Introduction
Telomeres have many functions within cells, such as
maintaining genomic stability and protecting the chromosomal ends
from degradation (1,2). Inhibition of telomerase activity or
shortened telomeres may increase the incidence of cancer in aged
cells (3). Pin2/TRF1-interacting
protein X1 (PinX1) which consisting of seven axons is located at
chromosome 8p23 (4). Its coding
protein that contains 328 amino acids has 2 domains, one is G-patch
found in RNA-binding proteins or damage repair agents, the second
is telomerase inhibitory domain (TID) (4). In previous studies, it has been
observed that PinX1 suppresses telomerase activity by its direct
interaction with hTERT and hTR (5).
Depletion of PinX1 results in lagging chromosome in mitosis and
micronuclei in interphase (6).
PinX1 is considered as a tumor suppressor in many tumors due to its
low expression in hepatocellular carcinoma (7), prostate cancer (8), gastric carcinoma (9) and medulloblastomas (10).
Although much is known regarding the role of PinX1
in regulating telomerase function, less is known concerning the
role of PinX1 without G-patch in colorectal cancer cells. The aim
of the present study was to ascertain whether PinX1 without G-patch
suppresses the growth of colorectal cancer cells.
Materials and methods
Cell culture and synchronization
SW480 cells [American Type Culture Collection
(ATCC), Manassas, VA, USA] were maintained as subconfluent
monolayers in Dulbecco’s modified Eagle’s medium (DMEM)
(Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS)
(HyClone, Logan, UT, USA) and 100 U/ml penicillin plus 100 μg/ml
streptomycin (Invitrogen) at 37°C with 5% CO2. Cells
were synchronized at G1/S with 5 mM thymidine for 12–16
h and then washed with phosphate-buffered saline (PBS) three times
and cultured in thymidine-free medium for 10 h.
Plasmid construction and
transfection
The cDNA encoding PinX1 was cloned into a pEGFP-A1
vector (reserved by our laboratory) to generate the pEGFP-A1-PinX1
plasmid. The PCR product for PinX1 69–328 was cut with restriction
enzymes and then inserted into the pEGFP-A1. Transfection of
plasmids into SW480 cells was performed using Lipofectamine 2000
(Invitrogen) according to the manufacturer’s instructions.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde,
permeabilized with 0.5% Triton X, then blocked with 2% BSA in PBS.
The primary PinX1 antibody (sc-376394, Epitope: 8–29; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) was used at a 1:100
concentration, and the secondary antibody was Alexa
Fluor® 594 rat anti-mouse IgG (H+L) antibody at a
concentration of 1:500. Cells were examined and photographed using
an Olympus CX71 fluorescence microscope (Olympus, Tokyo,
Japan).
MTT assay
Viability of the cells was determined using
3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma-Aldrich, Carlsbad, CA, USA). Cells were plated in
96-well plates (1,500 cells/well) and incubated under normal
culture conditions. After 48 h, the cells described above were
treated with 0.5 mg/ml of MTT for 4 h and lysed with dimethyl
sulfoxide (DMSO). Absorbance rates were measured at 550–560 nm
using a microplate reader (Bio-Rad, Hercules, CA, USA).
Annexin V-FITC and propidium iodide (PI)
double staining
Annexin V-FITC/PI double staining assays were
performed to detect apoptosis. Following the manufacturer’s
instructions (Apoptosis Detection kit; KeyGen, Nanjing, China),
cells were washed and resuspended in binding buffer prior to the
addition of FITC-labeled Annexin V and PI for 10 min. Suspensions
were immediately analyzed by flow cytometry using a FACSCalibur
machine (BD Biosciences, Baltimore, MD, USA).
Cell cycle study
Cells were suspended in PBS and then fixed with 70%
ethanol at 4°C. After centrifugation, the supernatants were
discarded, and cellular DNA was stained in a solution containing 10
μM of propidium iodide (KeyGen). Then the mixture was analyzed by a
FACSCalibur flow cytometer (Becton-Dickinson).
SA-β-gal analysis
For SA-β-gal staining, cells were washed twice in
PBS, fixed for 3–5 min at room temperature in 3% formaldehyde and
washed again with PBS. Then cells were incubated overnight at 37°C
without CO2 in a freshly prepared SA-β-gal-staining
solution as described (11).
TRAP telomerase activity assay
Cells were lysed in lysis buffer (10 mM Tris-HCl pH,
7.5, 1 mM MgCl2, 1 mM EGTA, 0.1 mM PMSF, 5 mM
2-mercaptoethanol, 0.5 % CHAPS, 10% glycerol) on ice for 30 min and
then centrifuged at high speed for 30 min, the suspension
containing telomerase was used for TRAP assay. Telomerase activity
was measured with the TRAPeze telomerase detection kit (Chemicon,
Temecula, CA, USA) according to the manufacturer’s instructions
(12).
Subjects
A total of 52 patients with colorectal cancer were
obtained from the Department of Colorectal Surgery, Liaoning Cancer
Hospital and Institute (January 2008 to December 2012). All
patients underwent standard laboratory tests (cytology and
histology). None of the patients underwent radiotherapy or
chemotherapy before the operation. Informed consent was provided by
all patients according to the Helsinki Declaration.
Extraction of total RNA and real-time
PCR
RNA was extracted from tissues using TRIzol solution
(Invitrogen) according to the protocols recommended by the
manufacturer. Total RNA concentration and quantity were assessed by
absorbency at 260 nm using a DNA/Protein Analyzer (DU 530; Beckman,
Fullerton, CA, USA). Real-time PCR was performed on a Rotor-Gene
RG-6000A apparatus (Corbett Research, Cambridge, UK) for 40 cycles
of 94°C for 10 sec, 60°C for 10 sec and 72°C for 15 sec. Reactions
(20 μl) included 2 μl of cDNA, target-specific primers, and the
QuantiTect SYBR-Green PCR kit (Qiagen, Valencia, CA, USA). The
temperature range for analysis of the melting curves was 55–99°C
over 30 sec. The primer sequences are listed in Table I. Relative quantitation was
calculated using the ΔΔCt method.
 | Table IThe primers used in the real-time PCR
analyses. |
Table I
The primers used in the real-time PCR
analyses.
| Gene | Sequence (5′-3′;
forward/reverse) |
|---|
| Pinx1 |
ATGTCTATGCTGGCTGAA/TCTGTGGCTCCTTGCT |
| Caspase 3 |
CTCGGTCTGGTACAGATGTCG/GGTTAACCCGGGTAAGAATG |
| Caspase 8 |
TCTGGAGCATCTGCTGTCTG/CCTGCCTGGTGTCTGAAGTT |
| Caspase 9 |
ATGGACGAAGCGGATCGG/CCCTGGCCTTATGATGTT |
| GAPDH |
TGGTATCGTGGAAGGACTCATGAC/ATGCCAGTGAGCTTCCCGTTCAGC |
Western blot analysis
Cells or tissues were lysed in lysis buffer (20 mM
Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) containing a
protease inhibitor cocktail (Sigma). Cell extract protein amounts
were quantified using the BCA protein assay kit. Equivalent amounts
of protein (30 μg) were separated using 12% SDS-PAGE and
transferred to a PVDF membrane (Millipore Corporation, Billerica,
MA, USA). Anti-Pinx1 antibody (Santa Cruz) was used to identify
transfection efficiency, and detection of β-actin was used as an
internal control. Cell apoptosis-related proteins were also probed
using the following antibodies (Santa Cruz): caspase 3, 8 and 9.
Binding of each specific antibody was detected with horseradish
peroxidase (HRP)-conjugated respective secondary antibodies and ECL
solutions (both from Amersham Biosciences, UK).
Immunohistochemical studies
Excised tumors were fixed in 4% paraformaldehyde for
24 h then embedded in paraffin. Sections (4-μm) were generated for
immunohistochemical staining. Endogenous peroxidase activity was
blocked by incubating sections in 3% hydrogen peroxide for 30 min.
Antigen retrieval was performed in citrate buffer (10 mM, pH, 6.0)
for 30 min at 95°C in a pressure cooker (YQ50-90A;
Peskoe®, Guangzhou, China). Antibodies described for
western blot analysis were applied, and sections were incubated at
4°C overnight. Sections were washed with PBS and then incubated
with a biotinylated secondary antibody at 37°C for 2 h and then
exposed to a streptavidin complex (HRP; Beyotime, Beijing).
Positive reactions were visualized using 3,3′-diaminobenzidine
tetrahydrochloride (DAB), followed by counterstaining with
hematoxylin (both from Beyotime).
Statistical analysis
The Statistical Package for the Social Sciences 16.0
software (SPSS, Chicago, IL, USA) was used for statistical
analyses. Kaplan-Meier survival plots were generated, and
comparisons were made with log-rank statistics. Data are expressed
as the means ± SD, and the intergroup difference was compared using
the Student’s t-test. A P-value <0.05 was considered to indicate
a statistically significant result.
Results
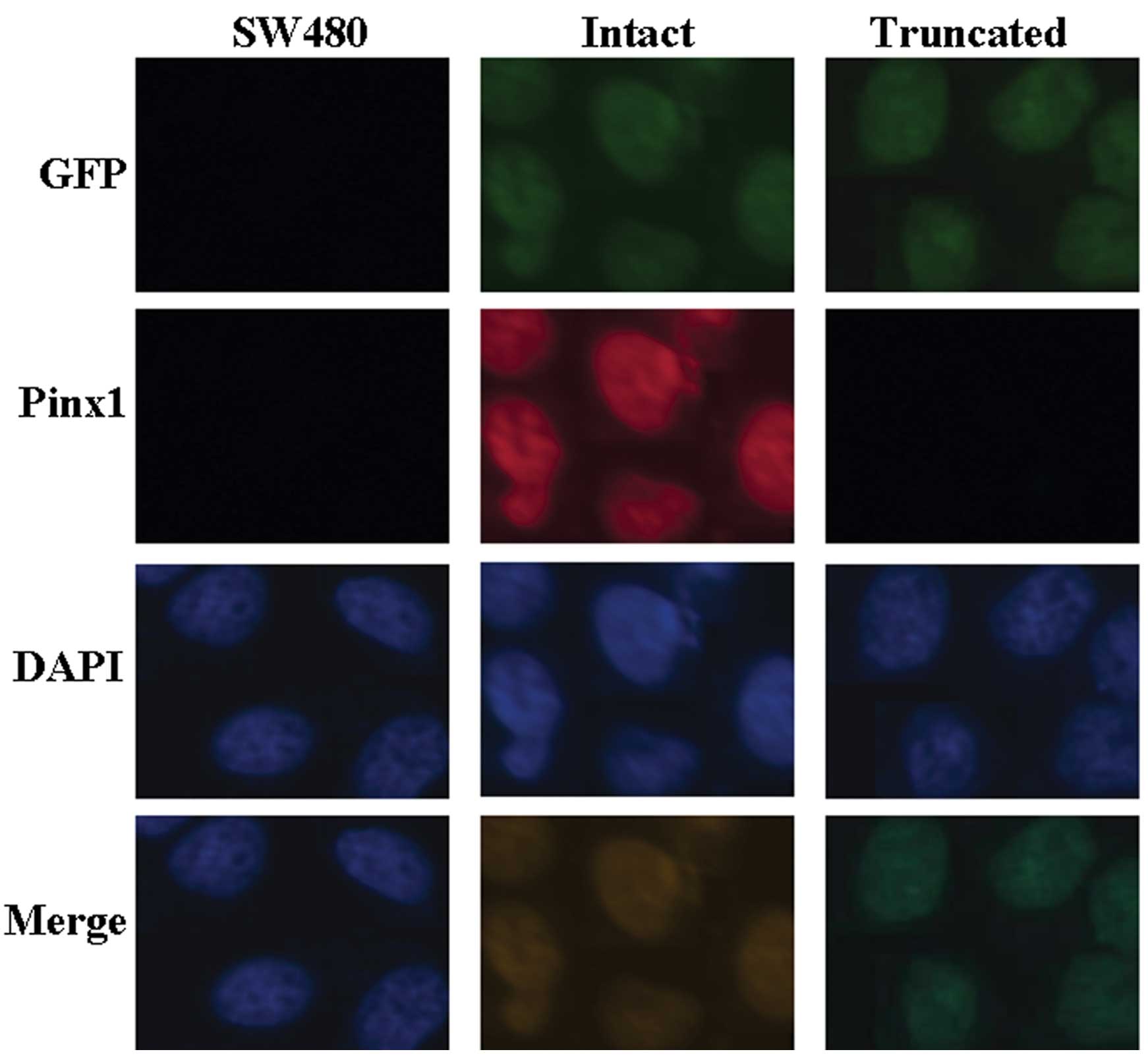
PinX1-expressing SW480 cell line
We investigated the consequence of exogenous PinX1
expression in SW480 cells. SW480 cells were transfected with
pEGFP-A1-PinX1 1–328 or pEGFP-A1-PinX1 69–328, and EGFP+
SW480 cells were measured by immunofluorescence analysis. As shown
in Fig. 1, immunofluorescence
analysis showed the localization of PinX1 and EGFP in the
PinX1-expressing SW480 cells.
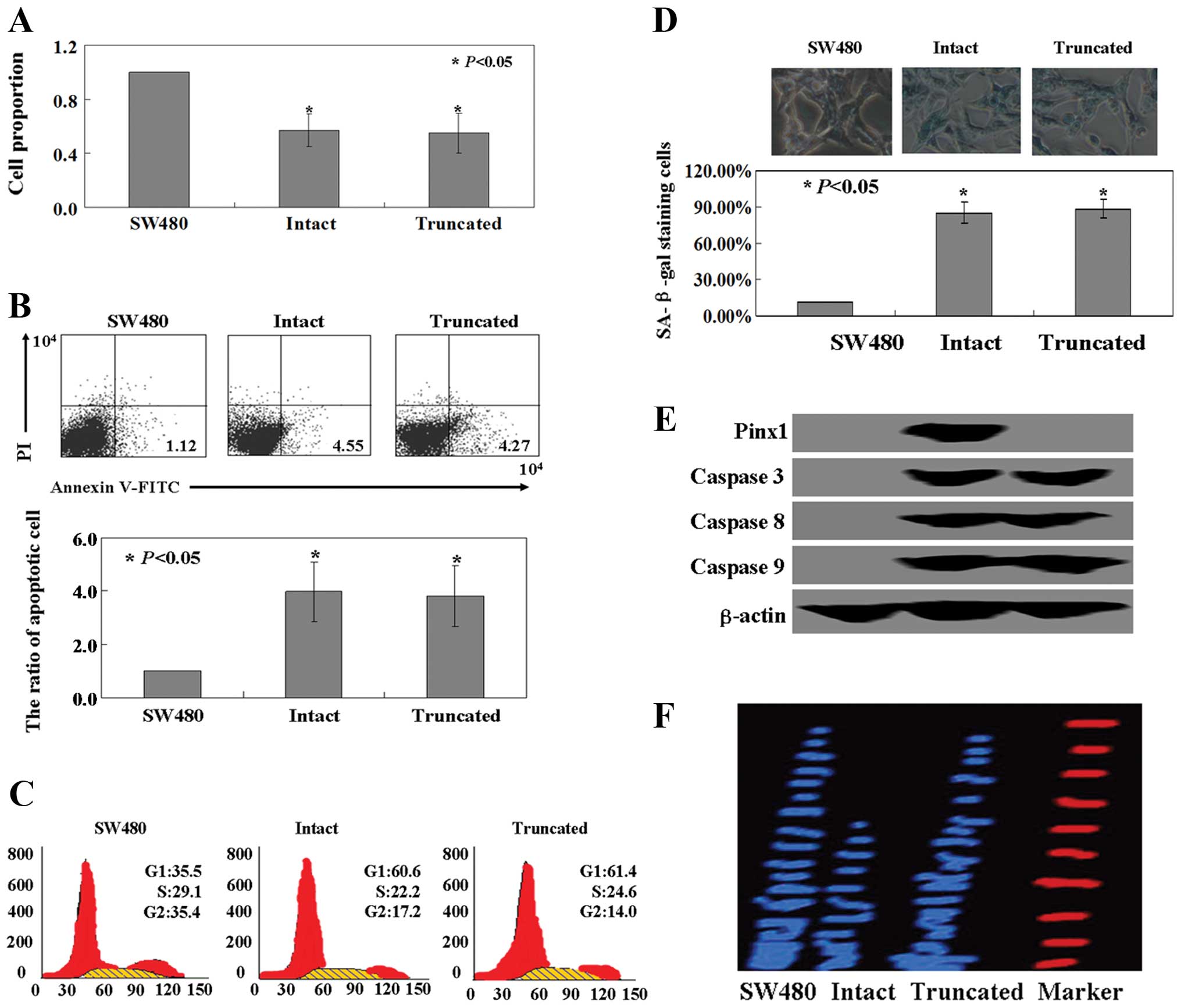
Effects of PinX1 or PinX1 69–328 on SW480
cells
Cell viability was monitored using an MTT assay, and
the results indicated that the proliferation ratio of SW480 cells
was inhibited by both PinX1 and PinX1 69–328 (P<0.05, Fig. 2A). We utilized Annexin V-FITC and PI
double staining to detect apoptotic cells. The apoptotic ratio was
3.8- to 4.2-fold higher in the cells expressing intact or truncated
PinX1 when compared with the ratio of the untransfected cells
(P<0.05, Fig. 2B). When the cell
cycle distribution of the transfected and untransfected cells was
examined using PI staining, the ratio of cells in the G1
phase was found to be increased in cells expressing intact or
truncated PinX1 vs. the untransfected cells (P<0.05, Fig. 2C). Intact or truncated
PinX1-transfected SW480 cells showed strong levels of blue SA-β-gal
staining when compared with the untransfected cells (P<0.05,
Fig. 2D). Levels of caspase 3, 8
and 9 protein were significantly increased in the SW480 cells
following PinX1 or PinX1 69–328 treatment (Fig. 2E). We next examined telomerase
activity in the intact or truncated PinX1-transfected SW480 cells
and found that the intact PinX1-transfected SW480 cells showed
reduced telomerase activity, while the truncated PinX1-transfected
cells displayed robust telomerase activity. In contrast, there was
no significant change in the telomerase activity in the
untransfected cells as observed by TRAP assay (Fig. 2F).
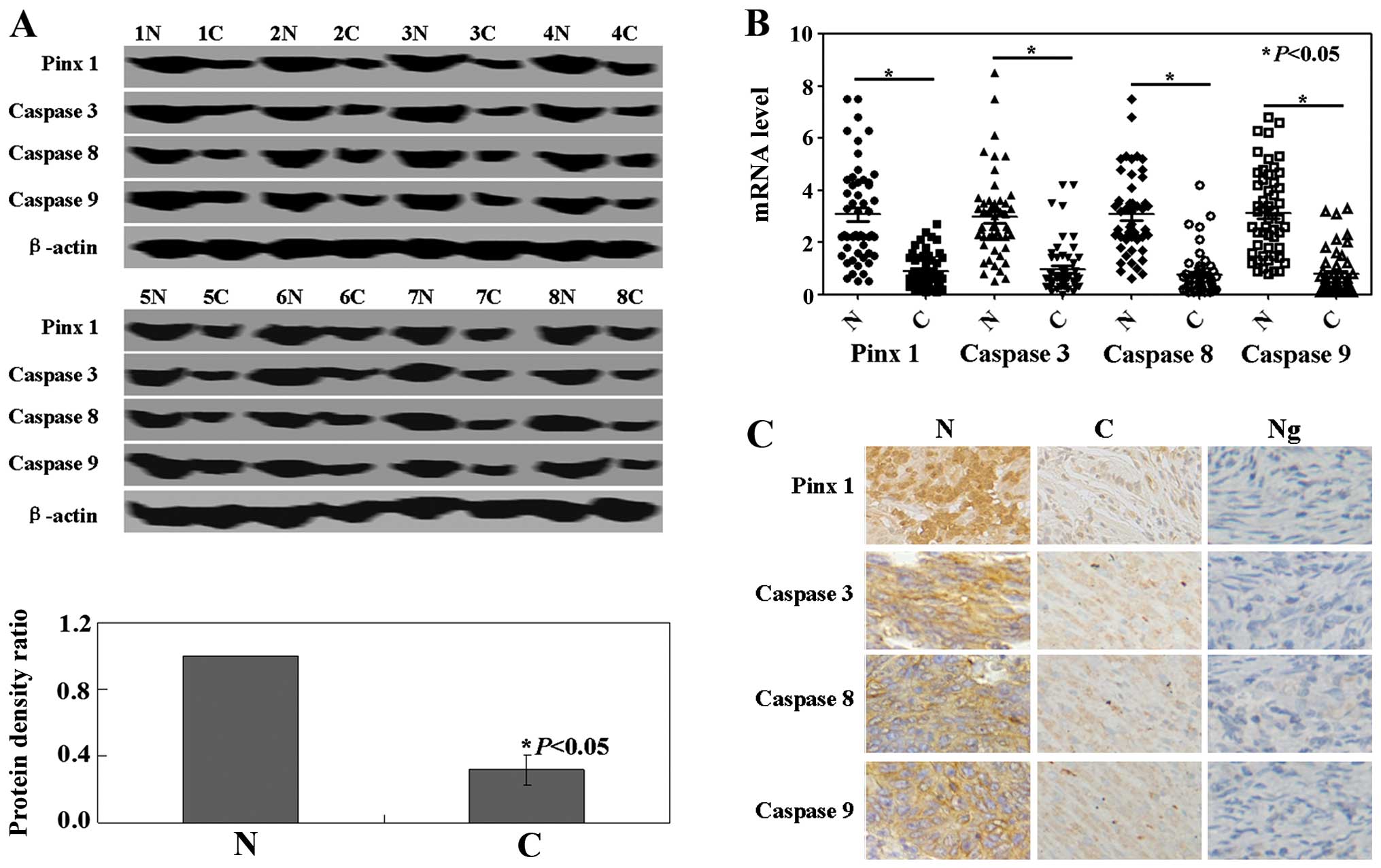
PinX1 or caspase expression in human
colorectal cancer specimens and the clinicopathological
variables
Western blot analysis was performed in order to
determine the protein expression levels of PinX1, caspase 3, 8 and
9. PinX1, caspase 3, 8 and 9 protein levels in the cancer tissues
were significantly lower than levels in the normal tissues
(P<0.05, Fig. 3A). Real-time PCR
analysis for PinX1 and caspase 3, 8 and 9 mRNA was performed in 52
colorectal cancer specimens. Results showed that the levels of
PinX1 and caspase 3, 8 and 9 mRNA in cancer tissues were
significantly lower than the levels in the normal tissues
(P<0.05, Fig. 3B). The results
of immunohistochemistry showed that caspase 3, 8 and 9 protein was
localized in the cytoplasm, while PinX1 protein was localized in
the nucleus of the cells (Fig. 3C).
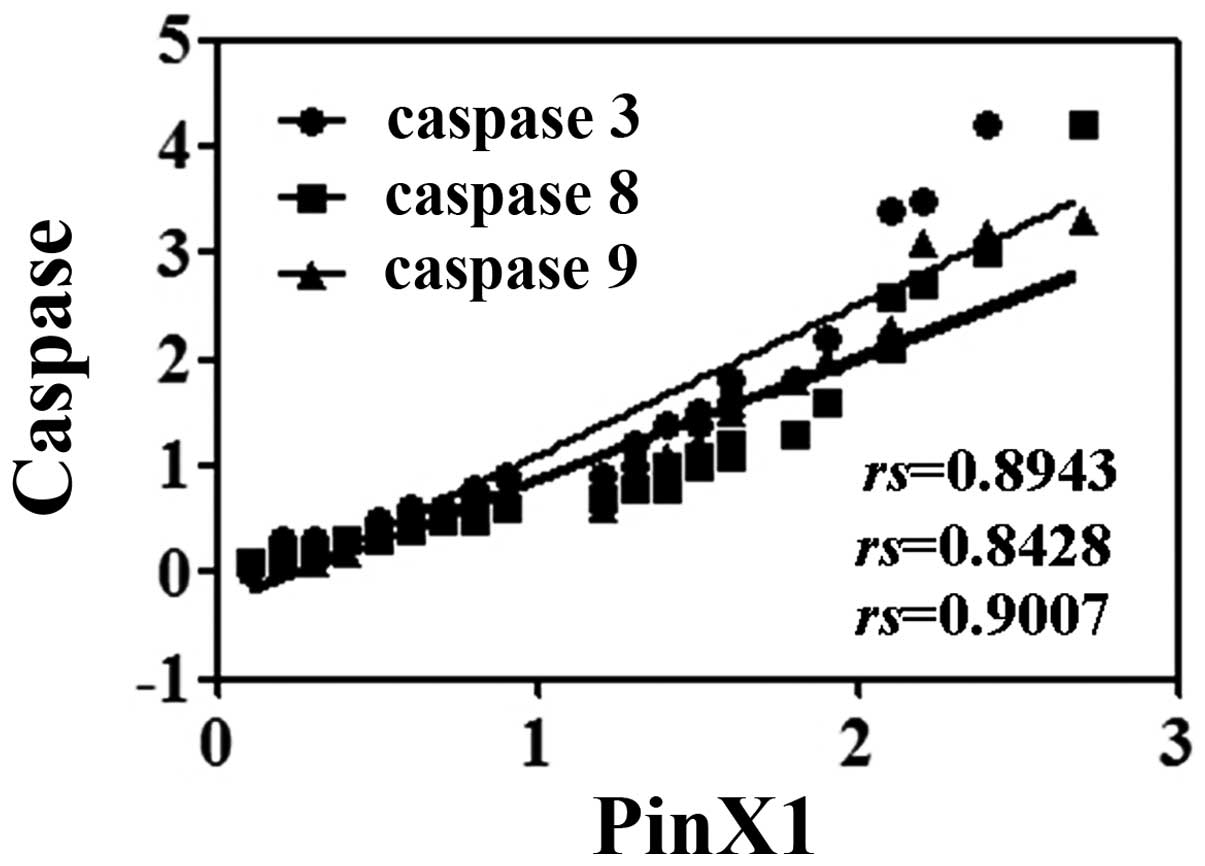
The protein levels of PinX1 and caspase 3, 8 and 9 were
significantly positively correlated (P<0.05, Fig. 4). We then analyzed the potential
relationship between the expression of PinX1 and caspase 3, 8 and 9
and the clinicopathological characteristics of these patients.
Unfortunately, we only found that caspase 3 was associated with
differentiation, and caspase 9 was associated with venous invasion
and tumor size (P<0.05, Table
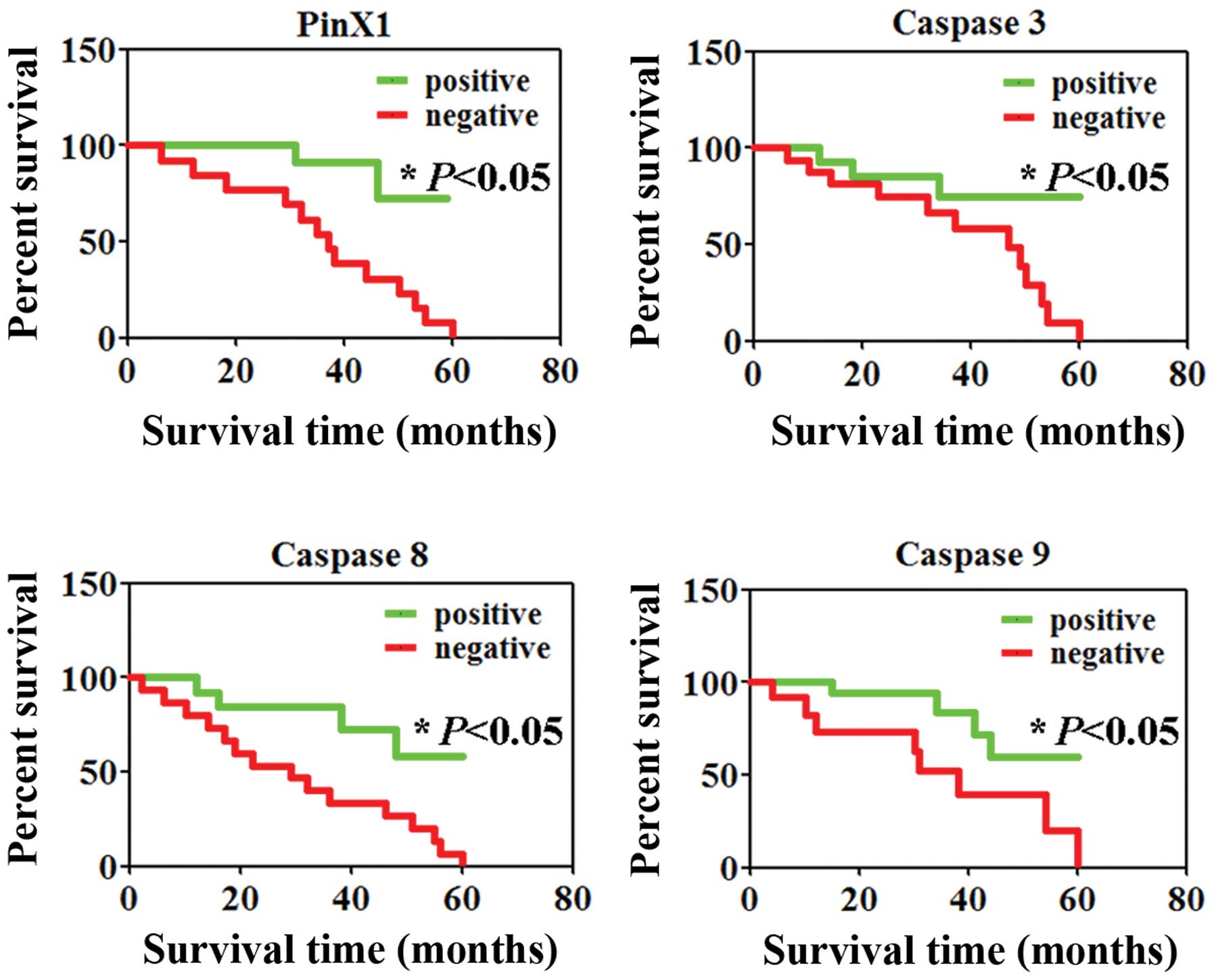
II). Finally, we investigated the level of PinX1 and caspase 3,
8 and 9 with patient survival. Comparison by the Kaplan-Meier
method for low vs. high PinX1 or caspase 3, 8 and 9 expression
showed a significant difference in the 5-year survival rate of the
patients with colorectal cancer (P<0.05, Fig. 5).
 | Table IIRelationship between Pinx1 or caspase
expression and clinicopathological parameters of the patients with
colorectal cancer. |
Table II
Relationship between Pinx1 or caspase
expression and clinicopathological parameters of the patients with
colorectal cancer.
| Pinx1 | Caspase 3 | Caspase 8 | Caspase 9 |
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | n | − | + | PR (%) | χ2 | P-value | − | + | PR (%) | χ2 | P-value | − | + | PR (%) | χ2 | P-value | − | + | PR (%) | χ2 | P-value |
|---|
| Gender | | | | | 0.1 | 0.74 | | | | 0.0 | 0.92 | | | | 0.0 | 0.95 | | | | 0.5 | 0.46 |
| Female | 21 | 16 | 5 | 23.8 | | | 17 | 4 | 19.0 | | | 18 | 3 | 14.3 | | | 15 | 6 | 28.6 | | |
| Male | 31 | 26 | 5 | 16.1 | | | 26 | 5 | 16.1 | | | 28 | 3 | 9.7 | | | 26 | 5 | 16.1 | | |
| Age (years) | | | | | 0.02 | 0.87 | | | | 0.6 | 0.45 | | | | 0.7 | 0.39 | | | | 0.2 | 0.68 |
| <50 | 14 | 12 | 2 | 14.3 | | | 13 | 1 | 7.1 | | | 11 | 3 | 21.4 | | | 10 | 4 | 28.6 | | |
| ≥50 | 38 | 30 | 8 | 21.1 | | | 30 | 8 | 21.1 | | | 35 | 3 | 7.9 | | | 31 | 7 | 18.4 | | |
|
Differentiation | | | | | 1.8 | 0.18 | | | | 5.9 | 0.01 | | | | 0.1 | 0.69 | | | | 0.13 | 0.71 |
| Well or
moderate | 33 | 29 | 4 | 12.1 | | | 31 | 2 | 6.1 | | | 31 | 2 | 6.1 | | | 25 | 8 | 24.2 | | |
| Poor | 19 | 13 | 6 | 31.6 | | | 12 | 7 | 36.8 | | | 15 | 4 | 21.1 | | | 16 | 3 | 15.8 | | |
| Lymphatic
invasion | | | | | 0.3 | 0.60 | | | | 0.3 | 0.61 | | | | 0.8 | 0.36 | | | | 0.0 | 0.92 |
| − | 22 | 19 | 3 | 13.6 | | | 17 | 5 | 22.7 | | | 21 | 1 | 4.5 | | | 17 | 5 | 22.7 | | |
| + | 30 | 23 | 7 | 23.3 | | | 26 | 4 | 13.3 | | | 25 | 5 | 16.7 | | | 24 | 6 | 20.0 | | |
| Venous
invasion | | | | | 0.2 | 0.63 | | | | 0.7 | 0.39 | | | | 0.3 | 0.59 | | | | 6.6 | 0.01 |
| − | 25 | 19 | 6 | 24.0 | | | 19 | 6 | 24.0 | | | 21 | 4 | 16.0 | | | 24 | 1 | 4.0 | | |
| + | 27 | 23 | 4 | 14.8 | | | 24 | 3 | 11.1 | | | 25 | 2 | 7.4 | | | 17 | 10 | 37.0 | | |
| Tumor size
(cm) | | | | | 1.6 | 0.21 | | | | 2.4 | 0.12 | | | | 1.4 | 0.24 | | | | 15.9 | 0.00 |
| <4 | 15 | 10 | 5 | 33.3 | | | 10 | 5 | 33.3 | | | 15 | 0 | 0.0 | | | 6 | 9 | 60.0 | | |
| ≥4 | 37 | 32 | 5 | 13.5 | | | 33 | 4 | 10.8 | | | 31 | 6 | 16.2 | | | 35 | 2 | 5.4 | | |
| pN category | | | | | 0.32 | 0.95 | | | | 0.2 | 0.97 | | | | 1.4 | 0.70 | | | | 2.4 | 0.50 |
| pN0 | 11 | 9 | 2 | 18.2 | | | 9 | 2 | 18.2 | | | 9 | 2 | 18.2 | | | 8 | 3 | 27.3 | | |
| pN1 | 9 | 7 | 2 | 22.2 | | | 7 | 2 | 22.2 | | | 8 | 1 | 11.1 | | | 6 | 3 | 33.3 | | |
| pN2 | 13 | 10 | 3 | 23.1 | | | 11 | 2 | 15.4 | | | 11 | 2 | 15.4 | | | 10 | 3 | 23.1 | | |
| pN3 | 19 | 16 | 3 | 15.8 | | | 16 | 3 | 15.8 | | | 18 | 1 | 5.3 | | | 17 | 2 | 10.5 | | |
Discussion
As noted in the Introduction, Pin2/TRF1-interacting
protein X1 (PINX1) is a co-regulating gene of telomerase and
telomere (7), which is located at
chromosome 8p23, consisting of seven axons with two transcription
pathways encoding 174 aa and 328 aa proteins (4). In the present study, we demonstrated
that PinX1 could induce apoptosis, G1 arrest, and
cellular senescence in colorectal cancer SW480 cells. In addition,
we revealed that intact PinX1 (1–328 aa) inhibited telomerase
activity in the SW480 cells. Previous studies also showed that
PinX1 is an internal telomerase inhibitor and suppresses tumor
growth both in vivo and in vitro (13–15).
Overexpression of PinX1 in tumor cells could inhibit telomerase
activity, shorten telomeres, and suppress tumor growth, while
depletion of endogenous PinX1 increased telomerase activity,
elongated telomeres, and enhanced tumorigenicity in a fibrosarcoma
cell line, HT1080 (4). However, to
our knowledge, no studies have shown the functions of the G-patch
motif in PinX1. Notably, in the present study, we found that PinX1
without G-patch (69–328 aa) induced apoptosis and G1
arrest but showed no effects on telomerase activity.
In budding yeast, the PinX1 homolog Gnop1 was shown
to bind the yeast telomerase catalytic protein Est2p and inhibit
telomerase (16). Gnop1 has
additional functions in processing ribosomal RNA and other small
nucleolar RNAs that are mediated, at least in part, through G-patch
(17,18). However, the effect of human PinX1 on
telomerase appears to be exclusive of the G-patch region and is
mediated instead by the C terminus of the protein (4). Thus, combined with our results, these
findings suggested that the interaction with hTR cannot be ascribed
to a general RNA binding feature of PinX1 mediated through G-patch.
However, why the PinX1 without G-patch has similar antitumor
activities as intact PinX1 remains unclear. The mechanisms of
G-patch warrant elucidation in further studies.
Acknowledgements
This study was supported by the Natural Science
Foundation of Liaoning Province, China (grant no. 2013020158).
References
|
1
|
Blackburn EH: Switching and signaling at
the telomere. Cell. 106:661–673. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Lange T: Protection of mammalian
telomeres. Oncogene. 21:532–540. 2002.
|
|
3
|
Smogorzewska A and de Lange T: Regulation
of telomerase by telomeric proteins. Annu Rev Biochem. 73:177–208.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou XZ and Lu KP: The
Pin2/TRF1-interacting protein PinX1 is a potent telomerase
inhibitor. Cell. 107:347–359. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Banik SS and Counter CM: Characterization
of interactions between PinX1 and human telomerase subunits hTERT
and hTR. J Biol Chem. 279:51745–51748. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan K, Li N, Jiang K, et al: PinX1 is a
novel microtubule-binding protein essential for accurate chromosome
segregation. J Biol Chem. 284:23072–23082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park WS, Lee JH, Park JY, et al: Genetic
analysis of the liver putative tumor suppressor (LPTS) gene
in hepatocellular carcinomas. Cancer Lett. 178:199–207. 2002.
|
|
8
|
Hawkins GA, Chang B, Zheng SL, et al:
Mutational analysis of PINX1 in hereditary prostate cancer.
Prostate. 60:298–302. 2004.
|
|
9
|
Kondo T, Oue N, Mitani Y, et al: Loss of
heterozygosity and histone hypoacetylation of the PINX1 gene
are associated with reduced expression in gastric carcinoma.
Oncogene. 24:157–164. 2005.PubMed/NCBI
|
|
10
|
Chang Q, Pang JC, Li J, Hu L, Kong X and
Ng HK: Molecular analysis of PinX1 in medulloblastomas. Int
J Cancer. 109:309–314. 2004. View Article : Google Scholar
|
|
11
|
Dimri GP, Lee X, Basile G, et al: A
biomarker that identifies senescent human cells in culture and in
aging skin in vivo. Proc Natl Acad Sci USA. 92:9363–9367. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim NW, Piatyszek MA, Prowse KR, et al:
Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qian D, Zhang B, He LR, et al: The
telomere/telomerase binding factor PinX1 is a new target to improve
the radiotherapy effect of oesophageal squamous cell carcinomas. J
Pathol. 229:765–774. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai XF, Shen CX, Wen Z, et al: PinX1
regulation of telomerase activity and apoptosis in nasopharyngeal
carcinoma cells. J Exp Clin Cancer Res. 31:122012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu JY, Qian D, He LR, et al: PinX1
suppresses bladder urothelial carcinoma cell proliferation via the
inhibition of telomerase activity and p16/cyclin D1 pathway. Mol
Cancer. 12:1482013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin J and Blackburn EH: Nucleolar protein
PinX1p regulates telomerase by sequestering its protein catalytic
subunit in an inactive complex lacking telomerase RNA. Genes Dev.
18:387–396. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guglielmi B and Werner M: The yeast
homolog of human PinX1 is involved in rRNA and small nucleolar RNA
maturation, not in telomere elongation inhibition. J Biol Chem.
277:35712–35719. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aravind L and Koonin EV: G-patch: a new
conserved domain in eukaryotic RNA-processing proteins and type D
retroviral polyproteins. Trends Biochem Sci. 24:342–344. 1999.
View Article : Google Scholar : PubMed/NCBI
|