Introduction
Colorectal cancer (CRC) is the fourth leading cause
of cancer-related death in the world (1). Epidemiologically, a Western diet,
alcohol, smoking and obesity are related to an increased risk of
CRC and mortality (2,3). Although screening and prevention
efforts have been promoted, a small fraction of CRC cases are still
discovered as unresectable tumors due to metastasis to critical
organs (4). Radiation therapy
(5) and chemotherapy (6) have major limitations to the treatment
of CRC patients. Therefore, a novel therapeutic approach is
required for CRC.
Antibody arrays represent a promising technology for
quantitative and exhaustive protein profiling (7). Sandwich-based arrays are frequently
used due to their high sensitivity, specificity and cost
effectiveness. However, a pair of antibodies could cross-react and
hamper the appropriate reaction with proteins in the array. Current
antibody protein arrays also have limited abilities to provide a
broad, panoramic view of protein expression levels. Recently, a new
antibody array with a relatively simple process, in which human
samples are biotinylated and dialyzed in preparation for
incubation, has become available using the largest available
antibody array. The expression levels of 507 human target proteins,
including angiogenic factors, cytokines and chemokines, can be
analyzed simultaneously. A broad, panoramic view of cytokine
expression can be obtained using this new technology.
Previously, we reported activated receptor tyrosine
kinase (RTK) arrays in CRC (8). The
downregulation of angiogenic factors such as vascular endothelial
growth factor (VEGF) and matrix metalloproteinases inhibits
invasiveness and tumor development in colon cancer (9). Since multistage carcinogenesis is
observed during colon cancer development, it is suggested that
colon cancer can be affected through multiple pathways. In the
present study, we examined 507 human target proteins simultaneously
using a new biotin label-based human antibody array in CRC
samples.
Materials and methods
Patients
Tissue samples of CRC and normal colon tissues were
obtained from 6 patients during surgery (2 males and 4 females;
mean age, 71±11 years; range, 56–92 years). The clinicopathological
data for the CRC patients are documented in Table I. Of these 6 patients, 1 had cancer
localized to the ascending colon, 2 had cancer localized to the
transverse colon, 1 had cancer localized to the sigmoid colon and 2
had cancer localized to the rectum. Histologically, 3 cancers were
well differentiated, 2 were moderately differentiated and 1 was
poorly differentiated. With respect to clinical stage, 4 were in
stage II, 1 was in stage IIIa and 1 was in stage IV. All
experimental protocols were approved by the Human Subjects
Committee of the Kagawa University School of Medicine. Informed
consent was obtained prior to participation in the study.
 | Table IClinical characteristics of the
colorectal cancer patients. |
Table I
Clinical characteristics of the
colorectal cancer patients.
| Age (years) | Gender | Region | Histologya | TNM stageb |
|---|
| 92 | Female | Ascending | WD | II |
| 85 | Female | Transverse | MD | IIIa |
| 63 | Male | Rectum | PD | IV |
| 69 | Male | Sigmoid | WD | II |
| 56 | Female | Transverse | MD | II |
| 63 | Female | Rectum | WD | II |
Sample preparation
CRC tissue samples were prepared using methods
previously described (10,11).
Materials for the protein array
The RayBio™ Biotin Label-based Human Antibody Array
I (catalog no. AAH-BLM-1–2) was purchased from RayBiotech, Inc.
(Norcross, GA, USA). This antibody array is a dot-blot based assay
that enables the detection and comparison of 507 different human
antibodies, including cytokines, chemokines, growth factors,
angiogenic factors, proteases and soluble receptors.
Biotin label-based antibody array
This array was used according to the manufacturer’s
protocol. Homogenized tissues were dialyzed using a dialyzer.
Biotin was added to the proteins that were derived from the colon
tissues. Antibody array membranes were blocked for 1 h and
incubated with 8 ml of lysate from colon tissues at room
temperature for 2 h. After a washing step, the array membranes were
incubated with HRP-conjugated streptavidin at room temperature for
2 h. Unbound HRP-conjugated streptavidin was washed, and each array
membrane was exposed to X-ray film using a chemiluminescence
detection system (Amersham Life Sciences, Tokyo, Japan). A
densitometric analysis was performed using ImageQuant TL (GE
Healthcare Bioscience, Tokyo, Japan).
Data analysis
The array data of CRC and normal colon tissues were
normalized using a positive control signal. The average local
background signal was subtracted from the average signal intensity
of duplicated spots for each antibody. A cluster analysis was
performed to evaluate the associations among members of the groups
or clusters.
Statistical analysis
All analyses were conducted using the
computer-assisted program JMP 8.0 (SAS Institute). A P-value of
0.05 represents a significant difference between groups.
Results
Enhanced expression of target proteins on
biotin label-based antibody arrays
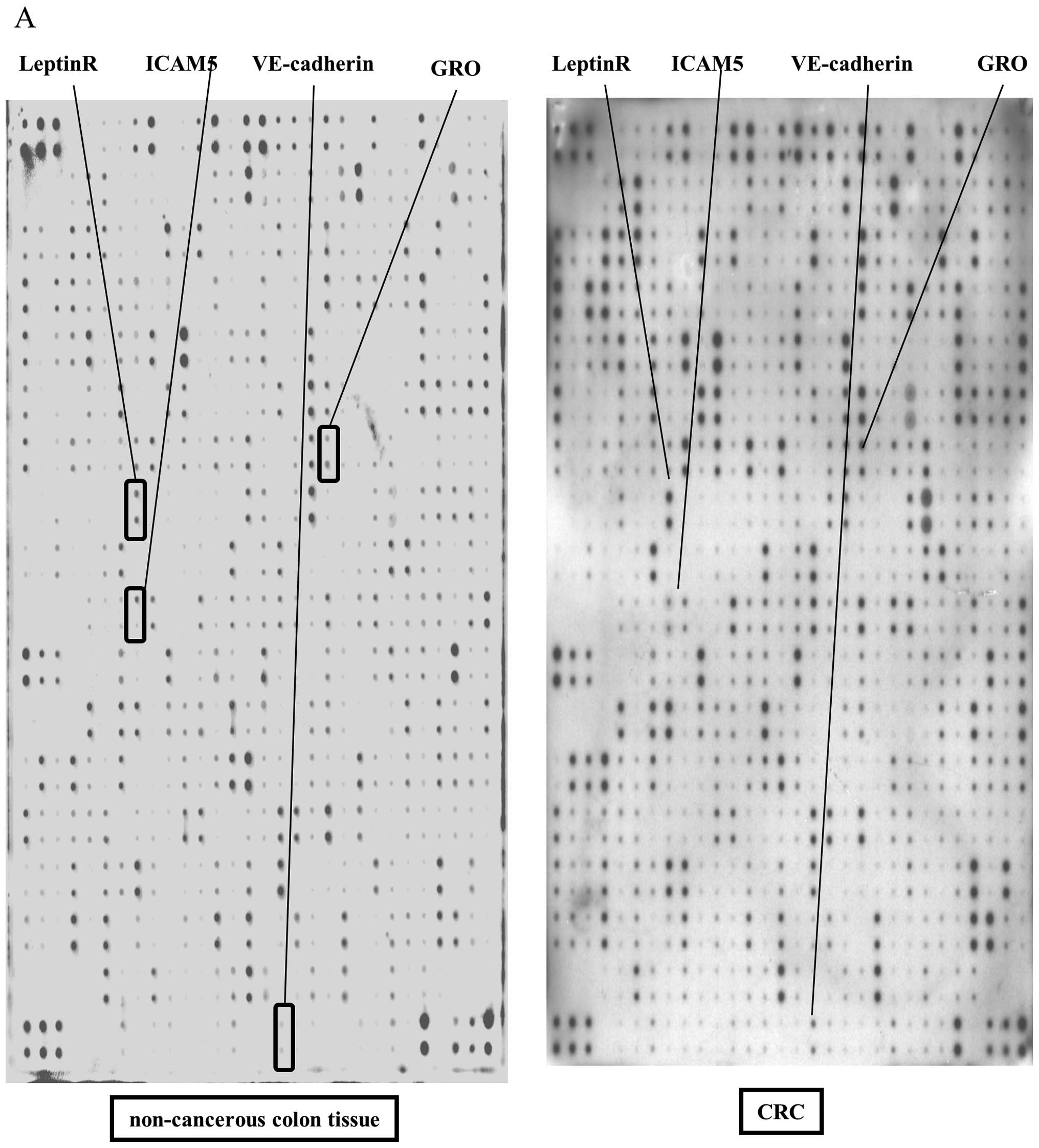
A total of 507 human proteins in CRC tissues were
analyzed simultaneously on a single array. Representative antibody
arrays between normal colon and CRC tissues are shown in Fig. 1A and B. IL-1α, GRO, Glut5, MIG,
ICAM5, VE-cadherin, uPA and Leptin R were upregulated in CRC
tissues when compared to these levels in normal colon tissues
(Fig. 1A and B). A densitometric
analysis was performed for each spot on the antibody array.
Unsupervised hierarchical clustering
analysis using Pearson’s correlation
To understand the results of the antibody protein
array, unsupervised hierarchical clustering analysis was performed
(Fig. 1C). IL-1α, GRO, Glut5, MIG,
ICAM5, VE-cadherin, uPA and Leptin R were statistically
significantly upregulated in CRC when compared to these levels in
normal colon tissues (P<0.05) (Table II). No statistically significant
protein downregulation was detected among the 507 human proteins on
the array in CRC samples when compared to the normal colon
tissues.
 | Table IIEnhanced protein expression levels in
colorectal cancer. |
Table II
Enhanced protein expression levels in
colorectal cancer.
| Ratio | P-value |
|---|
| IL-1α | 2.81 | 0.023 |
| GRO | 2.68 | 0.028 |
| Glut5 | 2.54 | 0.047 |
| MIG | 2.51 | 0.010 |
| ICAM5 | 2.29 | 0.046 |
| VE-cadherin | 2.28 | 0.026 |
| uPA | 2.26 | 0.017 |
| Leptin R | 2.13 | 0.045 |
Significant upregulation of 23 target
proteins in rectal cancer compared to non-rectal cancer
As shown in Table
II, although no statistically significant protein upregulation
was detected with respect to gender, age, histological grade or TNM
stage, 23 target proteins were significantly upregulated in rectal
cancer among CRC as compared to non-rectal cancer which consists of
ascending, transverse and sigmoid colon (P<0.01) (Table III). Among these 23 target
proteins, MPIF-1/CCL23, FGF R5 and MIP 2 were strongly upregulated
in rectal cancer (Table IV).
 | Table IIICharacteristics of the colorectal
cancer patients. |
Table III
Characteristics of the colorectal
cancer patients.
| No. | No. of upregulated
genes (P<0.01) |
|---|
| Gender | | 0 (in female vs.
male) |
| Male | 2 | |
| Female | 4 | |
| Age (years) | | 0 (in >65 vs.
≤65) |
| ≤65 | 3 | |
| >65 | 3 | |
| Histological
gradea | | 0 (in MD/PD vs.
WD) |
| WD | 3 | |
| MD/PD | 3 | |
| TNM stageb | | 0 (in IIIa/IV vs.
II) |
| II | 4 | |
| IIIa/IV | 2 | |
| Location | | 23 (in rectum vs.
non-rectum) |
| Non-rectum | 4 | |
| Rectum | 2 | |
 | Table IVTwenty-three target proteins in
non-rectum (ascending, transverse and sigmoid colon)/rectum (4/2)
(P<0.01) tissue samples. |
Table IV
Twenty-three target proteins in
non-rectum (ascending, transverse and sigmoid colon)/rectum (4/2)
(P<0.01) tissue samples.
| P-value |
|---|
| MPIF-1/CCL23 | 0.00035 |
| FGF R5 | 0.00045 |
| MIP 2 | 0.00072 |
| SAA | 0.0019 |
| IL-18 R β (AcPL) | 0.00199 |
| LFA-1 α | 0.00324 |
| G-CSF R/CD 114 | 0.00328 |
| IFN-γ R1 | 0.00501 |
| TRAIL
R4/TNFRSF10D | 0.00507 |
| FGF R3 | 0.0059 |
| IL-18 R α (IL-1
R5) | 0.00613 |
| GITR/TNFRF18 | 0.00658 |
| IL-22 R | 0.00666 |
| IL-18 BPa | 0.0075 |
| E-Selectin | 0.00786 |
| IL-17B | 0.00808 |
| GDF1 | 0.00837 |
| IL-1 F7/FIL1ζ | 0.00839 |
| ErbB4 | 0.00916 |
| FAM3B | 0.00943 |
| IL-27 | 0.00948 |
| CRIM 1 | 0.00954 |
| BMPR-II | 0.00955 |
Discussion
Various antibody protein arrays have been developed
in the last decade. In most antibody array methods, two detection
antibodies can recognize different epitopes of the same target
protein and target antigens are visualized. However, an antibody
cross-reaction can hamper accurate protein detection on the
membrane of a protein array. To avoid this complication, surface
plasmon resonance (SPR), which provides a label-free,
single-antibody approach (12), has
been developed. However, this approach also has additional
complications, such as low detection sensitivity and a long waiting
time to improve the instrumentation for high density detection.
Therefore, in the present study, a biotin label-based human
antibody array was used to perform the exhaustive analysis of
protein expression. This technology enabled the detection of 507
proteins simultaneously, and a broad, panoramic view of target
protein expression could be analyzed for each sample. Huang et
al (13) reported that a biotin
label-based antibody array could be used to profile the expression
levels of many proteins simultaneously in ovarian cancer. These
include cytokines, chemokines, adipokines, growth factors,
angiogenic factors, proteases, soluble receptors and soluble
adhesion molecules. Their finding supports the use of a biotin
label-based antibody array targeted for 507 proteins in the present
study.
In the present study, IL-1α, GRO, Glut, MIG and uPA
were upregulated in colon cancer. These proteins have previously
been reported as an angiogenic factor (14), anti-apoptotic factor, prognostic
marker (15,16), chemokine (17,18) or
in colon cancer. The present study data also demonstrated that
ICAM5, VE-cadherin and Leptin R were upregulated in colon cancer
(Table II). ICAM5 and VE-cadherin
are cell adhesion molecules that are related to the development of
breast cancer, prostate cancer and head and neck carcinoma
(19–21). This finding suggests that these
proteins could also be targetable molecules for colon cancer.
The array results also demonstrated that 23 target
proteins were enhanced in rectal cancer (Table III). However, none of these
proteins was a match to the target proteins that were presented in
Table II. Recently, the Cancer
Genome Atlas Network reported that hypermethylation was more common
in the right colon than in the rectum (22). These findings strongly support our
data that protein expression patterns differ between rectal cancer
and non-rectal cancer.
In conclusion, our findings demonstrated that IL-1α,
GRO, Glut5, MIG, ICAM-5, VE-cadherin, uPA and Leptin R were
upregulated in colon cancer when compared to levels in normal colon
tissues using a biotin label-based antibody array. These results
could aid in the simultaneous analysis of various types of
upregulated proteins and promising target proteins for
molecular-targeted therapies. The simplicity and ease of using the
biotin label-based antibody array suggests that this new array
method may be a powerful tool for detecting expression levels of a
wide range of proteins from each pathway that contributes to colon
carcinogenesis and for identifying new therapies for CRC.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paik SS, Jang SM, Jang KS, Lee KH, Choi D
and Jang SJ: Leptin expression correlates with favorable
clinicopathologic phenotype and better prognosis in colorectal
adenocarcinoma. Ann Surg Oncol. 16:297–303. 2009. View Article : Google Scholar
|
|
3
|
Calle EE, Rodriguez C, Walker-Thurmond K
and Thun MJ: Overweight, obesity, and mortality from cancer in a
prospectively studied cohort of U.S. adults. N Engl J Med.
24:1625–1638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Devine RM and Dozois RR: Surgical
management of locally advanced adenocarcinoma of the rectum. World
J Surg. 16:486–489. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Cummings B, Catton P, et al:
Primary radical external beam radiotherapy of rectal
adenocarcinoma: long term outcome of 271 patients. Radiother Oncol.
77:126–132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prados J, Melguizo C, Ortiz R, et al:
Colon cancer therapy: recent developments in nanomedicine to
improve the efficacy of conventional chemotherapeutic drugs.
Anticancer Agents Med Chem. Mar 4–2013.(Epub ahead of print).
|
|
7
|
Huang RP, Yang W, Yang D, Flowers L,
Horowitz IR, Cao X and Huang R: The promise of cytokine antibody
arrays in the drug discovery process. Expert Opin Ther Targets.
9:601–615. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Morishita A, Gong J, Nomura T, et al: The
use of protein array to identify targetable receptor tyrosine
kinases for treatment of human colon cancer. Int J Oncol.
37:829–835. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Auyeung KK, Law PC and Ko JK: Novel
anti-angiogenic effects of formononetin in human colon cancer cells
and tumor xenograft. Oncol Rep. 28:2188–2194. 2012.PubMed/NCBI
|
|
10
|
Yoshida S, Masaki T, Feng H, et al:
Enhanced expression of adaptor molecule p46 Shc in nuclei of
hepatocellular carcinoma cells: study of LEC rats. Int J Oncol.
25:1089–1096. 2004.PubMed/NCBI
|
|
11
|
Yukimasa S, Masaki T, Yoshida S, et al:
Enhanced expression of p46 Shc in the nucleus and p52 Shc in the
cytoplasm of human gastric cancer. Int J Oncol. 26:905–911.
2005.PubMed/NCBI
|
|
12
|
Boozer C, Kim G, Cong S, Guan H and
Londergan T: Looking towards label-free biomolecular interaction
analysis in a high-throughput format: a review of new surface
plasmon resonance technologies. Curr Opin Biotechnol. 17:400–405.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang R, Jiang W, Yang J, et al: A biotin
label-based antibody array for high-content profiling of protein
expression. Cancer Genomics Proteomics. 7:129–141. 2010.PubMed/NCBI
|
|
14
|
Matsuo Y, Sawai H, Ma J, et al: IL-1α
secreted by colon cancer cells enhances angiogenesis: the
relationship between IL-1α release and tumor cells’ potential for
liver metastasis. J Surg Oncol. 99:361–367. 2009.
|
|
15
|
Haber RS, Rathan A, Weiser KR, et al:
GLUT1 glucose transporter expression in colorectal carcinoma: a
marker for poor prognosis. Cancer. 83:34–40. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Langenskiold M, Holmdahl L, Angenete E,
Falk P, Nordgren S and Ivarsson ML: Differential prognostic impact
of uPA and PAI-1 in colon and rectal cancer. Tumour Biol.
30:210–220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ruehlmann JM, Xiang R, Niethammer AG, et
al: MIG (CXCL9) chemokine gene therapy combines with
antibody-cytokine fusion protein to suppress growth and
dissemination of murine colon carcinoma. Cancer Res. 61:8498–8503.
2001.PubMed/NCBI
|
|
18
|
Wen Y, Giardina SF, Hamming D, et al: GROα
is highly expressed in adenocarcinoma of the colon and
down-regulates fibulin-1. Clin Cancer Res. 12:5951–5959. 2006.
|
|
19
|
Maruya SI, Myers JN, Weber RS, Rosenthal
DI, Lotan R and El-Naggar AK: ICAM-5 (telencephalin) gene
expression in head and neck squamous carcinoma tumorigenesis and
perineural invasion! Oral Oncol. 41:580–588. 2005.PubMed/NCBI
|
|
20
|
Sulkowska M, Famulski W, Wincewicz A, et
al: Levels of VE-cadherin increase independently of VEGF in
preoperative sera of patients with colorectal cancer. Tumori.
92:67–71. 2006.PubMed/NCBI
|
|
21
|
Uddin S, Bavi PP, Hussain AR, et al:
Leptin receptor expression in Middle Eastern colorectal cancer and
its potential clinical implication. Carcinogenesis. 30:1832–1840.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cancer Genome Atlas Network. Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|