Introduction
Osteosarcoma, mainly arising from the metaphysis of
the long bones, is the most common primary malignancy of bone in
adolescents and young adults, with an estimated worldwide yearly
incidence rate of 4 million (1,2).
Despite current therapeutic strategies combining adjuvant
chemotherapy, surgery and sometimes radiotherapy, the prognosis of
osteosarcoma patients remains poor, since ~80% of patients
eventually develop recurrent metastatic osteosarcoma following
surgical treatment (3), and the
5-year survival rate of these patients is only 50–60% (4). Although recent developments in
molecular biology have provided insight into the molecular
mechanisms of osteosarcoma, the fundamental molecular mechanisms
underlying metastasis in osteosarcoma have not been fully
elucidated. Therefore, it is essential to identify
metastasis-associated molecules as effective drug targets and to
enhance the understanding of the mechanisms underlying the
metastasis of osteosarcoma.
MicroRNAs (miRNAs) are small non-coding RNAs 18–25
nucleotides in length, transcribed from non-protein-coding genes or
introns, which regulate gene expression through repressing
translation and cleaving their target mRNAs by binding to
complementary sites in their 3′-untranslated region (3′-UTR). It
has been demonstrated that aberrant expression of miRNAs cause them
to function as tumor suppressors or oncogenes according to the
roles of their target genes (5,6).
Particularly, miRNAs can regulate various cellular processes of
tumor cells, including differentiation, progression, apoptosis,
proliferation, migration and invasion (7). To date, several human miRNAs such as
miR-335, miR-145 and miR-128 have been shown to be dysregulated in
osteosarcoma (7–9), and contribute to the development and
progression of osteosarcoma.
Emerging data reveal that microRNA-26a (miR-26a) is
downregulated and may serve as a potential tumor suppressor in
several distinct cancer types including nasopharyngeal carcinoma,
breast cancer, thyroid anaplastic carcinomas and hepatocellular
carcinoma (5,10–14).
Importantly, miR-26a was found to be highly expressed in lymph node
metastatic tumors as compared with primary tumors and enhanced lung
cancer cell migration and invasion (15). miR-26a can suppress cell
differentiation, migration and invasion by targeting a number of
important genes such as SMAD1, MTDH, CDK6, CCNE1, CCNE2 CCND2,
PTEN, PB1, MAP3K2 and enhancer of zeste homolog 2 (EZH2) (13,15–20).
However, the roles of miR-26a and its regulated targets in
osteosarcoma have not yet been clarified.
In the present study, we investigated the potential
function of miR-26a in the development and progression of
osteosarcoma. We found that miR-26a expression was downregulated in
the majority of osteosarcoma tissues, and downregulation of miR-26a
was significantly associated with tumor recurrence, metastasis and
poor prognosis in osteosarcoma patients. Moreover, in vitro
assays showed that miR-26a was significantly associated with
suppressed tumor invasion and metastasis of osteosarcoma cells by
targeting its direct target EZH2. To the best of our knowledge,
this is the first study to examine the expression and role of
miR-26a in osteosarcoma prognosis and metastasis.
Materials and methods
Patients and tissue samples
The present study was approved by the Research
Ethics Committee of Xi’an Jiaotong University. Written informed
consent was obtained from all of the patients. All specimens were
handled and made anonymous according to the ethical and legal
standards. A total of 144 patients were enrolled in this study.
Patients received curative resection for osteosarcoma at the Second
Affiliated Hospital, Xi’an Jiaotong University (Xi’an, China)
between 2001 and 2008. No patients had received blood transfusion,
radiotherapy or chemotherapy prior to surgery. The
clinicopathological information of these patients is documented in
Table I. The follow-up information
of all participants was updated every 3 months by telephone. The
overall survival was defined as the time elapsed from surgery to
death. Information regarding the death of patients was ascertained
from their family. Patients were followed up after surgical
treatment until July 2012, with a median follow-up of 83 months
(range, 12–139 months). During the follow-up period, 65 patients
(45.1%) died of disease. Distant metastases developed in 37
patients at a mean of 15.1 months (range, 4–39 months) after
initial diagnosis. The median overall and disease-free survival of
patients was 43 and 37 months, respectively.
 | Table IAssociation of miR-26a expression with
the clinicopathological features of the osteosarcoma cases. |
Table I
Association of miR-26a expression with
the clinicopathological features of the osteosarcoma cases.
| | miR-26a
expression | |
|---|
| |
| |
|---|
| Clinicopathological
features | No. of cases | High n (%) | Low n (%) | P-value |
|---|
| Age (years) | | | | 0.865 |
| <55 | 62 | 34 (54.8) | 28 (45.2) | |
| ≥55 | 82 | 47 (57.3) | 35 (42.7) | |
| Gender | | | | 0.738 |
| Male | 76 | 44 (57.9) | 32 (42.1) | |
| Female | 68 | 37 (54.4) | 31 (45.6) | |
| Tumor size
(cm) | | | | 0.311 |
| >8 | 79 | 41 (51.9) | 38 (48.1) | |
| ≤8 | 65 | 40 (61.5) | 25 (38.5) | |
| Anatomic
location | | | | 0.611 |
| Femur | 78 | 44 (56.4) | 34 (43.6) | |
| Tibia | 42 | 21 (50.0) | 21 (50.0) | |
| Humeral bone | 17 | 11 (64.7) | 6 (35.3) | |
| Other | 7 | 5 (71.4) | 2 (28.6) | |
| Clinical stage | | | | 0.043a |
| IIA | 62 | 41 (66.1) | 21 (33.9) | |
| IIB/III | 82 | 40 (48.8) | 42 (51.2) | |
| Metastasis | | | | 0.004a |
| Present | 37 | 13 (35.1) | 24 (64.9) | |
| Absent | 107 | 68 (63.6) | 39 (36.4) | |
| Response to
chemotherapy | | | | 0.044a |
| Favorable | 69 | 45 (65.2) | 24 (34.8) | |
| Poor | 75 | 36 (48.0) | 39 (52.0) | |
Quantitative reverse transcriptase PCR
(qRT-PCR) assay
The expression of miR-26a in the osteosarcoma and
corresponding non-cancer tissues was determined by qRT-PCR assay.
Briefly, total RNA was extracted from the tissues using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s protocol. miRNA expression levels were then
quantitated using the TaqMan miRNA real-time RT-PCR kit (Applied
Biosystems) according to the manufacturer’s protocol. Data were
analyzed using 7500 software v.2.0.1 (Applied Biosystems), with the
automatic Ct setting for adapting baseline and threshold for Ct
determination. The universal small nuclear RNA U6 (RNU6B) was used
as an endogenous control for miRNAs. Each sample was examined in
triplicate, and the amounts of PCR products produced were
non-neoplasticized to RNU6B.
Cell culture
Human osteosarcoma cell lines MG-63 and U20S were
obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China), where they were characterized by mycoplasma
detection, DNA-fingerprinting, isozyme detection and cell vitality
detection. They were cultured in Dulbecco’s modified Eagle’s medium
(DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS)
(HyClone, Logan, UT, USA) and cultured in a humidified incubator at
37°C in 5% CO2.
In vitro migration and invasion
assays
Cell migration and invasion capacities were assessed
in vitro using Transwell migration assays (Millipore,
Billerica, MA, USA). The osteosarcoma cells were transfected with
miR-26a mimics, inhibitor or scramble for 48 h and suspended in
DMEM with 10 g/l BSA at a density of 50 cells/ml. Then, cell
suspensions (200 μl) were seeded into the upper chamber with a
porous membrane coated with (for the Transwell invasion assay) or
without (for the migration assay) Matrigel (BD Biosciences, San
Diego, CA, USA). To attract the cells, 500 μl of DMEM with 10%
serum was added to the bottom chamber. After allowing the cells to
migrate for 24 h or to invade for 48 h, the penetrated cells on the
filters were fixed in dried methanol and stained in 4 g/l crystal
violet. The numbers of migrated or invasive cells were determined
from five random fields using a microscope (Olympus) at ×10
magnification.
Oligonucleotide transfection
miR-26a mimics and inhibitor were chemically
synthesized by Shanghai GenePharma (Shanghai, China). When the
cells achieved 80% confluence, miR-26a mimics or the inhibitor was
transfected into the osteosarcoma cells with Lipofectamine 2000
(Invitrogen) according to the manufacturer’s instructions. Cells
were also transfected with a scramble oligonucleotide as a negative
control (NC). The expression level of miR-26a in the transfected
osteosarcoma cells was determined by qRT-PCR.
Luciferase reporter assay
Osteosarcoma cells were seeded into a 96-well plate
at 60% confluence. After 24 h, cells were transfected with 120 ng
of miR-26a expression vector or the negative control. Cells were
transfected with 30 ng of wild-type (WT) or mutant (MT) 3′-UTR of
EZH2 mRNA. Cells were collected 48 h after transfection, and the
luciferase activity was measured using a dual-luciferase reporter
assay system according to the manufacturer’s protocol
(Promega).
Western blotting
Cells were harvested in lysis buffer (50 mM NaCl, 50
mM EDTA, 1% Triton X-100) containing protease inhibitor cocktail
(Roche, Indianapolis, IN, USA). The cell lysates (30 μg) were
separated using 10% SDS-PAGE gels and then transferred onto
nitrocellulose membranes (Millipore, Bedford, MA, USA). The
membranes were blocked with 5% non-fat milk diluted in PBS for 2 h
at room temperature before the addition of the appropriate primary
antibody. The antibodies used in the present study included
anti-EZH2 (1:1000; ab3748) and anti-β-actin (1:1,000; ab14128)
(both from Abcam). The membranes were then washed with PBS
containing 0.05% Tween and incubated with the appropriate
HRP-conjugated secondary antibody (1:10,000; Abcam) for 1 h at room
temperature. The bands were visualized using a chemiluminescence
reagent (New England Nuclear, Boston, MA, USA).
Statistical analysis
Statistical analysis was performed using IBM SPSS
statistical software (version 20.0). Survival curves were estimated
using the Kaplan-Meier method, and distributions were evaluated by
the log-rank test. Cox proportional hazard models of factors
related to survival were used to calculate risk ratios (RRs) and
identify the factors that affected survival. The differences in
characteristics between two groups were examined by the
χ2 test and Fisher’s exact test. All P-values were
determined from 2-sided tests, and statistical significance was
based on a P-value of 0.05.
Results
Downregulation of miR-26a is associated
with metastasis and recurrence of osteosarcoma
Expression of miR-26a was analyzed in 144 pairs of
osteosarcoma and corresponding non-cancerous bone tissues by
quantitative real-time polymerase chain reaction (qRT-PCR) and
normalized against an endogenous control (U6 RNA). As shown in
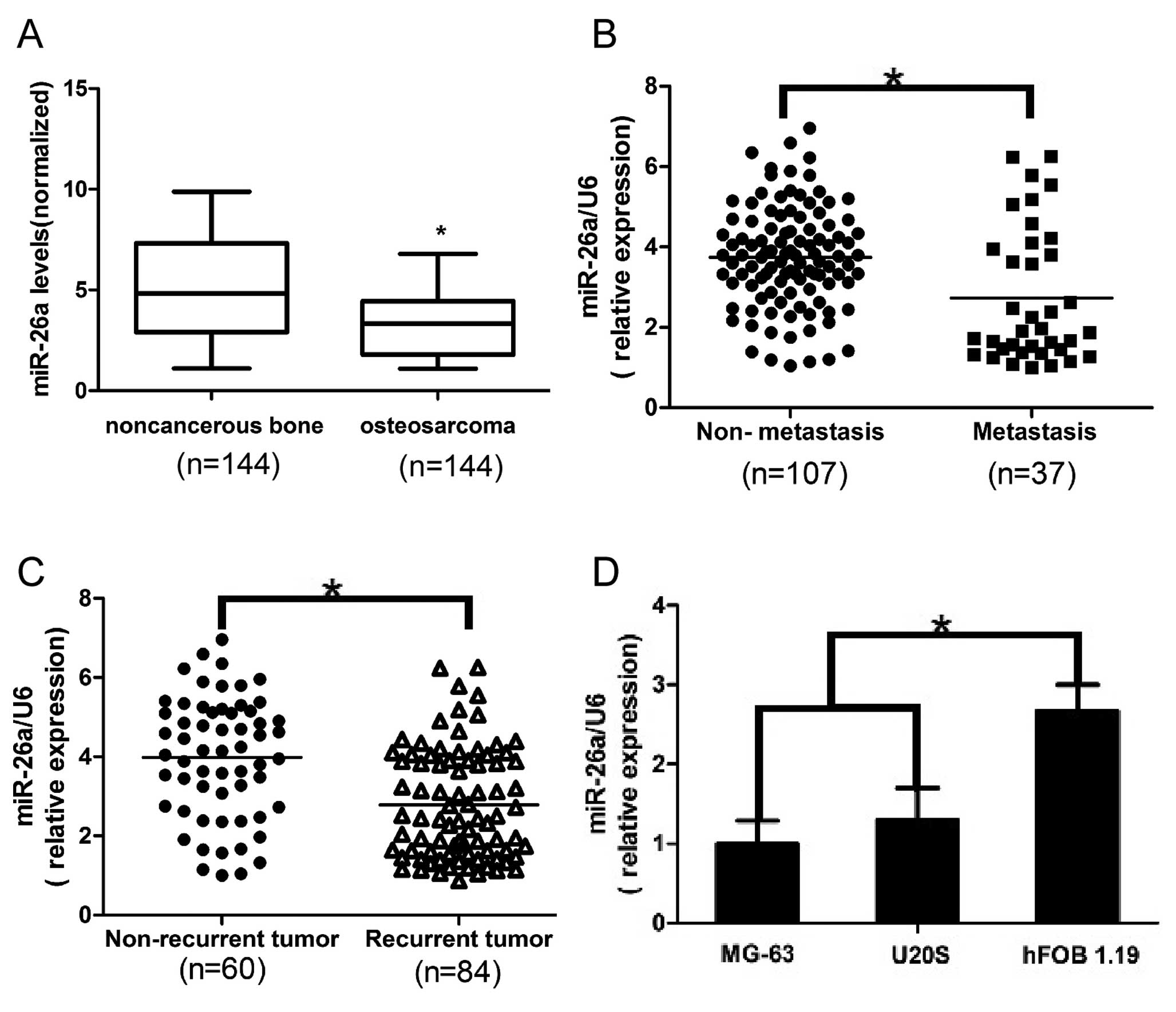
Fig. 1A, the expression level of
miR-26a in the osteosarcoma tissues was found to be distinctly
downregulated when compared to that in the non-cancerous bone
tissues. Furthermore, in comparison to the non-metastatic
osteosarcoma tissues, the miR-26a levels were significantly lower
in the metastatic osteosarcoma tissues (Fig. 1B). Moreover, miR-26a levels were
decreased in the tumor tissues obtained from the patients who
suffered osteosarcoma recurrence in comparison with patients who
did not have tumor recurrence (Fig.
1C). These data indicate that significant downregulation of
miR-26a expression occurred in osteosarcoma and was correlated with
osteosarcoma relapse and metastasis. To further evaluate the
association of miR-26a with osteosarcoma metastasis, we analyzed
miR-26a levels in human osteosarcoma cell lines, MG-63 and U20S,
and human osteoblast cell line hFOB 1.19. Similarly, we found that
expression of miR-26a was much lower in the osteosarcoma cell lines
than that in the osteoblast cell line (Fig. 1D). Collectively, the above findings
suggest that loss of miR-26a expression is correlated with
increased metastatic potential of osteosarcoma cells.
Downregulation of miR-26a is associated
with advanced clinicopathological features of osteosarcoma
To determine the clinical significance of miR-26a in
osteosarcoma, we analyzed the association of miR-26a expression
with various clinicopathological parameters of osteosarcoma
tissues. The median miR-26a expression level in all 144 patients
with osteosarcoma was 3.37. The patients were divided into two
groups according to their expression levels of miR-26a, using the
median level as a cut off: the high miR-26a expression group (n=81)
and the low miR-26a expression group (n=63). As shown in Table I, miR-26a was significantly
downregulated in osteosarcoma patients with advanced clinical stage
(P=0.043) and positive distant metastasis (P=0.004). Taking into
consideration the relationship between miR-26a expression and the
response to chemotherapy, we found that patients with low miR-26a
had a poorer response to chemotherapy than those with high miR-26a
expression (P=0.044), whereas, miR-26a expression was not
significantly correlated with gender, age, tumor size, anatomic
location or alkaline phosphatase level.
Downregulation of miR-26a is associated
with poor prognosis in patients with osteosarcoma
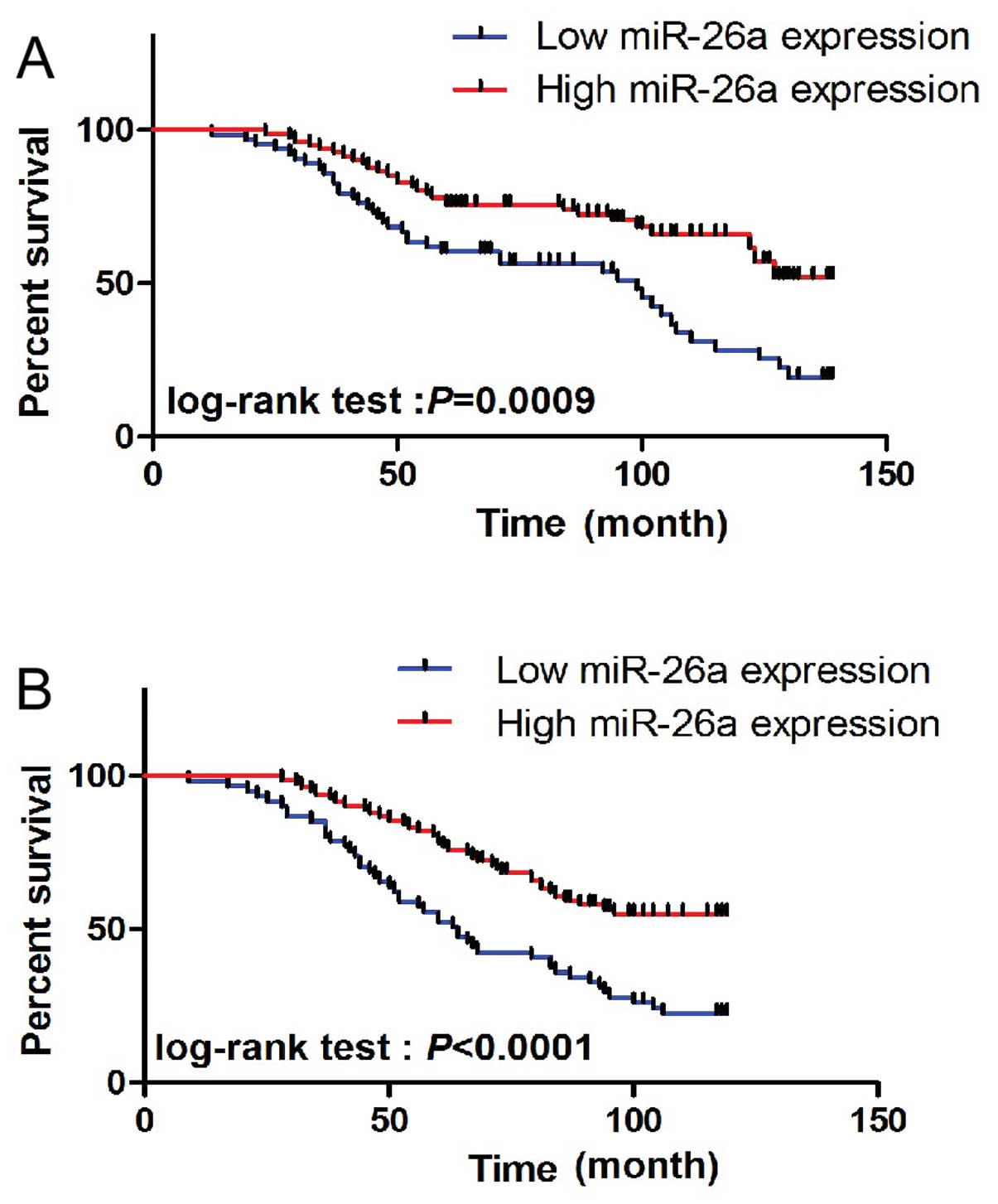
To determine the prognostic value of miR-26a
expression in osteosarcoma, we analyzed the relationship between
the miR-26a expression and clinical outcome. The relationship
between miR-26a expression and overall survival or disease-free
survival was investigated using Kaplan-Meier analysis and log-rank
test. A statistically significant difference in overall survival
and disease-free survival was found between the high miR-26a
expression group and the low miR-26a expression group (Fig. 2A and B; log-rank test, P=0.0009 and
P<0.0001, respectively). The patients with low miR-26a
expression tended to have a shorter overall and disease-free
survival time when compared to patients with high miR-26a
expression. In the multivariate analysis, we found that low miR-26a
expression was associated with a decreased overall and disease-free
survival. The adjusted risk ration (RR) was 5.724 (95% CI,
1.008–10.991; P=0.007) and 3.972 (95% CI, 1.191–9.871; P=0.014),
respectively, indicating that the expression of miR-26a may be a
prognostic factor for overall and disease-free survival independent
of these adjusted clinicopathologic characteristics (Table II).
 | Table IIMultivariate survival analysis of
overall survival and disease-free survival in the 144 patients with
osteosarcoma. |
Table II
Multivariate survival analysis of
overall survival and disease-free survival in the 144 patients with
osteosarcoma.
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|---|
| Variables | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| miR-26a
expression | 5.724 | 1.008–10.941 | 0.007a | 3.972 | 1.191–9.871 | 0.014a |
| Clinical stage | 2.541 | 1.157–4.214 | 0.031a | 1.915 | 1.219–5.412 | 0.041a |
| Status of
metastasis | 5.192 | 1.221–9.481 | 0.011a | 4.021 | 1.092–10.327 | 0.009a |
| Response to
chemotherapy | 2.198 | 1.871–6.517 | 0.029a | 2.109 | 0.983–5.284 | 0.037a |
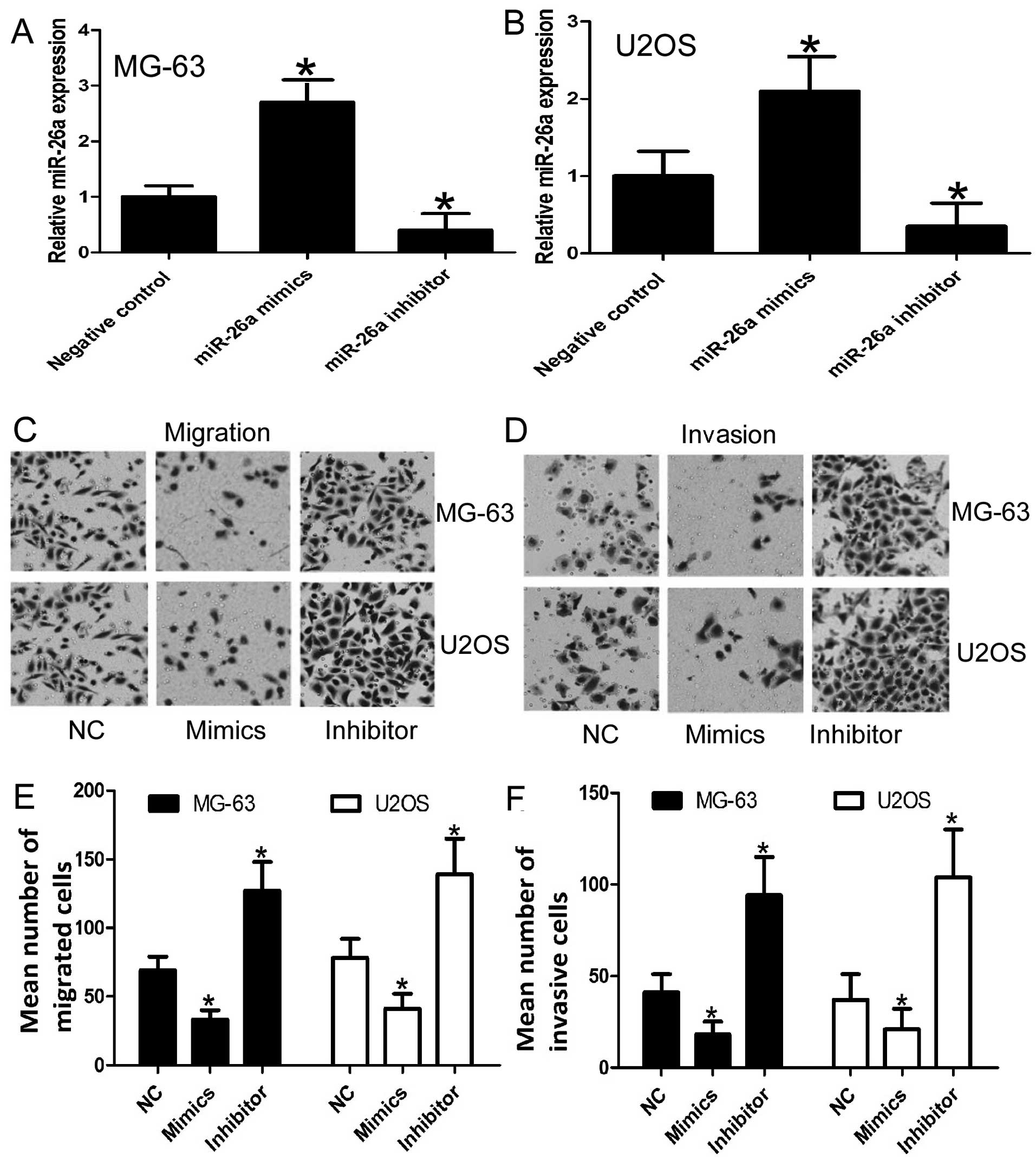
miR-26a reduces the migration and
invasion of osteosarcoma cells
As patients with recurrent metastatic osteosarcoma
present with a poor outcome and based on our previous results
indicating that miR-26a is significantly associated with
disease-free survival and prognosis, we aimed to ascertain whether
miR-26a affects the cell migration and invasion of osteosarcoma
cells. To confirm the effect of miR-26a on cell migration and
invasion, MG-63 and U2OS cells were transiently transfected with
miR-26a mimics or the miR-26a inhibitor, respectively. As expected,
transfection of miR-26a mimics or the miR-26a inhibitor resulted in
an increase and decrease, respectively, in miR-26a expression when
compared to the negative control (Fig.
3A and B). Moreover, the results of the migration and invasion
assays revealed that miR-26a restoration resulted in reduced
migration and invasion of MGG-63 and U2OS cells when compare to the
control, whereas, anti-miR-26a promoted cell migration and invasion
(Fig. 3C–F). These results indicate
that miR-26a functions as a tumor suppressor miRNA and contributes
to the inhibition of migration and invasion of osteosarcoma
cells.
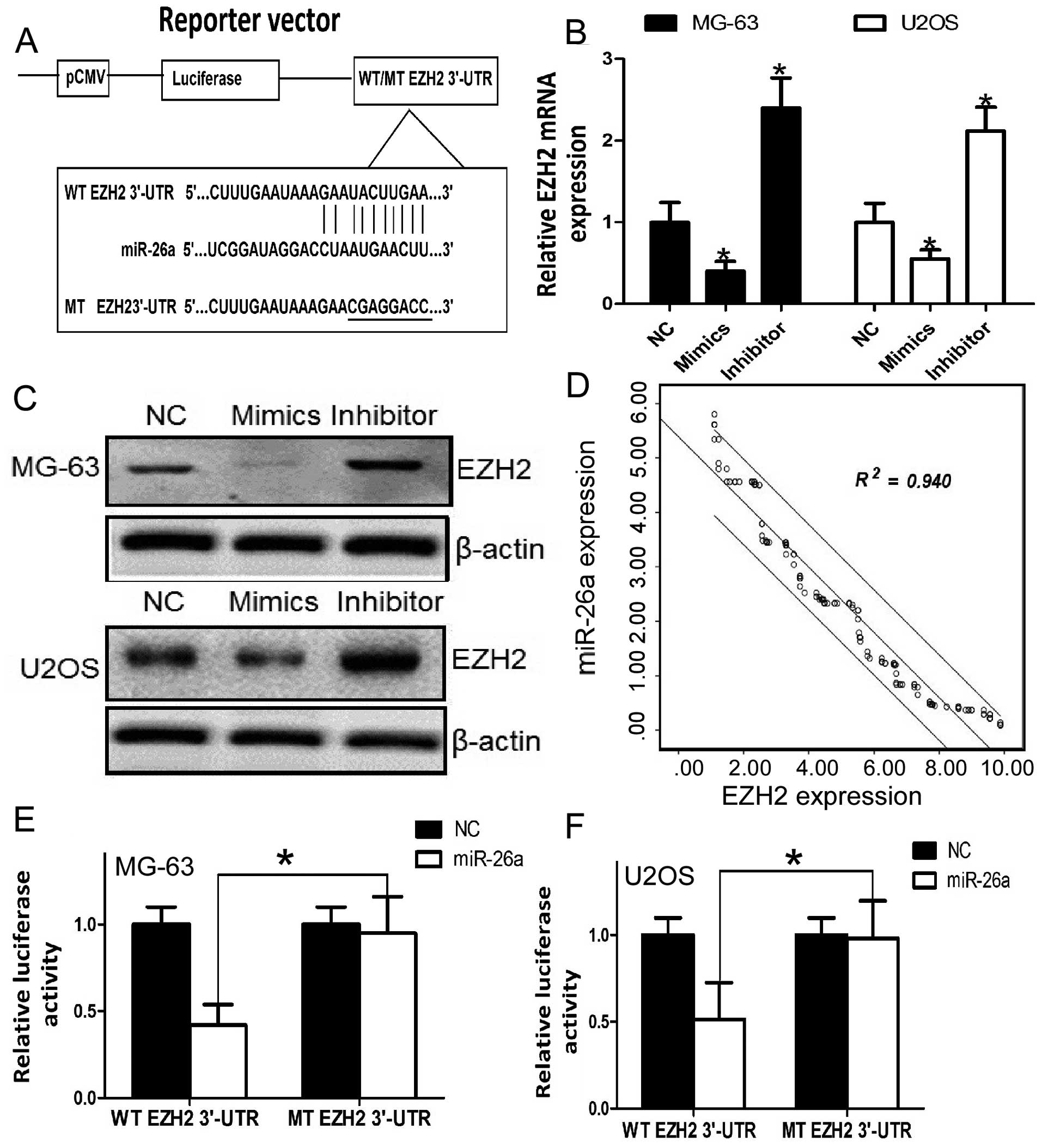
EZH2 is a direct target of miR-26a in
osteosarcoma cells
To elucidate the mechanism of inhibition involved in
the migration and invasion in human osteosarcoma by miR-26a, we
searched for candidate genes for miR-26a using publicly available
databases, including TargetScan, PicTar and miRanda. EZH2 was
selected for further experimental validation, since the
complementary sequence of miR-26a was identified in the 3′-UTR of
EZH2 mRNA by miRanda analysis (Fig.
4A). To further confirm EZH2 as a target gene for miR-26a,
qRT-PCR and western blot analysis were used to detect the
expression of EZH2 which was regulated by miR-26a in the MG-63 and
U2OS cells. The expression of EZH2 was significantly downregulated
after overexpression of miR-26a at the mRNA and protein levels in
the osteosarcoma cells (Fig. 4B and
C). Furthermore, we assessed the significance of the miR-26a
and EZH2 correlation in osteosarcoma tissues. We determined the
EZH2 mRNA and miR-26a expression in the same osteosarcoma specimens
by qRT-PCR. As shown in Fig. 4D, a
statistically significant inverse correlation was revealed by
Spearman’s correlation analysis between mRNA levels of miR-26a and
EZH2 (r=−0.969; P<0.001). Taken together, our results suggest
that miR-26a negatively regulates the expression of its target gene
EZH2.
We further performed a luciferase reporter assay to
verify whether miR-26a directly targets the 3′-UTR of EZH2 in
osteosarcoma cells. The target sequence of EZH2 3′-UTR (WT 3′-UTR)
or the mutant sequence (MT 3′-UTR) was cloned into a luciferase
reporter vector (Fig. 4A). MG-63
and U2OS cells were then transfected with the WT or MT 3′-UTR
vector and the miR-26a mimic. As shown in Fig. 4E, a significant decrease in
luciferase activity was noted between the EZH2 WT 3′-UTR group and
the negative control group (P<0.05). The repressive effect was
abrogated by point mutation in the core binding sites of the EZH2
3′-UTR. A similar trend was also found in the U2OS cells (Fig. 4F). These results indicate that
miR-26a exerts an inhibitory effect on EZH2 expression via
interaction with the 3′-UTR of EZH2 in osteosarcoma cells.
Discussion
Metastasis is a major concern in the clinical
treatment of osteosarcoma. The cure rate of osteosarcoma is ~65%
for localized osteosarcoma patients, whereas for patients
presenting with metastases at the time of diagnosis, the survival
rate is 25% (21,22). Several miRNAs have been reported to
modulate tumor metastasis (23)
including osteosarcoma (9).
However, research concerning the aberrant expression and function
of miRNAs in the progression and metastasis of osteosarcoma is
still in its infancy.
In the present study, we demonstrated that miR-26a
was weakly expressed in the osteosarcoma tissues when compared with
that in the in non-cancerous bone tissues. The decreased expression
of miR-26a in osteosarcoma tissues was also found to be
significantly associated with adverse clinicopathological features.
Moreover, we demonstrated that the miR-26a expression level was
also predictive of disease progression and cancer-specific death.
Our results showed that the patients with weak miR-26a expression
had a significantly higher risk of cancer progression,
cancer-specific death and shorter disease-free and overall survival
time. Multivariate analysis demonstrated that low expression of
miR-26a was a statistically significant risk factor affecting both
disease-free survival and overall survival in osteosarcoma
patients, which indicated that miR-26a expression may be an
independent predictor of osteosarcoma progression and prognosis.
The results provide initial evidence supporting miR-26a as a
predictor of poor prognosis in osteosarcoma carcinoma.
To reveal the role of miR-26a in osteosarcoma cells,
we assessed the effect of miR-26a on cell migration and invasion.
Our results showed that miR-26a inhibited cell migration and
invasion. Thus, miR-26a may be a novel tumor suppressor and may
play an important role in the regulation of tumor metastasis of
osteosarcoma. Next, we found the following evidence that miR-26a
inhibits tumor metastasis in part by suppressing EZH2. (i) miRanda
analysis showed that the 3′-UTR region of EZH2 mRNA contained the
complementary sequence of miR-26a (Fig.
4A). (ii) Upregulation of miR-26a significantly reduced EZH2
expression at the mRNA and protein levels in osteosarcoma cells
(Fig. 4B and C). (iii) The mRNA
levels of miR-26a were inversely correlated with EZH2 levels in
osteosarcoma tissues (Fig. 4D).
(iv) Overexpression of miR-26a decreased the luciferase reporter
activity of WT 3′-UTR but not MT 3′-UTR of EZH2 (Fig. 4E and F). These data support EZH2 as
a downstream mediator of miR-26a function in osteosarcoma.
Downregulated miR-26a was reported to play an
important role in the progression of tumor, and miR-26a functions
as a potential tumor suppressor in several distinct cancer types
(5,10,13,14,24),
but not including osteosarcoma. The underlying mechanisms
responsible for the low expression of miR-26a in tumors have been
investigated by several groups. Cellular homologue of avian
myelocytomatosis virus oncogene (MYC) was reported to regulate
miR-26a expression in thyroid anaplastic carcinoma and Burkitt’s
lymphoma (19,24,25).
In addition, miR-26a was shown to be a tumor suppressor and may
cause the inhibition of the precancerous molecule EZH2 in many
types of cancer (26,27). However the relationship between
miR-26a and EZH2 in the tumorigenesis of human osteosarcoma has not
been proven. Our study found that EZH2 is closely related to the
expression of miR26a which may inhibit the metastasis of
osteosarcoma cells. These results suggest that miR-26a regulates
the process of metastasis and invasion by downregulating EZH2.
EZH2 which belongs to the family of polycomb group
(PcG) proteins inhibits gene transcription through histone
methylation (19) and plays a
master regulatory role in many important cellular processes
(28). Mounting evidence has shown
that EZH2 is overexpressed in multiple cancer types and is
associated with tumor invasive growth and aggressive clinical
behavior, in prostate, renal, lung and breast cancer (29–32).
Moreover, EZH2 has been proven to enhance cell
metastasis and neoplastic transformation (29,33–35).
Consistent with our results, a previous study found that expression
of EZH2 at the protein level was upregulated in osteosarcoma
patient biopsy specimens when compare to normal bone, yet they also
found that knockdown of EZH2 did not prevent osteosarcoma growth
in vitro and in vivo (36). The discrepancies between this study
and our research may be due to the different osteosarcoma cell
lines selected by each study and the different focus on the
functions of EZH2. In the previous study, the authors wanted to
investigate the effect of EZH2 on osteosarcoma cell proliferation,
while in our study, we aimed to prove that EZH2 was a downstream
target of miR-26a. We did confirm that miR-26a affects the capacity
of metastasis of osteosarcoma cells, thus EZH2 should have an
effect on cell migration and invasion of osteosarcoma cells.
Metastasis is the most important factor affecting the prognosis and
progression of osteosarcoma. Thus, we suggest that overexpression
of EZH2 in osteosarcoma holds significant promise for the
advancement of cancer therapy, either in terms of improving
diagnosis or predicting prognosis. Considering that EZH2
overexpression has been proven to be associated with poor prognosis
in both metastatic breast and prostate cancer (29,33) as
well as in our research results, we believe that miR-26a plays an
important role in the process of metastasis of osteosarcoma, and
EZH2 is also involved in this process. The role of miR-26a and EZH2
in the proliferation of osteosarcoma warrants further detailed
experiments with a higher number of samples.
In conclusion, our results demonstrate that miR-26a
is downregulated in osteosarcoma, and reduced expression of miR-26a
more frequently occurs in osteosarcoma tissues with adverse
clinical stage and the presence of distant metastasis. Multivariate
survival analyses demonstrated that loss of miR-26a is an
independent prognostic factor for both disease-free and overall
survival in osteosarcoma. In addition, miR-26a inhibited the
invasion and migration of osteosarcoma cells, and miR-26a directly
inhibited EZH2 expression by targeting its 3′-UTR. Moreover, EZH2
was upregulated and inversely correlated with miR-26a in the
osteosarcoma tissue samples. For the first time, the current data
offer convincing evidence that the downregulation of miR-26a may be
associated with tumor aggressiveness and tumor metastasis of
osteosarcoma, and that miR-26a is an independent prognostic
predictor for osteosarcoma patients. Our data suggest an important
role for miR-26a in the molecular etiology of human osteosarcoma
and implicate the potential application of miR-26a in the therapy
of osteosarcoma.
Acknowledgements
The authors thank the local doctors and the patients
who participated in the present study.
Abbreviations:
|
miR-26a
|
miRNA-26a
|
|
EZH2
|
enhancer of zeste homolog 2
|
|
PcG
|
polycomb group
|
|
WT
|
wild-type
|
|
MT
|
mutant
|
|
3′-UTR
|
3′-untranslated region
|
|
qRT-PCR
|
quantitative real-time polymerase
chain reaction
|
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Teicher BA: Searching for molecular
targets in sarcoma. Biochem Pharmacol. 84:1–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gorlick R: Current concepts on the
molecular biology of osteosarcoma. Cancer Treat Res. 152:467–478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang M, Lin L, Cai H, Tang J and Zhou Z:
MicroRNA-145 downregulation associates with advanced tumor
progression and poor prognosis in patients suffering osteosarcoma.
Onco Targets Ther. 6:833–838. 2013.PubMed/NCBI
|
|
8
|
Shen L, Chen XD and Zhang YH: MicroRNA-128
promotes proliferation in osteosarcoma cells by downregulating
PTEN. Tumour Biol. Oct 15–2013.(Epub ahead of print).
|
|
9
|
Wang Y, Zhao W and Fu Q: miR-335
suppresses migration and invasion by targeting ROCK1 in
osteosarcoma cells. Mol Cell Biochem. 384:105–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kota J, Chivukula RR, O’Donnell KA, et al:
Therapeutic microRNA delivery suppresses tumorigenesis in a murine
liver cancer model. Cell. 137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji J, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ciarapica R, Russo G, Verginelli F, et al:
Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma.
Cell Cycle. 8:172–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Liu XX, He JR, et al:
Pathologically decreased miR-26a antagonizes apoptosis and
facilitates carcinogenesis by targeting MTDH and EZH2 in breast
cancer. Carcinogenesis. 32:2–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cabrera R and Szabo G: Another armed
CD4+ T cell ready to battle hepatocellular carcinoma.
Hepatology. 58:1–3. 2013.
|
|
15
|
Liu B, Wu X, Liu B, et al: MiR-26a
enhances metastasis potential of lung cancer cells via AKT pathway
by targeting PTEN. Biochim Biophys Acta. 1822:1692–1704. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huse JT, Brennan C, Hambardzumyan D, et
al: The PTEN-regulating microRNA miR-26a is amplified in high-grade
glioma and facilitates gliomagenesis in vivo. Genes Dev.
23:1327–1337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang J, Han C and Wu T: MicroRNA-26a
promotes cholangiocarcinoma growth by activating β-catenin.
Gastroenterology. 143:246–256. 2012.PubMed/NCBI
|
|
18
|
Lu J, He ML, Wang L, et al: MiR-26a
inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma
through repression of EZH2. Cancer Res. 71:225–233. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sander S, Bullinger L, Klapproth K, et al:
MYC stimulates EZH2 expression by repression of its negative
regulator miR-26a. Blood. 112:4202–4212. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luzi E, Marini F, Sala SC, Tognarini I,
Galli G and Brandi ML: Osteogenic differentiation of human adipose
tissue-derived stem cells is modulated by the miR-26a targeting of
the SMAD1 transcription factor. J Bone Miner Res. 23:287–295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gorlick R, Anderson P, Andrulis I, et al:
Biology of childhood osteogenic sarcoma and potential targets for
therapeutic development: meeting summary. Clin Cancer Res.
9:5442–5453. 2003.PubMed/NCBI
|
|
22
|
Wittig JC, Bickels J, Priebat D, et al:
Osteosarcoma: a multidisciplinary approach to diagnosis and
treatment. Am Fam Physician. 65:1123–1132. 2002.PubMed/NCBI
|
|
23
|
Sreekumar R, Sayan BS, Mirnezami AH and
Sayan AE: MicroRNA control of invasion and metastasis pathways.
Front Genet. 2:582011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang TC, Yu D, Lee YS, et al: Widespread
microRNA repression by Myc contributes to tumorigenesis. Nat Genet.
40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dews M, Homayouni A, Yu D, et al:
Augmentation of tumor angiogenesis by a Myc-activated microRNA
cluster. Nat Genet. 38:1060–1065. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu H, Yao Y, Smith LP and Nair V:
MicroRNA-26a-mediated regulation of interleukin-2 expression in
transformed avian lymphocyte lines. Cancer Cell Int. 10:152010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dang X, Ma A, Yang L, et al: MicroRNA-26a
regulates tumorigenic properties of EZH2 in human lung
carcinoma cells. Cancer Genet. 205:113–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sparmann A and van Lohuizen M: Polycomb
silencers control cell fate, development and cancer. Nat Rev
Cancer. 6:846–856. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kleer CG, Cao Q, Varambally S, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Raaphorst FM, Meijer CJ, Fieret E, et al:
Poorly differentiated breast carcinoma is associated with increased
expression of the human polycomb group EZH2 gene. Neoplasia.
5:481–488. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu C, Hou Z, Zhan P, et al: EZH2 regulates
cancer cell migration through repressing TIMP-3 in non-small cell
lung cancer. Med Oncol. 30:7132013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wagener N, Macher-Goeppinger S, Pritsch M,
et al: Enhancer of zeste homolog 2 (EZH2) expression is an
independent prognostic factor in renal cell carcinoma. BMC Cancer.
10:5242010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Varambally S, Dhanasekaran SM, Zhou M, et
al: The polycomb group protein EZH2 is involved in progression of
prostate cancer. Nature. 419:624–629. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Richter GH, Plehm S, Fasan A, et al: EZH2
is a mediator of EWS/FLI1 driven tumor growth and metastasis
blocking endothelial and neuro-ectodermal differentiation. Proc
Natl Acad Sci USA. 106:5324–5329. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Croonquist PA and Van Ness B: The polycomb
group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene
that influences myeloma cell growth and the mutant ras phenotype.
Oncogene. 24:6269–6280. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sasaki H, Setoguchi T, Matsunoshita Y, Gao
H, Hirotsu M and Komiya S: The knock-down of overexpressed EZH2 and
BMI-1 does not prevent osteosarcoma growth. Oncol Rep. 23:677–684.
2010.PubMed/NCBI
|