Introduction
Hepatocellular carcinoma (HCC) is one of the most
prevalent and malignant cancers in the world (1). Over 600,000 new HCC cases are
diagnosed annually (2). Despite the
improvements in diagnosis and treatment, the prognosis of patients
with HCC remains poor. Therefore, the exploration of promising
therapies and prognostic factors for HCC is of great clinical
significance.
Autophagy is an evolutionarily conserved cellular
pathway which degrades and recycles cytoplasmic components via the
lysosomal system (3,4). Deregulated autophagy is related to
several physiological defects, including liver injury, muscular
disorder, neurodegeneration, pathogen infections and cancer
(5,6). The relationship between autophagy and
cancer development has been studied in various types of cancer
(7–10). Autophagy-related genes are reported
to be cancer repressor genes (11–14).
Specifically, in HCC, it was reported that the expression of
ATG5, ATG7 and BECN1, and the autophagic
activity was decreased in HCC cell lines (15). The expression of BECN1 was
decreased in HCC tissues compared to the adjacent liver tissues,
and it was shown to be a prognostic factor in Bcl-xL+
patients (15). However, the
expression of other autophagy-related genes in HCC and their
correlation with HCC development remain largely unknown.
GABARAPL1 (also known as GEC1 or
ATG8L) was first identified as an early estrogen-induced
gene in quiescent guinea-pig endometrial glandular epithelial cells
(16,17). Previous studies showed that
GABARAPL1 is one of the six human Atg8 family proteins which locate
in autophagic vesicles after post-translational modification. They
mediate the cargo recognition and the autophagosome formation
(18,19). Recently, GABARAPL1 was
reported to be downregulated in breast adenocarcinoma and the
expression of GABARAPL1 is associated with the risk of
metastasis, specifically for lymph node-positive patients (20).
In the present study, we examined the mRNA
expression of autophagy-related genes in the Oncomine database and
dissected tissue samples from HCC patients. We found that both the
mRNA and protein expression of the GABARAPL1 gene was
decreased in HCC tissues. Overexpression or knockdown of
GABARAPL1 in HCC cell lines affected their growth rates. In
addition, we found a significant association between low
GABARAPL1 expression and poor prognosis in HCC patients.
Materials and methods
Patients and tissue specimens
Seventy-three pairs of HCC tissues and adjacent
liver tissues were collected from patients undergoing resection
from 2006 to 2009 at the Liver Cancer Institute, Zhongshan
Hospital, Fudan University, Shanghai. Tumor specimens were obtained
from the areas of the tumor, necrotic tissues were avoided. The
specimens were then snap frozen in liquid nitrogen and stored at
−80°C. Patients were monitored after surgery until March 2010.
Overall survival was defined as the interval between surgery and
mortality or the last observation. The histological grade of tumor
differentiation was determined according to the classification
proposed by Edmondson and Steine, as described by Wittekind
(21). TNM stage was determined
according to the 6th edition of Tumor-node-metastasis
classification of the International Union against Cancer. Ethics
approval for the present study was obtained from the Research
Ethics Committee of Zhongshan Hospital, and informed consent was
obtained from each patient.
Oncomine data analysis
Oncomine (http://www.oncomine.com) is an integrated cancer
microarray database which contains unified bio-informatics
resources from 715 datasets (version 4.4.4.3 after Q2 update 2013)
(22). We compared the mRNA
expression of autophagy-related genes from liver cancer datasets
which contain data from both HCC tissues and normal liver tissues.
Four datasets were included in our study, Chen et al
(23), Roessler et al
(24), Wurmbach et al
(25) and Mas et al
(26). The differentiated
expression for each gene between HCC tissues and normal liver
tissues was analyzed and their fold-change values and statistical
significance determined by P-value were collected.
RNA extraction and quantitative real-time
PCR (qRT-PCR)
Total RNA was extracted from tissues using TRIzol
reagent (Invitrogen). Two micrograms of total RNA were applied for
reverse transcription using oligo(dT) primer and reverse
transcriptase (Invitrogen). qRT-PCR was performed with SYBR-Green
Supermix kit (Takara) and LightCycler® 480 system
(Roche). The primer for each gene was designed with Beacon Designer
(Bio-Rad). The amplification conditions for each gene were
optimized by using melting curve analysis and gel electrophoresis.
Relative gene expression was calculated using the formula
2−ΔCt and GAPDH was used as internal gene for
normalization. ΔCt (critical threshold) = Ct of genes of interest -
Ct of GAPDH. Relative gene expression between HCC tissues
and adjacent liver tissues was calculated using the
2−ΔΔCt as previously described (27).
Western blot analysis
Protein samples were subjected to 12% SDS-PAGE
followed by standard western blotting protocols. The anti-GABARAPL1
antibody was purchased from Proteintech. Anti-myc antibody and
anti-β-actin antibody were purchased from Sigma-Aldrich.
Stable cell line and siRNA knockdown
The GABARAPL1 gene was cloned from HeLa cell
cDNA. The GABARAPL1-G116A point mutation was prepared using a
mutated primer. Then, GABARAPL1 and the G116A mutant were
subcloned into a pcDNA3.1 vector. Stable cell lines which
overexpress GABARAPL1 or the G116A mutant protein and control cell
lines were prepared as previously described (28).
The siRNA duplexes were purchased from Shanghai
GeneChem Co., Ltd. (Shanghai, China). The siRNA sequences targeting
GABARAPL1 were 1, 5′-GGACCAUCCCUUUGA GUAUUU-3′ and 2,
5′-GAAAAGAUCCGGAAGAAAUUU-3′. Control siRNA was
5′-UAAGGCUAUGAAGAGAUACUU-3′. siRNAs were transfected into the cells
using oligofectamine (Invitrogen) according to the manufacturer’s
instructions.
Cell proliferation assay
Control cells and cells stably expressing GABARAPL1
or the G116A mutant protein were plated in 96-well plates at a
density of 1,500 cells/well in the Dulbecco’s modified Eagle’s
medium with 10% fetal bovine serum (FBS) and 800 μg/ml G418
(Invitrogen). At the indicated time, cell proliferation was
determined by using the Cell Counting Kit-8 assay according to the
manufacturer’s instructions. For the cell proliferation assay after
siRNA treatment, cells were first transfected with siRNA duplexes.
After 24 h, cells were plated in 96-well plates as day 0. At days 2
and 6, cells were again subjected to siRNA treatment in 96-well
plates. Cell proliferation was determined by the Cell Counting
Kit-8 assay at the indicated time.
Statistical analysis
Statistical analyses were performed using SPSS 12.0
for Windows. The P-values and gene fold-change values from Oncomine
were previously described (22,29).
The χ2 test, Fisher’s exact probability, and Student’s
t-test were used for comparison between groups. Cumulative survival
time was calculated by the Kaplan-Meier method and analyzed by the
log-rank test.
Results
Oncomine datasets suggest GABARAPL1 is
downregulated in HCC tissues
To explore the potential differentially expressed
autophagy-related genes in HCC, we firstly analyzed Oncomine, the
integrated cancer microarray database. Four liver cancer data sets
(23–26), which contain both normal and HCC
tissue data, were selected to analyze the mRNA expression of 29
autophagy-related genes. These genes function in different steps of
the autophagy process, including protein kinase initiation step,
vesicle nucleation, vesicle elongation and autophagosome assembly.
We compared their mRNA expression between HCC tissues and normal
liver tissues, and collected their fold-change values and P-value.
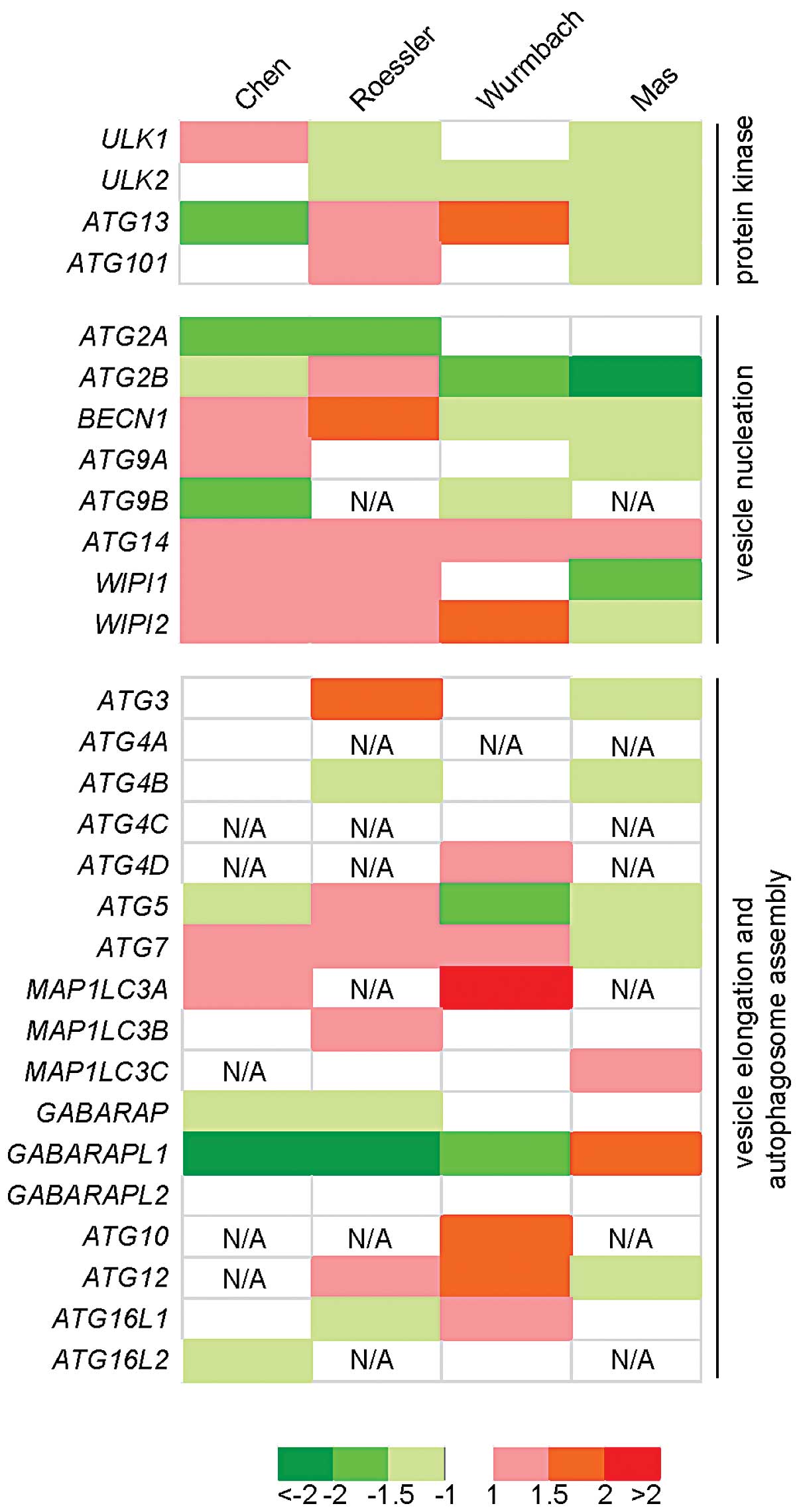
As shown in Fig. 1, ATG14,
WIPI2 and ATG7 were upregulated in HCC tissues in 3
of these 4 data sets. ULK2, ATG2B and
GABARAPL1 were downregulated in HCC tissues also in 3 of
these 4 data sets. Among these genes, the fold-change values of
GABARAPL1 were −2.65 and −2.646 in Chen_Liver and
Roessler_Liver data sets, respectively, suggesting that
GABARAPL1 was greatly downregulated in HCC tissues compared
to the normal liver tissues.
GABARAPL1 is downregulated in HCC
tissues
To corroborate the gene expression patterns
experimentally, we randomly selected 24 pairs of HCC tissues and
adjacent liver tissues and detected the mRNA expression of 21
autophagy-related genes by qRT-PCR. We found that GABARAP,
another member of the human Atg8 family, was slightly but
significantly decreased in HCC tissues (averagely 20% decrease in
HCC tissues as compared to adjacent liver tissues, P=0.003), and
the expression of GABARAPL1 was largely decreased in HCC
tissues (78.2% decrease in HCC tissues as compared to adjacent
liver tissues P<0.0001).
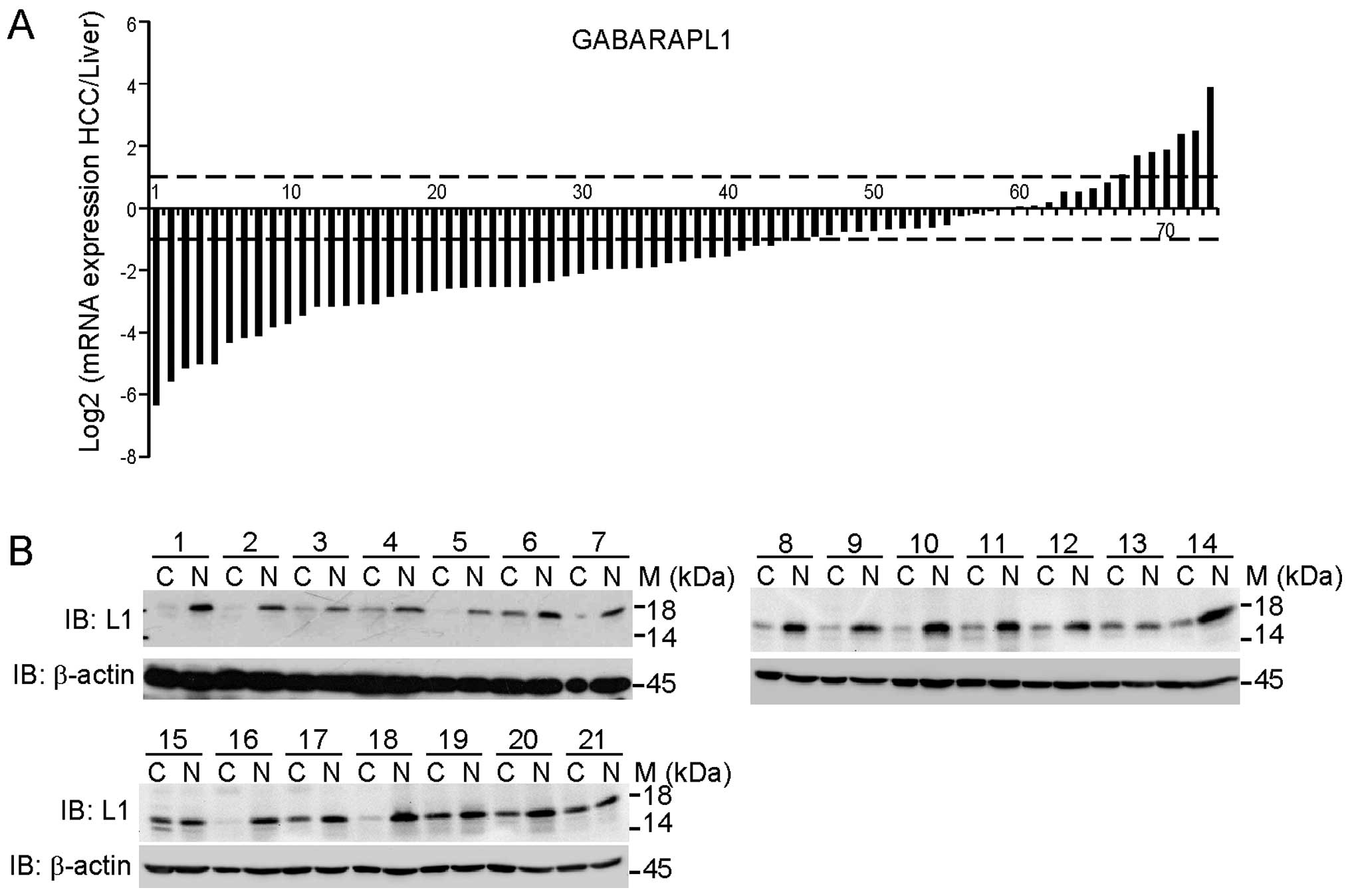
To confirm this result in a larger sample size, we
detected the GABARAPL1 transcript expression in 73 pairs of HCC
tissues and adjacent liver tissues. Fig. 2A presents the log2 transformed
fold-change of GABARAPL1 mRNA expression ratio of
tumor/adjacent liver tissue. Forty-five of 73 cases (61.6%) showed
significant reduction of GABARAPL1 expression in HCC tissues
(log2 transformed fold-change ≤ −1); 21/73 cases (28.8%) showed no
alteration (−1<log2 transformed fold-change <1); and only
7/73 cases (9.6%) showed upregulation (log2 transformed fold-change
≥1). The average expression of GABARAPL1 mRNA in HCC was
33.7% of adjacent liver tissues. To detect the protein expression
of GABARAPL1, we randomly selected 21 pairs of samples and carried
out immunoblotting analysis. As shown in Fig. 2B, the protein expression of
GABARAPL1 was largely decreased in HCC tissues as compared to
adjacent liver tissues, consistent with the mRNA expression
pattern.
The expression of GABARAPL1 in HCC cell
lines affects cellular growth rates
Based on the pronounced down-regulated expression of
GABARAPL1 in HCC tissues, we next investigated whether the
expression of GABARAPL1 affects cell growth. We first
detected the mRNA and protein expression of GABARAPL1 in 9
HCC cell lines. We found that Hep3B, SMMC-7721, Focus and PLC/PRF/5
cell lines have low GABARAPL1 expression, while HepG2, Huh7,
QGY-7703, YY-8103 and SK-Hep1 have moderate to high expression of
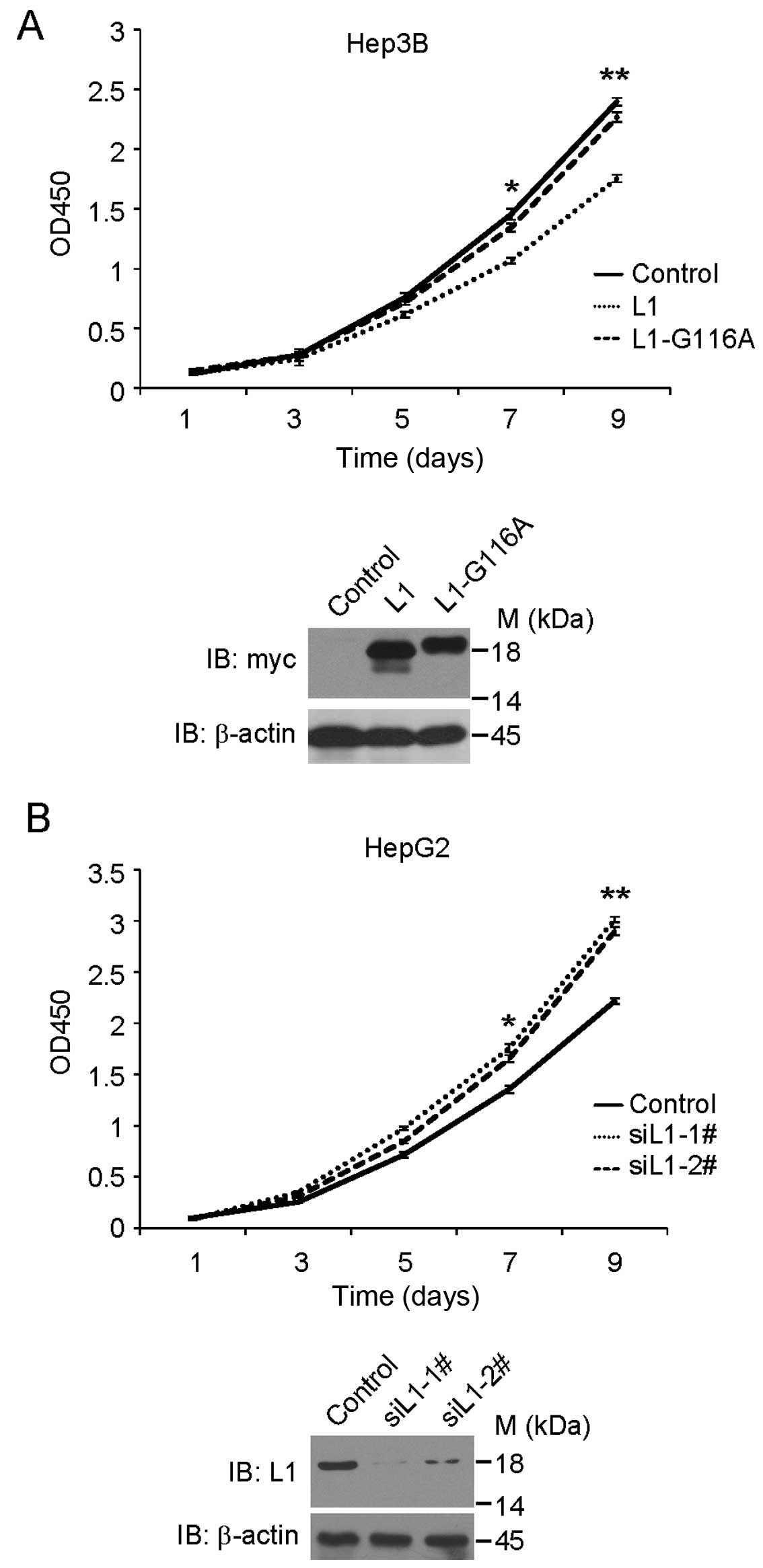
GABARAPL1 (data not shown). We then established stable Hep3B cell
lines that expressed pcDNA3.1 empty vector, pcDNA3.1-myc-GABARAPL1,
or pcDNA3.1-myc-GABARAPL1-G116A mutant. For each vector, we chose
three single clones which expressed indicated protein and mixed
them as a pool. As shown in Fig.
3A, cell growth assay suggested that overexpression of
GABARAPL1 inhibited the cellular growth rate in Hep3B cells.
However, overexpression of GABARAPL1-G116A mutant, which cannot be
functionally located to the autophagosomes (18), did not inhibit the cellular growth
rate in Hep3B cells, suggesting that GABARAPL1 inhibits cellular
growth via the autophagy-related pathway. Similar results were
observed in SMMC-7721 cells (data not shown).
We chose HepG2 cells to carry out siRNA treatment
with control or GABARAPL1 siRNAs. Two different siRNAs were
designed to knock down GABARAPL1 expression. As shown in
Fig. 3B, after siRNA treatment,
cells treated with GABARAPL1 siRNA grew faster than those
treated with control siRNA. Similar data were observed in Sk-Hep1
cells (data not shown). These results suggest that GABARAPL1
expression may affect the cellular growth rate in HCC cell
lines.
Low expression of GABARAPL1 in HCC
tissues is associated with poor outcome of HCC patients
To explore the clinicopathological correlation of
GABARAPL1 downregulation in HCC, we analyzed the
GABARAPL1 mRNA expression with various clinical parameters
in 73 HCC patients. The patients were divided into low or high
expression groups according to their GABARAPL1 transcript
expression levels. As shown in Table
I, we found that GABARAPL1 was significantly associated
with pathological differentiation (P=0.018) and tumor encapsulation
(P=0.047). No correlation was found with other variables. These
correlations suggest that the downregulation of GABARAPL1
may serve as a prognosis indicator of HCC.
 | Table ICorrelation between GABARAPL1
mRNA expression and clinicopathological variables. |
Table I
Correlation between GABARAPL1
mRNA expression and clinicopathological variables.
| GABARAPL1
mRNA expression |
|---|
|
|
|---|
| Variables | Low | High | P-value |
|---|
| Age (years) |
| ≤57 | 18 | 20 | 0.555 |
| >57 | 19 | 16 | |
| Gender |
| Female | 7 | 8 | 0.727 |
| Male | 30 | 28 | |
| Hepatitis
history |
| No | 17 | 14 | 0.542 |
| Yes | 20 | 22 | |
| AFP (ng/ml) |
| ≤20 | 19 | 17 | 0.724 |
| >20 | 18 | 19 | |
| Liver
cirrhosis |
| No | 33 | 31 | 0.736a |
| Yes | 4 | 5 | |
| Tumor size
(cm) |
| ≤5 | 11 | 16 | 0.193 |
| >5 | 26 | 20 | |
| Tumor
multiplicity |
| Single | 31 | 28 | 0.515 |
| Multiple | 6 | 8 | |
|
Differentiation |
| I+II | 19 | 28 | 0.018 |
| III+IV | 18 | 8 | |
| TNM stage |
| I | 3 | 1 | 0.615a |
| II+III | 34 | 35 | |
| Tumor capsule |
| Present | 13 | 21 | 0.047 |
| Absent | 24 | 15 | |
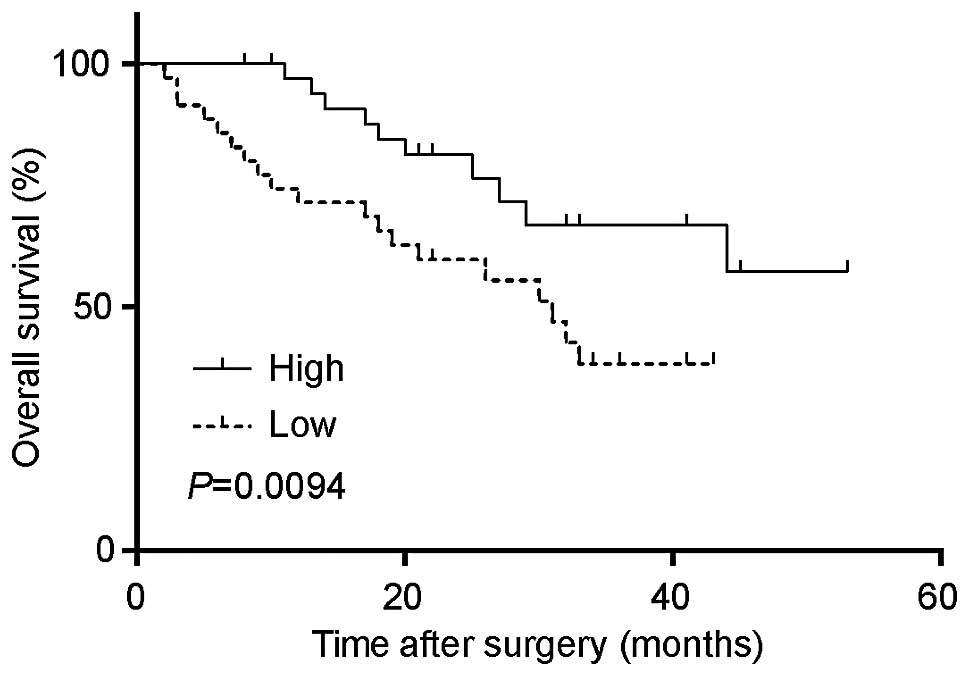
Furthermore, we analyzed the survival time with the
GABARAPL1 expression. As shown in Fig. 4, the low expression of
GABARAPL1 was significantly associated with a poorer
outcome, while patients with a higher expression of
GABARAPL1 were more likely to have a longer overall survival
time (P=0.0094).
Discussion
Previous studies revealed that defective autophagy
was related to poor prognosis in various cancers, including HCC
(15,30), breast cancer (31) and bladder cancer (32). In the present study, we combined
both meta-analysis and experimental data to search differentially
regulated autophagy-related genes in HCC and liver tissues. We
found that the expression of GABARAPL1 was significantly
downregulated in HCC tissues, both in mRNA and protein expression
levels. Also, GABARAPL1 might be a potential biomarker for
HCC patients, as the low expression of GABARAPL1 in HCC
tissue correlated the poor survival of HCC patients. However, the
sample size of the present study was relatively small (n=73), and
these patients were mainly from South and East China. Therefore, in
a future study, a lager sample size should be used to evaluate the
extent of GABARAPL1 as a predictive biomarker of patient
survival.
In addition to GABARAPL1, GABARAP was
also found to be downregulated from both meta-analysis and
experimental data. These two proteins are members of the GABARAP
sub-family and their functions in autophagy were reported to be
distinct from those of LC3 sub-family proteins (including
MAP1LC3A, MAP1LC3B and MAP1LC3C), although
they have high sequence similarity. Specifically, in the initiation
step of autophagy, the GABARAP sub-family proteins are much
preferred in the ULK complex assembly (33). LC3 sub-family proteins are involved
in the following elongation of the phagophore membrane. Then, at
the late stage of autophagosome formation, the GABARAP sub-family
proteins are essential for autophagosome completion (34). Thus, downregulation of these two
members may decrease the cellular autophagic activity, and then
affect the tumorigenesis in HCC development. The same regulation
defect was reported in breast cancer (31) and neuroblastoma (35) for GABARAP, and in breast
cancer (20) for
GABARAPL1.
Another notable finding was that the expression of
GABARAPL1 was related to the cell growth rate in HCC cell
lines. This function of GABARAPL1 was dependent on its role
in the autophagy process. Recent research identified that GABARAPL1
may negatively regulate the Wnt signaling pathway by mediating Dvl2
degradation through autophagy (36). Thus, it is possible that
downregulation of GABARAPL1 may inhibit the selective
autophagy mediated by GABARAPL1, thereby affecting the cell growth
or tumorigenesis process. The detailed mechanism of GABARAPL1 in
cell growth control requires further investigation.
Collectively, our data suggest that
GABARAPL1, one of the proteins functioning during the
autophagosome formation, is downregulated in HCC and its expression
is associated with the survival of HCC patients and can be used as
a prognostic factor in HCC.
Acknowledgements
We thank Dr Jie Zuo, Dr Haijie Ma and Dr He-Xi Ge
Sai-Yin (Fudan University) for technical assistance. The authors
are grateful to Matthew D. Pauly (University of Michigan Medical
School) for proofreading. The present study was supported by the
National Natural Science Foundation of China for Creative Research
Groups (30024001 to L.Y.), the National Key Sci-Tech Special
Project of China (2013ZX10002010 and 2008ZX10002-020 to L.Y.), the
Project of the Shanghai Municipal Science and Technology Commission
(03dz14086 to L.Y.), and the National Natural Science Foundation of
China (31071193 to L.Y.).
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
qRT-PCR
|
quantitative real-time PCR
|
References
|
1
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
3
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Z and Klionsky DJ: Eaten alive: a
history of macroautophagy. Nat Cell Biol. 12:814–822. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shintani T and Klionsky DJ: Autophagy in
health and disease: a double-edged sword. Science. 306:990–995.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Zhang J, Chen X, et al: Molecular
machinery of autophagy and its implication in cancer. Am J Med Sci.
343:155–161. 2011. View Article : Google Scholar
|
|
7
|
Rautou PE, Mansouri A, Lebrec D, Durand F,
Valla D and Moreau R: Autophagy in liver diseases. J Hepatol.
53:1123–1134. 2010. View Article : Google Scholar
|
|
8
|
Pandey S and Chandravati: Autophagy in
cervical cancer: an emerging therapeutic target. Asian Pac J Cancer
Prev. 13:4867–4871. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaza N, Kohli L and Roth KA: Autophagy in
brain tumors: a new target for therapeutic intervention. Brain
Pathol. 22:89–98. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cook KL, Shajahan AN and Clarke R:
Autophagy and endocrine resistance in breast cancer. Expert Rev
Anticancer Ther. 11:1283–1294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang XH, Jackson S, Seaman M, et al:
Induction of autophagy and inhibition of tumorigenesis by beclin 1.
Nature. 402:672–676. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qu X, Yu J, Bhagat G, et al: Promotion of
tumorigenesis by heterozygous disruption of the beclin 1 autophagy
gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Takahashi Y, Coppola D, Matsushita N, et
al: Bif-1 interacts with Beclin 1 through UVRAG and regulates
autophagy and tumorigenesis. Nat Cell Biol. 9:1142–1151. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding ZB, Shi YH, Zhou J, et al:
Association of autophagy defect with a malignant phenotype and poor
prognosis of hepatocellular carcinoma. Cancer Res. 68:9167–9175.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pellerin I, Vuillermoz C, Jouvenot M,
Ordener C, Royez M and Adessi GL: Identification and
characterization of an early estrogen-regulated RNA in cultured
guinea-pig endometrial cells. Mol Cell Endocrinol. 90:R17–R21.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jouvenot M, Pellerin I, Alkhalaf M,
Marechal G, Royez M and Adessi GL: Effects of 17 beta-estradiol and
growth factors on c-fos gene expression in endometrial epithelial
cells in primary culture. Mol Cell Endocrinol. 72:149–157. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chakrama FZ, Seguin-Py S, Le Grand JN, et
al: GABARAPL1 (GEC1) associates with autophagic vesicles.
Autophagy. 6:495–505. 2010. View Article : Google Scholar
|
|
19
|
Pankiv S, Clausen TH, Lamark T, et al:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–24145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Berthier A, Seguin S, Sasco AJ, et al:
High expression of gabarapl1 is associated with a better
outcome for patients with lymph node-positive breast cancer. Br J
Cancer. 102:1024–1031. 2010.
|
|
21
|
Wittekind C: Pitfalls in the
classification of liver tumors. Pathologe. 27:289–293. 2006.(In
German).
|
|
22
|
Rhodes DR, Yu J, Shanker K, et al:
ONCOMINE: a cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Cheung ST, So S, et al: Gene
expression patterns in human liver cancers. Mol Biol Cell.
13:1929–1939. 2002. View Article : Google Scholar PubMed/NCBI
|
|
24
|
Roessler S, Jia HL, Budhu A, et al: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wurmbach E, Chen YB, Khitrov G, et al:
Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mas VR, Maluf DG, Archer KJ, et al: Genes
involved in viral carcinogenesis and tumor initiation in hepatitis
C virus-induced hepatocellular carcinoma. Mol Med. 15:85–94.
2009.PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
28
|
Wu Y, Zuo J, Ji G, et al: Proapoptotic
function of integrin β3 in human hepatocellular
carcinoma cells. Clin Cancer Res. 15:60–69. 2009.
|
|
29
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, et al: Oncomine 3.0: genes, pathways, and networks in a
collection of 18,000 cancer gene expression profiles. Neoplasia.
9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Inoue D, Suzuki T, Mitsuishi Y, et al:
Accumulation of p62/SQSTM1 is associated with poor prognosis in
patients with lung adenocarcinoma. Cancer Sci. 103:760–766. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klebig C, Seitz S, Arnold W, et al:
Characterization of γ-aminobutyric acid type A receptor-associated
protein, a novel tumor suppressor, showing reduced expression in
breast cancer. Cancer Res. 65:394–400. 2005.
|
|
32
|
Liu GH, Zhong Q, Ye YL, et al: Expression
of beclin 1 in bladder cancer and its clinical significance. Int J
Biol Markers. 28:56–62. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Alemu EA, Lamark T, Torgersen KM, et al:
ATG8 family proteins act as scaffolds for assembly of the ULK
complex: sequence requirements for LC3-interacting region (LIR)
motifs. J Biol Chem. 287:39275–39290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weidberg H, Shvets E, Shpilka T, Shimron
F, Shinder V and Elazar Z: LC3 and GATE-16/GABARAP subfamilies are
both essential yet act differently in autophagosome biogenesis.
EMBO J. 29:1792–1802. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roberts SS, Mori M, Pattee P, et al:
GABAergic system gene expression predicts clinical outcome in
patients with neuroblastoma. J Clin Oncol. 22:4127–4134. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Y, Wang F, Han L, et al: GABARAPL1
negatively regulates Wnt/β-catenin signaling by mediating Dvl2
degradation through the autophagy pathway. Cell Physiol Biochem.
27:503–512. 2011.PubMed/NCBI
|