-221/miR-222,
which strongly increased cell growth potential by inducing a G1-S
shift in the cell cycle subsequently enhancing tumorigenicity in
SCID mice (63). In prostate
cancer, miR-21 was found to be elevated in PC3 and DU145 cells.
Blocking miR-21 by antisense oligonucleotides did not affect
proliferation, but it sensitized cells to staurosporine-induced
apoptosis and impaired cell motility and invasion (64). Both miR-143 and -145 have been
reported to be associated with bone metastasis of prostate cancer
and are involved in the regulation of EMT (65). H3K27me3 methyltransferase EZH2 is
shown to be enriched due to miR-101 decrease during prostate cancer
progression, thus, leading to widespread gene silencing. miR-34
activation can recapitulate the elements of p53 activity, inducing
cell cycle arrest and apoptosis by the down-modulation of proteins
such as CDK4, CDK6, cyclin D1, cyclin E2, E2F3 and BCL2 (66,67).
Notably, miR-34 also inhibits SIRT1, a gene that hinders
p53-dependent apoptosis, promoting survival under genotoxic and
oxidative stress. Likewise, by targeting glutaminase, miR-23 has
been found to participate in the pro-tumorigenic network resulting
from MYC overexpression, which is thought to be the most common
alteration in prostate cancer. In addition to belonging to the
group of reduced miRs, the contribution of miR-146 to prostate
cancer progression has been identified in its capacity to repress
ROCK1 expression, a downstream effector of hyaluronan-mediated
signaling on the CD168 receptor (68).
Lung cancer is a major worldwide health threat and
is the leading cause of cancer-related mortality. Global
hypomethylation and regional hypermethylation in normally
unmethylated CpG islands have all been implicated in lung cancer
(69). Loss of imprinting of the
H19, IGF2 and MEST genes has been found in lung cancer cells due to
genome-wide DNA hypomethylation, which may result in deregulated
cell growth. In addition, upregulation of cancer testis antigens
(CTAs) including the melanoma-associated antigen family as a result
of global hypomethylation has also been observed (70). However, a number of tumor-suppressor
genes has been shown to be aberrantly methylated and associated
with different cellular processes, such as cell cycle regulation
(p16) (71), DNA repair (MGMT)
(72), apoptosis (DAPK, caspase 8,
ARF, FAS and TRAILR1) (72,73), RAS signaling (RASSF1A, NORE1A and
G0S2) and invasion (cadherins, TIMP3 and laminin family) (74,75).
Different histone modifications may play crucial
roles in the epigenetic alterations in lung cancer. Gain of H4K5ac
and H4K8ac and loss of H4K12ac, H4K16ac and H4K20me3 have been
found in lung cancer cells (76).
In addition, low cellular levels of both H3K4me2 and H3K18ac
predict poor clinical outcome in lung cancer patients (77). HDACs have been reported to repress
critical gene pathways involved in protection against lung cancer
and, therefore, reduction in lung HDACs may promote tumorigenesis
(78). Previous studies have
demonstrated that expression of HDACs is significantly increased in
various lung cancer cells and is associated with poor prognosis
after surgery. The abnormal overexpression of HDACs may result in
the downregulation of critical tumor-suppressor genes which
promotes tumorigenesis. For example, transcription factor ZBP-89,
which has been implicated in the induction of growth arrest and
apoptosis, can recruit HDAC3 to the promoter of p16, and thus
downregulates p16 expression by altering the histone modification
status. In addition, FEZ1 and MYO18B have been suggested to be
related to tumorigenesis of lung cancer through repression as the
result of histone deacetylation (79,80).
In addition to DNA methylation, alteration of histone modification
is another crucial mechanism leading to the silencing of TGF β RII,
MAGE-3, Ep-CAM and MYO18B (81).
These results suggest that histone deacetylation contributes to
gene silencing in lung cancer cells and is involved in lung
carcinogenesis.
Previous studies have demonstrated that miRNA
alterations occur as an early event in response to environmental
carcinogens ahead of the onset of cancer. The expression of let-7
miRNA, which correlates with shorter survival and is an independent
prognostic factor, is observed to be reduced in primary lung
tumors. It has also been observed that overexpression of miRNA
let-7 in A549 lung adenocarcinoma cell lines inhibited cancer cell
growth. Further studies have shown that let-7 negatively regulates
the expression of RAS and MYC by targeting their mRNAs for
translational repression (82,83).
Downregulation of miR-128b, which is a direct negative regulator of
the EGFR oncogene, is found in lung tumors. In addition, expression
of miR-124a is epigenetically silenced by DNA hypermethylation in
lung cancer (84). In contrast to
the above miRNAs, the expression of miRNA cluster miR-17 is
markedly amplified in lung cancer, and stimulates cell
proliferation. The predicted targets of the miR-17 cluster include
PTEN, E2F1 and RB2 that are known to play important roles in lung
cancer (85). In addition, abnormal
amplifications of miR-155 and miR-21 have been correlated with poor
prognosis and reduced survival of patients diagnosed with lung
cancer (86).
Colon cancer is one of the most common types of
cancer and is a leading cause of cancer-related mortality
worldwide. It has been more than 25 years since an extensive loss
of DNA hypomethylation was reported in colon cancer cells. Various
studies have confirmed this initial finding not only in colon
cancer but also in a number of other cancer types. This widespread
hypomethylation may include different epithelial cells, increasing
genome instability, overexpression of a number of genes and loss of
imprint of specific genes. Hypermethylation targeting promoters of
specific genes has also frequently been detected in colon cancer.
Numerous genes influenced by DNA hypermethylation are correlated
with diverse biological functions including cell cycle control
(p16, p15, MINT1, MINT2 and MINT31), DNA damage repair (MLH1, MSH2
and MGMT), apoptosis (DAPK), tumor cell invasion (APC and LKB1),
cell proliferation (IGF2) and tumor angiogenesis and metastasis
(COX-2) (87–89).
Histone modifications are necessary for the
regulation of gene expression, but levels of these covalent changes
and modification enzymes are usually altered in colon cancer. Colon
cancer cells exhibit increased HDAC activity compared with
non-malignant cells. HDACs are upregulated in colon cancer cells
and in primary colon cancer. For example, overexpression of HDAC1
and HDAC3 may silence SLC5A8, the gene coding for the Na(+)-coupled
pyruvate transporter (90);
upregulation of HDAC1 may repress P21, the gene involved in cell
cycle regulation (91); and
amplification of HDAC3 which is determined in approximately half of
all colon adenocarcinomas alters the epigenetic programming of
colon cancer cells to impact intracellular wnt signaling and their
sensitivity to external growth regulation by vitamin D (92). In addition, YPEL3 and NDRG1, members
of the secreted frizzle-related proteins (SFRPs) and the GATA
family of transcription factors which are demonstrated to be
silenced in specific colon cancer cell lines are occupied by
inactive histone modifications (93,94).
Enrichment of H3K27me3, HDAC1 and EZH2 are found at the promoters
of RUNX3 and PTPRR-1 in cancer cells, which may downregulate these
genes and are associated with tumor progression (95). It has also been reported that
overexpression of hSET1 in colon cancer promotes cell proliferation
and cancer cell survival (96).
Furthermore, HIF recruits JMJD1A to regulate the expression of
adrenomedullin (ADM) and growth and differentiation factor 15
(GDF15), ultimately enhancing tumor growth (97). In addition, RGC-32 may contribute to
the development of colon cancer by regulating chromatin assembly
(98).
miRNAs are negative regulators of target genes
through post-transcriptional inhibition of specific mRNAs. Both
overexpression and suppression of miRNAs have been found to be
involved in the tumorigenesis of colon cancer. Overexpressed miRNAs
such as miR-20, miR-21, miR-17-5p, miR-15b, miR-181b, miR-191 and
miR-200c have been found in colon cancer cells. miRNAs function by
targeting and inhibiting different tumor-suppressor genes such as
E2F1, tropomyosin 126, PTEN and Pdcd4 (99). Lower levels of mature miRNAs such as
let7, miR-22, miR-34a, miR-126, miR-143, miR-145, miR-342 and
miR-345 are also found in colon cancers, suggesting that they act
as tumor-suppressor miRNAs (100).
The loss of such miRNAs may lead to overactivity of oncogenes and
deregulation of signaling pathways finally promoting cell growth
and invasion in colon cancer. For example, the putative identified
targets of miR-145 are transforming growth factor receptor II and
insulin receptor substrate 1 (IRS-1), which promote
tumor-suppressor activity (101).
Repression of miR-22 upregulates HIF-1α expression, promoting VEGF
production during hypoxia (102).
miR-345 may play an important antineoplastic role; a growth
inhibitor in the development of colon cancer through downregulation
of BCL2-associated athanogene 3 (BAG3) (103).
Human tumors are a group of diseases triggered by
various causes, including progressive genetics and abnormal
epigenetics. More and more studies have demonstrated that
epigenetic changes are main factors in tumorigenesis and cancer
development. Epigenetic abnormalities occurring in tumors have led
to the development of epigenetic treatment in cancer. Epigenetic
therapy aims to reverse the epigenetic alterations occurring in
tumors, thus, restoring the normal epigenome.
Remarkable progress has been made during the past
few decades on DNA methylation and histone modifications in gene
transcription, yet the role of epigenetic events in cancer has not
been fully explained. However, great progress has been accomplished
in regards to epigenetic drugs targeting chromatin and
histone-modifying enzymes. Many epigenetic drugs, including two DNA
methyltransferase enzyme (DNMT) inhibitors and a deacetylase
(HDACs) inhibitor have been approved by the FDA as effective drugs
for cancer treatment. Meanwhile, various inhibitor drugs, such as
FK228, SAHA and MS-275, have already been the focus of phase III
clinical experiments. Nevertheless, there is still a long way to go
until the succesful epigenetic treatment of cancer. The main
strategy of recent epigenetic treatment is to inhibit abnormal
DNMTs and HDACs using specific inhibitors. More specific and
effective inhibitors should be developed to reduce unwanted
side-effects as much as possible since epigenetic modifying enzymes
function in a wide range of organs in the body; in addition,
epigenetic changes occurring in tumors have not been completely
studied. Research on detailed epigenetic changes in cancer, and the
in-depth study of tumor pathology are expected to enhance the
ability to diagnose and treat cancer.
|
1
|
Luger K, Mader AW, Richmond RK, Sargent DF
and Richmond TJ: Crystal structure of the nucleosome core particle
at 2.8 A resolution. Nature. 389:251–260. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar
|
|
3
|
Suzuki MM and Bird A: DNA methylation
landscapes: provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rijnkels M, Kabotyanski E,
Montazer-Torbati MB, Beauvais CH, Vassetzky Y, Rosen JM and Devinoy
E: The epigenetic landscape of mammary gland development and
functional differentiation. J Mammary Gland Biol Neoplasia.
15:85–100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bernstein BE, Meissner A and Lander ES:
The mammalian epigenome. Cell. 128:669–681. 2007. View Article : Google Scholar
|
|
6
|
Watt F and Molloy PL: Cytosine methylation
prevents binding to DNA of a HeLa cell transcription factor
required for optimal expression of the adenovirus major late
promoter. Genes Dev. 2:1136–1143. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nan X, Ng HH, Johnson CA, Laherty CD,
Turner BM, Eisenman RN and Bird A: Transcriptional repression by
the methyl-CpG-binding protein MeCP2 involves a histone deacetylase
complex. Nature. 393:386–389. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weber M, Hellmann I, Stadler MB, Ramos L,
Paabo S, Rebhan M and Schubeler D: Distribution, silencing
potential and evolutionary impact of promoter DNA methylation in
the human genome. Nat Genet. 39:457–466. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamada Y, Shirakawa T, Taylor TD, et al: A
comprehensive analysis of allelic methylation status of CpG islands
on human chromosome 11q: comparison with chromosome 21q. DNA Seq.
17:300–306. 2006.PubMed/NCBI
|
|
10
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheung P, Allis CD and Sassone-Corsi P:
Signaling to chromatin through histone modifications. Cell.
103:263–271. 2000. View Article : Google Scholar
|
|
12
|
Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau
LA, Whetstine JR and Price BD: Histone H3 methylation links DNA
damage detection to activation of the tumour suppressor Tip60. Nat
Cell Biol. 11:1376–1382. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li B, Carey M and Workman JL: The role of
chromatin during transcription. Cell. 128:707–719. 2007. View Article : Google Scholar
|
|
14
|
Li Y, Sun L, Zhang Y, et al: The histone
modifications governing TFF1 transcription mediated by estrogen
receptor. J Biol Chem. 286:13925–13936. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rouhi A, Mager DL, Humphries RK and
Kuchenbauer F: MiRNAs, epigenetics, and cancer. Mamm Genome.
19:517–525. 2008. View Article : Google Scholar
|
|
17
|
Lim LP, Glasner ME, Yekta S, Burge CB and
Bartel DP: Vertebrate microRNA genes. Science. 299:15402003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lai EC, Tomancak P, Williams RW and Rubin
GM: Computational identification of Drosophila microRNA
genes. Genome Biol. 4:R422003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Griffiths-Jones S: The microRNA Registry.
Nucleic Acids Res. 32:D109–D111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iorio MV, Piovan C and Croce CM: Interplay
between microRNAs and the epigenetic machinery: an intricate
network. Biochim Biophys Acta. 1799:694–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Friedman JM, Liang G, Liu CC, et al: The
putative tumor suppressor microRNA-101 modulates the cancer
epigenome by repressing the polycomb group protein EZH2. Cancer
Res. 69:2623–2629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan W, Zhu S, Yuan M, et al: MicroRNA-21
and microRNA-148a contribute to DNA hypomethylation in lupus
CD4+ T cells by directly and indirectly targeting DNA
methyltransferase 1. J Immunol. 184:6773–6781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barber BA and Rastegar M: Epigenetic
control of Hox genes during neurogenesis, development, and disease.
Ann Anat. 192:261–274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martin GS: The road to Src. Oncogene.
23:7910–7917. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dietrich D, Lesche R, Tetzner R, et al:
Analysis of DNA methylation of multiple genes in microdissected
cells from formalin-fixed and paraffin-embedded tissues. J
Histochem Cytochem. 57:477–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yballe CM, Vu TH and Hoffman AR:
Imprinting and expression of insulin-like growth factor-II and H19
in normal breast tissue and breast tumor. J Clin Endocrinol Metab.
81:1607–1612. 1996.PubMed/NCBI
|
|
28
|
Li S, Rong M and Iacopetta B: DNA
hypermethylation in breast cancer and its association with
clinicopathological features. Cancer Lett. 237:272–280. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Parrella P, Poeta ML, Gallo AP, et al:
Nonrandom distribution of aberrant promoter methylation of
cancer-related genes in sporadic breast tumors. Clin Cancer Res.
10:5349–5354. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Swift-Scanlan T, Vang R, Blackford A,
Fackler MJ and Sukumar S: Methylated genes in breast cancer:
associations with clinical and histopathological features in a
familial breast cancer cohort. Cancer Biol Ther. 11:853–865. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fackler MJ, McVeigh M, Evron E, et al: DNA
methylation of RASSF1A, HIN-1, RAR-β,
Cyclin D2 and Twist in in situ and invasive
lobular breast carcinoma. Int J Cancer. 107:970–975. 2003.
|
|
32
|
Yan L, Yang X and Davidson NE: Role of DNA
methylation and histone acetylation in steroid receptor expression
in breast cancer. J Mammary Gland Biol Neoplasia. 6:183–192. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lehmann U, Hasemeier B, Romermann D,
Muller M, Langer F and Kreipe H: Epigenetic inactivation of
microRNA genes in mammary carcinoma. Verh Dtsch Ges Pathol.
91:214–220. 2007.(In German).
|
|
34
|
Kapoor-Vazirani P, Kagey JD, Powell DR and
Vertino PM: Role of hMOF-dependent histone H4 lysine 16 acetylation
in the maintenance of TMS1/ASC gene activity. Cancer Res.
68:6810–6821. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang X, Karuturi RK, Sun F, et al: CDKN1C
(p57) is a direct target of EZH2 and suppressed by multiple
epigenetic mechanisms in breast cancer cells. PLoS One.
4:e50112009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Luo Q, Yuan Y, et al: Pygo2
associates with MLL2 histone methyltransferase and GCN5 histone
acetyltransferase complexes to augment Wnt target gene expression
and breast cancer stem-like cell expansion. Mol Cell Biol.
30:5621–5635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shi L, Sun L, Li Q, et al: Histone
demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes
hormonally responsive breast carcinogenesis. Proc Natl Acad Sci
USA. 108:7541–7546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Zhang H, Chen Y, et al: LSD1 is a
subunit of the NuRD complex and targets the metastasis programs in
breast cancer. Cell. 138:660–672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sieuwerts AM, Mostert B, Bolt-de Vries J,
et al: mRNA and microRNA expression profiles in circulating tumor
cells and primary tumors of metastatic breast cancer patients. Clin
Cancer Res. 17:3600–3618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
O’Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010.
|
|
41
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Walter BA, Gomez-Macias G, Valera VA,
Sobel M and Merino MJ: miR-21 expression in pregnancy-associated
breast cancer: a possible marker of poor prognosis. J Cancer.
2:67–75. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim SJ, Kelly WK, Fu A, Haines K, Hoffman
A, Zheng T and Zhu Y: Genome-wide methylation analysis identifies
involvement of TNF-α mediated cancer pathways in prostate cancer.
Cancer Lett. 302:47–53. 2011.
|
|
46
|
Bedford MT and van Helden PD:
Hypomethylation of DNA in pathological conditions of the human
prostate. Cancer Res. 47:5274–5276. 1987.PubMed/NCBI
|
|
47
|
Pakneshan P, Szyf M and Rabbani SA:
Hypomethylation of urokinase (uPA) promoter in breast and prostate
cancer: prognostic and therapeutic implications. Curr Cancer Drug
Targets. 5:471–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cho B, Lee H, Jeong S, et al: Promoter
hypomethylation of a novel cancer/testis antigen gene CAGE is
correlated with its aberrant expression and is seen in premalignant
stage of gastric carcinoma. Biochem Biophys Res Commun. 307:52–63.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tokizane T, Shiina H, Igawa M, et al:
Cytochrome P450 1B1 is overexpressed and regulated by
hypomethylation in prostate cancer. Clin Cancer Res. 11:5793–5801.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Laner T, Schulz WA, Engers R, Muller M and
Florl AR: Hypomethylation of the XIST gene promoter in prostate
cancer. Oncol Res. 15:257–264. 2005.PubMed/NCBI
|
|
51
|
Reibenwein J, Pils D, Horak P, et al:
Promoter hypermethylation of GSTP1, AR, and 14-3-3σ in serum of
prostate cancer patients and its clinical relevance. Prostate.
67:427–432. 2007.PubMed/NCBI
|
|
52
|
Dammann R, Schagdarsurengin U, Liu L, et
al: Frequent RASSF1A promoter hypermethylation and K-ras
mutations in pancreatic carcinoma. Oncogene. 22:3806–3812.
2003.PubMed/NCBI
|
|
53
|
Woodson K, Hayes R, Wideroff L, Villaruz L
and Tangrea J: Hypermethylation of GSTP1, CD44, and
E-cadherin genes in prostate cancer among US Blacks and Whites.
Prostate. 55:199–205. 2003.
|
|
54
|
Phe V, Cussenot O and Roupret M: Interest
of methylated genes as biomarkers in urothelial cell carcinomas of
the urinary tract. BJU Int. 104:896–901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pulukuri SM, Patibandla S, Patel J, Estes
N and Rao JS: Epigenetic inactivation of the tissue inhibitor of
metalloproteinase-2 (TIMP-2) gene in human prostate tumors.
Oncogene. 26:5229–5237. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Henrique R, Costa VL, Cerveira N, et al:
Hypermethylation of Cyclin D2 is associated with loss of mRNA
expression and tumor development in prostate cancer. J Mol Med.
84:911–918. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Seligson DB, Horvath S, Shi T, Yu H, Tze
S, Grunstein M and Kurdistani SK: Global histone modification
patterns predict risk of prostate cancer recurrence. Nature.
435:1262–1266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ellinger J, Kahl P, von der Gathen J, et
al: Global levels of histone modifications predict prostate cancer
recurrence. Prostate. 70:61–69. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Muller I, Wischnewski F, Pantel K and
Schwarzenbach H: Promoter- and cell-specific epigenetic regulation
of CD44, Cyclin D2, GLIPR1 and PTEN by methyl-CpG binding proteins
and histone modifications. BMC Cancer. 10:2972010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yu J, Cao Q, Mehra R, et al: Integrative
genomics analysis reveals silencing of β-adrenergic signaling by
polycomb in prostate cancer. Cancer Cell. 12:419–431. 2007.
|
|
61
|
Sikand K, Slaibi JE, Singh R, Slane SD and
Shukla GC: miR 488* inhibits androgen receptor expression in
prostate carcinoma cells. Int J Cancer. 129:810–819. 2011.
|
|
62
|
Majid S, Dar AA, Saini S, et al:
MicroRNA-205-directed transcriptional activation of tumor
suppressor genes in prostate cancer. Cancer. 116:5637–5649. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mercatelli N, Coppola V, Bonci D, et al:
The inhibition of the highly expressed miR-221 and miR-222 impairs
the growth of prostate carcinoma xenografts in mice. PLoS One.
3:e40292008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dong Q, Meng P, Wang T, et al: MicroRNA
let-7a inhibits proliferation of human prostate cancer cells in
vitro and in vivo by targeting E2F2 and CCND2. PLoS One.
5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Peng X, Guo W, Liu T, et al:
Identification of miRs-143 and -145 that is associated with bone
metastasis of prostate cancer and involved in the regulation of
EMT. PLoS One. 6:e203412011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fujita Y, Kojima K, Hamada N, et al:
Effects of miR-34a on cell growth and chemoresistance in prostate
cancer PC3 cells. Biochem Biophys Res Commun. 377:114–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lodygin D, Tarasov V, Epanchintsev A, et
al: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lin SL, Chiang A, Chang D and Ying SY:
Loss of mir-146a function in hormone-refractory prostate cancer.
RNA. 14:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rauch TA, Zhong X, Wu X, et al:
High-resolution mapping of DNA hypermethylation and hypomethylation
in lung cancer. Proc Natl Acad Sci USA. 105:252–257. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Glazer CA, Smith IM, Ochs MF, et al:
Integrative discovery of epigenetically derepressed cancer testis
antigens in NSCLC. PLoS One. 4:e81892009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Otterson GA, Khleif SN, Chen W, Coxon AB
and Kaye FJ: CDKN2 gene silencing in lung cancer by DNA
hypermethylation and kinetics of p16INK4 protein induction by 5-aza
2′deoxycytidine. Oncogene. 11:1211–1216. 1995.PubMed/NCBI
|
|
72
|
Paz MF, Avila S, Fraga MF, et al:
Germ-line variants in methyl-group metabolism genes and
susceptibility to DNA methylation in normal tissues and human
primary tumors. Cancer Res. 62:4519–4524. 2002.PubMed/NCBI
|
|
73
|
Pereira MA, Tao L, Liu Y, Li L, Steele VE
and Lubet RA: Modulation by budesonide of DNA methylation and mRNA
expression in mouse lung tumors. Int J Cancer. 120:1150–1153. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Katayama H, Hiraki A, Fujiwara K, et al:
Aberrant promoter methylation profile in pleural fluid DNA and
clinicopathological factors in patients with non-small cell lung
cancer. Asian Pac J Cancer Prev. 8:221–224. 2007.PubMed/NCBI
|
|
75
|
Licchesi JD, Westra WH, Hooker CM and
Herman JG: Promoter hypermethylation of hallmark cancer genes in
atypical adenomatous hyperplasia of the lung. Clin Cancer Res.
14:2570–2578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Barski A, Cuddapah S, Cui K, et al:
High-resolution profiling of histone methylations in the human
genome. Cell. 129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Seligson DB, Horvath S, McBrian MA, et al:
Global levels of histone modifications predict prognosis in
different cancers. Am J Pathol. 174:1619–1628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Feng Y, Wang X, Xu L, et al: The
transcription factor ZBP-89 suppresses p16 expression through a
histone modification mechanism to affect cell senescence. FEBS J.
276:4197–4206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Nonaka D, Fabbri A, Roz L, et al: Reduced
FEZ1/LZTS1 expression and outcome prediction in lung cancer. Cancer
Res. 65:1207–1212. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nishioka M, Kohno T, Tani M, et al:
MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1,
deleted, mutated, and methylated in human lung cancer. Proc Natl
Acad Sci USA. 99:12269–12274. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen MW, Hua KT, Kao HJ, et al: H3K9
histone methyltransferase G9a promotes lung cancer invasion and
metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer
Res. 70:7830–7840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Grosshans H, Johnson T, Reinert KL,
Gerstein M and Slack FJ: The temporal patterning microRNA let-7
regulates several transcription factors at the larval to adult
transition in C. elegans. Dev Cell. 8:321–330. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lujambio A and Esteller M: CpG island
hypermethylation of tumor suppressor microRNAs in human cancer.
Cell Cycle. 6:1455–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Saito M, Schetter AJ, Mollerup S, et al:
The association of microRNA expression with prognosis and
progression in early-stage, non-small cell lung adenocarcinoma: a
retrospective analysis of three cohorts. Clin Cancer Res.
17:1875–1882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Fang JY, Lu R, Mikovits JA, Cheng ZH, Zhu
HY and Chen YX: Regulation of hMSH2 and hMLH1
expression in the human colon cancer cell line SW1116 by DNA
methyltransferase 1. Cancer Lett. 233:124–130. 2006.
|
|
88
|
Goel A, Arnold CN, Niedzwiecki D, et al:
Frequent inactivation of PTEN by promoter hypermethylation in
microsatellite instability-high sporadic colorectal cancers. Cancer
Res. 64:3014–3021. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yuan BZ, Durkin ME and Popescu NC:
Promoter hypermethylation of DLC-1, a candidate tumor suppressor
gene, in several common human cancers. Cancer Genet Cytogenet.
140:113–117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thangaraju M, Carswell KN, Prasad PD and
Ganapathy V: Colon cancer cells maintain low levels of pyruvate to
avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J.
417:379–389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Spurling CC, Godman CA, Noonan EJ,
Rasmussen TP, Rosenberg DW and Giardina C: HDAC3 overexpression and
colon cancer cell proliferation and differentiation. Mol Carcinog.
47:137–147. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Godman CA, Joshi R, Tierney BR, et al:
HDAC3 impacts multiple oncogenic pathways in colon cancer cells
with effects on Wnt and vitamin D signaling. Cancer Biol Ther.
7:1570–1580. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tuttle R, Simon M, Hitch DC, et al:
Senescence-associated gene YPEL3 is downregulated in human colon
tumors. Ann Surg Oncol. 18:1791–1796. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li Q and Chen H: Transcriptional silencing
of N-Myc downstream-regulated gene 1 (NDRG1) in metastatic colon
cancer cell line SW620. Clin Exp Metastasis. 28:127–135. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Menigatti M, Cattaneo E, Sabates-Bellver
J, et al: The protein tyrosine phosphatase receptor type R gene is
an early and frequent target of silencing in human colorectal
tumorigenesis. Mol Cancer. 8:1242009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yadav S, Singhal J, Singhal SS and Awasthi
S: hSET1: a novel approach for colon cancer therapy. Biochem
Pharmacol. 77:1635–1641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Krieg AJ, Rankin EB, Chan D, Razorenova O,
Fernandez S and Giaccia AJ: Regulation of the histone demethylase
JMJD1A by hypoxia-inducible factor 1α enhances hypoxic gene
expression and tumor growth. Mol Cell Biol. 30:344–353. 2010.
|
|
98
|
Vlaicu SI, Tegla CA, Cudrici CD, et al:
Epigenetic modifications induced by RGC-32 in colon cancer. Exp Mol
Pathol. 88:67–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yin Q, Wang X, Fewell C, et al: MicroRNA
miR-155 inhibits bone morphogenetic protein (BMP) signaling and
BMP-mediated Epstein-Barr virus reactivation. J Virol.
84:6318–6327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu C, Kelnar K, Liu B, et al: The
microRNA miR-34a inhibits prostate cancer stem cells and metastasis
by directly repressing CD44. Nat Med. 17:211–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li T, Li D, Sha J, Sun P and Huang Y:
MicroRNA-21 directly targets MARCKS and promotes apoptosis
resistance and invasion in prostate cancer cells. Biochem Biophys
Res Commun. 383:280–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tang JT, Wang JL, Du W, et al:
MicroRNA-345, a methylation-sensitive microRNA is involved in cell
proliferation and invasion in human colorectal cancer.
Carcinogenesis. 32:1207–1215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Schneider AC, Heukamp LC, Rogenhofer S, et
al: Global histone H4K20 trimethylation predicts cancer-specific
survival in patients with muscle-invasive bladder cancer. BJU Int.
1082:E290–E296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Elsheikh SE, Green AR, Rakha EA, et al:
Global histone modifications in breast cancer correlate with tumor
phenotypes, prognostic factors, and patient outcome. Cancer Res.
69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Van Den Broeck A, Brambilla E,
Moro-Sibilot D, et al: Loss of histone H4K20 trimethylation occurs
in preneoplasia and influences prognosis of non-small cell lung
cancer. Clin Cancer Res. 14:7237–7245. 2008.PubMed/NCBI
|
|
107
|
Ellinger J, Kahl P, Mertens C, et al:
Prognostic relevance of global histone H3 lysine 4 (H3K4)
methylation in renal cell carcinoma. Int J Cancer. 127:2360–2366.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ke XS, Qu Y, Rostad K, et al: Genome-wide
profiling of histone h3 lysine 4 and lysine 27 trimethylation
reveals an epigenetic signature in prostate carcinogenesis. PLoS
One. 4:e46872009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Li Q, Wang X, Lu Z, et al: Polycomb CBX7
directly controls trimethylation of histone H3 at lysine 9 at the
p16 locus. PLoS One. 5:e137322010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
McGarvey KM, Van Neste L, Cope L, et al:
Defining a chromatin pattern that characterizes DNA-hypermethylated
genes in colon cancer cells. Cancer Res. 68:5753–5759. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wei Y, Xia W, Zhang Z, et al: Loss of
trimethylation at lysine 27 of histone H3 is a predictor of poor
outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog.
47:701–706. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Pogribny IP, Tryndyak VP, Muskhelishvili
L, Rusyn I and Ross SA: Methyl deficiency, alterations in global
histone modifications, and carcinogenesis. J Nutr. 137:216S–222S.
2007.PubMed/NCBI
|
|
113
|
Canaani E, Nakamura T, Rozovskaia T, Smith
ST, Mori T, Croce CM and Mazo A: ALL-1/MLL1, a homologue of
Drosophila TRITHORAX, modifies chromatin and is directly
involved in infant acute leukaemia. Br J Cancer. 90:756–760.
2004.
|
|
114
|
Liu H, Takeda S, Kumar R, et al:
Phosphorylation of MLL by ATR is required for execution of
mammalian S-phase checkpoint. Nature. 467:343–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Scacheri PC, Davis S, Odom DT, et al:
Genome-wide analysis of menin binding provides insights into MEN1
tumorigenesis. PLoS Genet. 2:e512006. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Seigne C, Fontaniere S, Carreira C, et al:
Characterisation of prostate cancer lesions in heterozygous Men1
mutant mice. BMC Cancer. 10:3952010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Feng ZJ, Gao SB, Wu Y, Xu XF, Hua X and
Jin GH: Lung cancer cell migration is regulated via repressing
growth factor PTN/RPTP β/ζ signaling by menin. Oncogene.
29:5416–5426. 2010.PubMed/NCBI
|
|
118
|
Wang J, Zhou Y, Yin B, et al: ASH2L:
alternative splicing and downregulation during induced
megakaryocytic differentiation of multipotential leukemia cell
lines. J Mol Med. 79:399–405. 2001. View Article : Google Scholar
|
|
119
|
Magerl C, Ellinger J, Braunschweig T, et
al: H3K4 dimethylation in hepatocellular carcinoma is rare compared
with other hepatobiliary and gastrointestinal carcinomas and
correlates with expression of the methylase Ash2 and the
demethylase LSD1. Hum Pathol. 41:181–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kobayashi Y, Absher DM, Gulzar ZG, et al:
DNA methylation profiling reveals novel biomarkers and important
roles for DNA methyltransferases in prostate cancer. Genome Res.
21:1017–1027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Velichutina I, Shaknovich R, Geng H,
Johnson NA, Gascoyne RD, Melnick AM and Elemento O: EZH2-mediated
epigenetic silencing in germinal center B cells contributes to
proliferation and lymphomagenesis. Blood. 116:5247–5255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Tell R, Rivera CA, Eskra J, Taglia LN,
Blunier A, Wang QT and Benya RV: Gastrin-releasing peptide
signaling alters colon cancer invasiveness via heterochromatin
protein 1Hsβ. Am J Pathol. 178:672–678. 2011.PubMed/NCBI
|
|
124
|
Xi Y, Formentini A, Nakajima G, Kornmann M
and Ju J: Validation of biomarkers associated with 5-fluorouracil
and thymidylate synthase in colorectal cancer. Oncol Rep.
19:257–262. 2008.PubMed/NCBI
|
|
125
|
Wang XQ, Miao X, Cai Q, Garcia-Barcelo MM
and Fan ST: SMYD3 tandem repeats polymorphism is not associated
with the occurrence and metastasis of hepatocellular carcinoma in a
Chinese population. Exp Oncol. 29:71–73. 2007.PubMed/NCBI
|
|
126
|
Oue N, Mitani Y, Motoshita J, et al:
Accumulation of DNA methylation is associated with tumor stage in
gastric cancer. Cancer. 106:1250–1259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Fang W, Piao Z, Buyse IM, Simon D, Sheu
JC, Perucho M and Huang S: Preferential loss of a polymorphic RIZ
allele in human hepatocellular carcinoma. Br J Cancer. 84:743–747.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhao Q, Caballero OL, Levy S, et al:
Transcriptome-guided characterization of genomic rearrangements in
a breast cancer cell line. Proc Natl Acad Sci USA. 106:1886–1891.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lucio-Eterovic AK, Singh MM, Gardner JE,
Veerappan CS, Rice JC and Carpenter PB: Role for the nuclear
receptor-binding SET domain protein 1 (NSD1) methyltransferase in
coordinating lysine 36 methylation at histone 3 with RNA polymerase
II function. Proc Natl Acad Sci USA. 107:16952–16957. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Berdasco M, Ropero S, Setien F, et al:
Epigenetic inactivation of the Sotos overgrowth syndrome gene
histone methyltransferase NSD1 in human neuroblastoma and glioma.
Proc Natl Acad Sci USA. 106:21830–21835. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Nimura K, Ura K, Shiratori H, Ikawa M,
Okabe M, Schwartz RJ and Kaneda Y: A histone H3 lysine 36
trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome.
Nature. 460:287–291. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Taketani T, Taki T, Nakamura H, Taniwaki
M, Masuda J and Hayashi Y: NUP98-NSD3 fusion gene in
radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15)
and expression pattern of NSD family genes. Cancer Genet Cytogenet.
190:108–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Morishita M and di Luccio E: Cancers and
the NSD family of histone lysine methyltransferases. Biochim
Biophys Acta. 1816:158–163. 2011.PubMed/NCBI
|
|
134
|
Watanabe H, Soejima K, Yasuda H, et al:
Deregulation of histone lysine methyltransferases contributes to
oncogenic transformation of human bronchoepithelial cells. Cancer
Cell Int. 8:152008. View Article : Google Scholar
|
|
135
|
Visakorpi T, Suikki HE, Kujala PM, Tammela
TLJ, van Weerden WM and Vessella RL: Genetic alterations and
changes in expression of histone demethylases in prostate cancer.
Prostate. 70:889–898. 2010.PubMed/NCBI
|
|
136
|
Fukuda T, Tokunaga A, Sakamoto R and
Yoshida N: Fbxl10/Kdm2b deficiency accelerates neural progenitor
cell death and leads to exencephaly. Mol Cell Neurosci. 46:614–624.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Vinatzer U, Gollinger M, Mullauer L,
Raderer M, Chott A and Streubel B: Mucosa-associated lymphoid
tissue lymphoma: novel translocations including rearrangements of
ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 14:6426–6431. 2008.
View Article : Google Scholar
|
|
138
|
Yang ZQ, Imoto I, Fukuda Y, et al:
Identification of a novel gene, GASC1, within an amplicon at
9p23–24 frequently detected in esophageal cancer cell lines. Cancer
Res. 60:4735–4739. 2000.PubMed/NCBI
|
|
139
|
Zeng J, Ge Z, Wang L, et al: The histone
demethylase RBP2 is overexpressed in gastric cancer and its
inhibition triggers senescence of cancer cells. Gastroenterology.
138:981–992. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Rao M, Chinnasamy N, Hong JA, et al:
Inhibition of histone lysine methylation enhances cancer-testis
antigen expression in lung cancer cells: implications for adoptive
immunotherapy of cancer. Cancer Res. 71:4192–4204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liggins AP, Lim SH, Soilleux EJ, Pulford K
and Banham AH: A panel of cancer-testis genes exhibiting
broad-spectrum expression in haematological malignancies. Cancer
Immun. 10:82010.PubMed/NCBI
|
|
142
|
Jankowska A, Makishima H, Tiu RV, et al:
Mutational spectrum analysis of chronic myelomonocytic leukemia
includes genes associated with epigenetic regulation: UTX, EZH2,
and DNMT3A. Blood. 116:268–269. 2010.PubMed/NCBI
|
|
143
|
Xiang Y, Zhu Z, Han G, Lin H, Xu L and
Chen CD: JMJD3 is a histone H3K27 demethylase. Cell Res.
17:850–857. 2007. View Article : Google Scholar : PubMed/NCBI
|