Introduction
Deregulating cellular metabolism is one of the
hallmarks of cancer, first observed by Otto Warburg in the 20th
century, and now known as the Warburg effect (1–4). The
Warburg effect in cancer cells is partially induced by rapid cell
division, proliferation and anaerobic conditions as well as the
requirement of small molecules for the subcellular construction of
new cells (1). Under anaerobic
conditions, cancer cells convert glucose to pyruvate and then the
pyruvate is transformed to lactic acid for the production of ATP
and other cellular construction materials.
High glycolytic activity and inadequate glucose
intake are contradictory factors in cancer cells. To handle this
stress, the expression of glucose transporters,
glucose-metabolizing enzymes and pyruvate dehydrogenase kinase are
upregulated to promote glucose uptake; however, there is
insufficient adaptive angiogenesis and blood supply (5). To attenuate nutrient starvation,
cancer cells utilize several compensatory approaches, such as
autophagy (6). Autophagy may be
induced by high temperature, hormonal stimulation, low oxygen or
nutrient starvation (7,8). In cancer cells, a lack of nutrient
sources triggers autophagy, which can sustain the survival of
malignant cells (6). Cancer cells
may undergo autophagy for back-up energy reserve to support
self-survival (6,9), although autophagy may increase
apoptosis and cell death (10–12).
The non-vascularized and metabolically stressed sites of tumors are
usually accompanied by autophagy (9).
During the early neonatal stages of newborns,
starvation occurs in most mammals (13). To overcome this life-threatening
problem, autophagy is activated and self-holding proteins are
degraded by autophagy to produce a pool of amino acids with which
to attenuate nutrient starvation (13). Meanwhile, nitrogen deprivation also
triggers autophagy-dependent amino acid production (14). Amino acids are enzymatically
degraded and become nutrient molecules, which may sustain cancer
cell survival while cancer cells are under conditions of low oxygen
and blood supply.
To investigate the relationship between amino acids
and cancer cells under low glucose conditions, we designed and
performed a series of experiments. The present study determined
whether amino acids can attenuate nutrient starvation of gastric
cancer cells.
Materials and methods
Cell lines and materials
Gastric cancer cells SGC7901, AGS and MGC803 were
purchased from Shanghai Cell Collection (China) and cultured in
RPMI-1640 medium (Gibco, USA) containing 2 mM L-glutamine, 100 U/ml
penicillin, and 100 μg/ml streptomycin and supplemented with 10%
fetal bovine serum (FBS; HyClone, USA). The cell culture flasks
were maintained in a humidified incubator at 37°C with a 5%
CO2 atmosphere until they reached appropriate
confluence. RPMI-1640 [−] (no glucose, cat. 11879-020), standard
RPMI-1640, MEM [100X, non-essential amino acids (NEAs); Invitrogen,
cat. 11140-050], and MEM amino acid solution (cat. 11130-051) were
obtained from Life Technologies (USA). The JC-1 kit was obtained
from Beyotime Biotech Inc. (China).
Cell viability assay
When grown to 60–80% confluence, gastric cells were
trypsinized and seeded on 96-well plates at a density of
1×104 cells/well. The next day, complete medium
containing 2,000, 1,000 or 500 mg/ml glucose was added into
microplate wells, and a NEA solution was added into corresponding
wells. Twenty microliters of a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma Chemical Co., St. Louis, MO, USA) solution (5 mg/ml) were
added to each well at intervals of 24, 48, 72 and 96 h after NEA
infection. Plates were incubated at 37°C for 4 h, and 100 μl of a
lysis buffer DMSO was then added to each well and mixed thoroughly
for 10 min. Finally, absorbance values were determined at 570 nm by
a microplate reader (Bio-Rad, USA) (15).
Flow cytometry for apoptosis
To measure low-glucose-induced apoptosis, gastric
cancer cells were pelleted in 6-well tissue plates and 16 h later,
complete medium containing 2,000, 1,000 or 500 mg/ml glucose or
complete medium containing 2,000, 1,000 or 500 mg/ml glucose with
2× NEAs (20 μl of 100× NEAs in 1 ml of experimental treatment
medium) was added. Forty-eight hours later, gastric cancer cells
were washed with phosphate-buffered saline (PBS), trypsinized with
no-EDTA trypsin, centrifuged at 1,000 rpm for 5 min, repeatedly
washed with PBS, and resuspended in 500 μl binding
buffer/106 cells. Cells were then incubated with
FITC-labeled Annexin V for 15 min on ice, followed by incubation
with propidium iodide (PI) for another 15 min (BD Biosciences, USA)
(16). The apoptosis of gastric
cancer cells was determined by flow cytometry.
Mitochondrial membrane potential
detection by JC-1 staining
Gastric cancer cells were seeded in 6-well tissue
plates at a density of 1×105 cells/well and grown to
60–70% confluence. Cells were then stimulated by experimental
treatment medium for 48 h. The cells were washed twice with
serum-free medium and incubated with a final concentration of 1 μM
JC-1
(5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine
iodide) in DMEM medium for 30 min at 37°C and 5% CO2
(17,18). After the medium was removed, each
treatment group of cells was washed with PBS 3 times, and 100 μl of
PBS was added to each well. Finally, the changes in the
mitochondria membrane potential were determined by
fluorescence-activated cell sorting (FACS; BD Biosciences) at
excitation and emission wavelengths of 544 and 590 nm,
respectively.
Total RNA isolation and quantitative
real-time PCR
Total RNA from different treatment groups was
extracted using TRIzol reagent (Invitrogen, USA), and the quality
and concentration of RNAs were determined by UV spectrophotometry
(Bio-Rad). Complimentary DNA (cDNA) was obtained, and quantitative
real-time PCR (qRT-PCR) was then performed using a StepOne Plus
instrument (Applied Biosystems, USA) with the following procedures:
95°C for 10 min, followed by 95°C for 15 sec, and 60°C for 1 min
for 40 cycles. The relative expressions of the mRNAs of amino acid
transporter genes (Lat1, Lat2 and h4F2hc) and cell apoptosis genes
(Bcl-2, Bcl-xL, Bik and Bax) were detected by qRT-PCR, and the
GAPDH gene was used as an internal control. The comparative CT
method was used to calculate the relative expression level of these
genes, and qRT-PCR was performed according to a previously
described protocol (19). Primers
for these genes are listed in Table
I.
 | Table IPrimers for amino acid transporter
and apoptosis-related genes used in qRT-PCR. |
Table I
Primers for amino acid transporter
and apoptosis-related genes used in qRT-PCR.
| Gene names | Primers |
|---|
| LAT1 | F:
TGTGCTGGCATTATACAGCG |
| LAT1 | R:
AGGTGATAGTTCCCGAAGTC |
| LAT2 | F:
TTTCCAGGAACCTGACATCG |
| LAT2 | R:
ACATTGCAGTGACATAAGCG |
| h4F2hc | F:
CTCAGGCAAGGCTCCTGACT |
| h4F2hc | R:
GGCAGGGTGAAGAGCATCA |
| Bax | F:
GATGCGTCCACCAAGAAGCT |
| Bax | R:
CGGCCCCAGTTGAAGTTG |
| Bcl-xL | F:
ACCCCAGGGACAGCATATCA |
| Bcl-xL | R:
TGCGATCCGACTCACCAATA |
| Bcl-2 | F:
TCCGCATCAGGAAGGCTAGA |
| Bcl-2 | R:
AGGACCAGGCCTCCAAGCT |
| Bik | F:
CTTGATGGAGACCCTCCTGTATG |
| Bik | R:
AGGGTCCAGGTCCTCTTCAGA |
Mitochondrial DNA copy number
detection
mtDNA copy number from experimental treatment
medium-stimulated gastric cancer cells was determined by a standard
protocol, as described in Current Protocols in Human Genetics
(20). Total genomic DNA was
isolated with a QIAamp kit (Qiagen, Germany), according to the
manufacturer’s instructions. The method for mtDNA copy number
detection based on qRT-PCR involved the utilization of a
107-bp-sized amplicon of mtDNA tRNALeu(UUR) (forward
primer, CACCCAAGAACAGGGTT TGT and reverse primer,
TGGCCATGGGTATGTTGTTA); and an 86-bp-sized amplicon of
β2-microglobulin (forward primer, TGCTGTCTCCATGTTTGATGTATCT and
reverse primer, TCTCTGCTCCCCACCTCTAAGT) of nuclear DNA (nDNA) was
used as an internal control (20).
The PCR procedure was as follows: 95°C for 10 min, followed by 95°C
for 15 sec, and 60°C for 1 min for 40 cycles. PCR assays were
performed in triplicate for each DNA sample. The expression of
mtDNA copy number relative to the expression of nDNA was determined
using the formula: 2×2(ΔCT), where ΔCT is the difference
in CT values between the β2-microglobulin gene and
tRNALeu(UUR) (20).
Statistical analysis
The statistical significance of the data between
different groups was evaluated by performing an analysis of
variance and Student’s t-test using the SPSS 1 statistical
software. p<0.05 was considered to indicate a statistically
significant difference.
Results
Amino acids help gastric cancer cells
survive during glucose starvation
In solid tumors, low blood supply, glucose
starvation and hypoxia are common due to the rapid proliferation of
malignant cells (21). Therefore,
glucose starvation leads to apoptosis or necrosis of fast-growth
cancer cells. Gastric cancer cells MGC803, AGS and SGC7901 were
cultured in medium containing glucose concentrations of 2,000,
1,000 or 500 mg/ml in the presence or absence of 2× MEM amino
acids. Cell viability detection assays were performed 24, 48 and 72
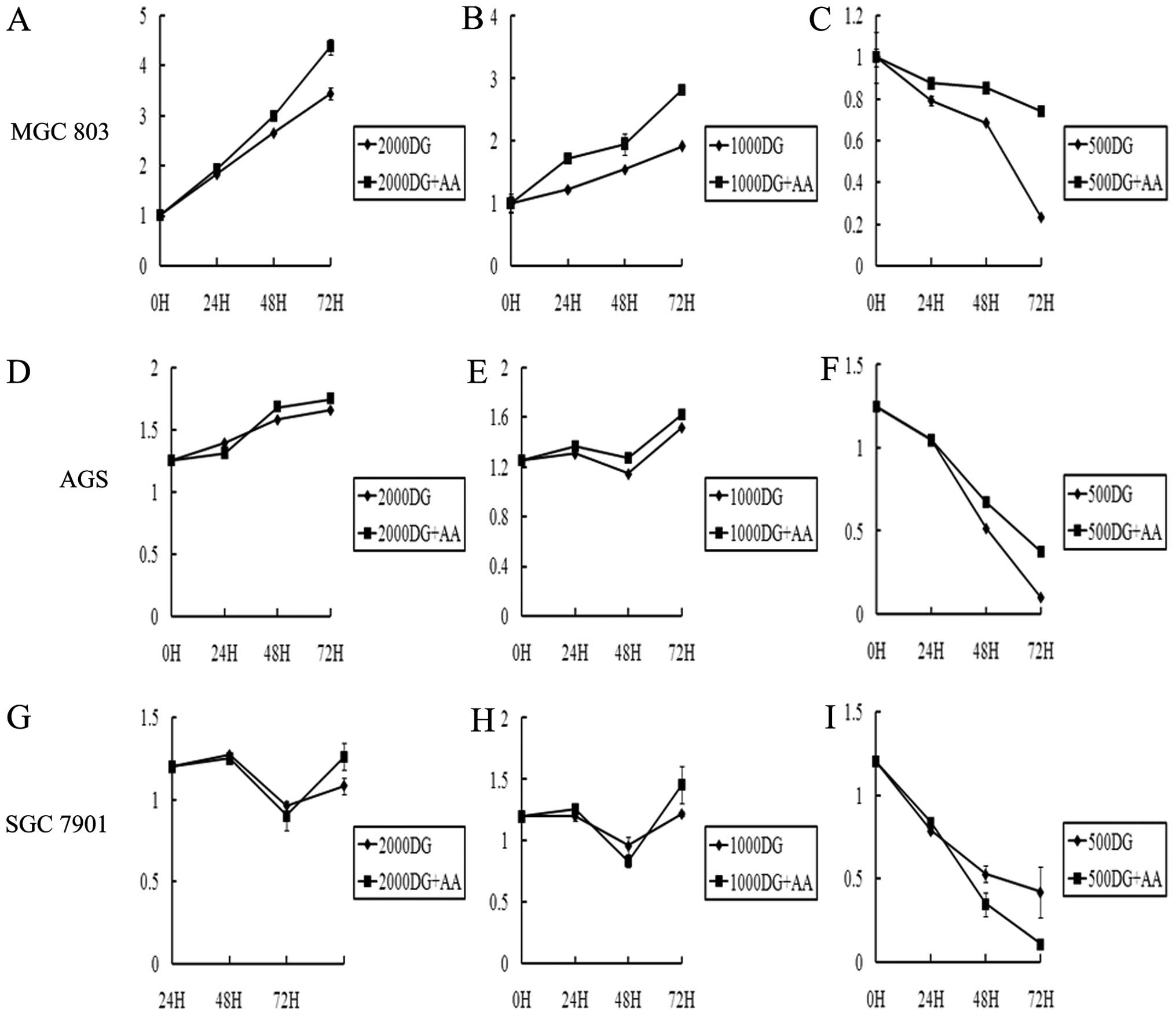
h later. As shown in Fig. 1, the
cell viability at concentrations of 2,000 and 1,000 D-glucose (DG)
was not significantly different between samples with and without
NEA (Fig. 1A, B, D, E, G and H),
but cells in 500 DG medium with NEA had a significantly higher
survival rate compared with those in medium without NEA (Fig. 1C, F and I). Amino acids, therefore,
promote gastric cancer cell survival under glucose starvation.
NEAs inhibit apoptosis caused by glucose
starvation
To further investigate the effects of NEA on gastric
cancer, we utilized flow cytometry to confirm the results of
NEA-assisted gastric cancer cells surviving glucose starvation.
Forty-eight hours after stimulation with conditional medium (medium
containing glucose at a concentration of 2,000, 1,000 or 500 mg/ml
with or without 2× MEM amino acids), cells were trypsinized and
stained with PI and FITC-labeled Annexin V (22). An obvious increase in the apoptosis
rate was detected in the 500 DG groups (15.9–44.1%) compared with
the 500 DG with NEA group in both the FITC/PI-positive (31.2–54.3%)
and FITC-positive/PI-negative (19.2–27.6%) cells (Fig. 2A-c and -f; Fig. 2B-c and -f; Fig. 2C-c and -f). In contrast, no
significant differences were found between the 2,000 and 1,000 DG
groups (Fig. 2A-a, -b, -d and -e;
Fig. 2B-a, -b, -d and -e; Fig. 2C-a, -b, -d and -e). NEA therefore
decreased the apoptosis rate of gastric cancer cells in
glucose-deficient medium.
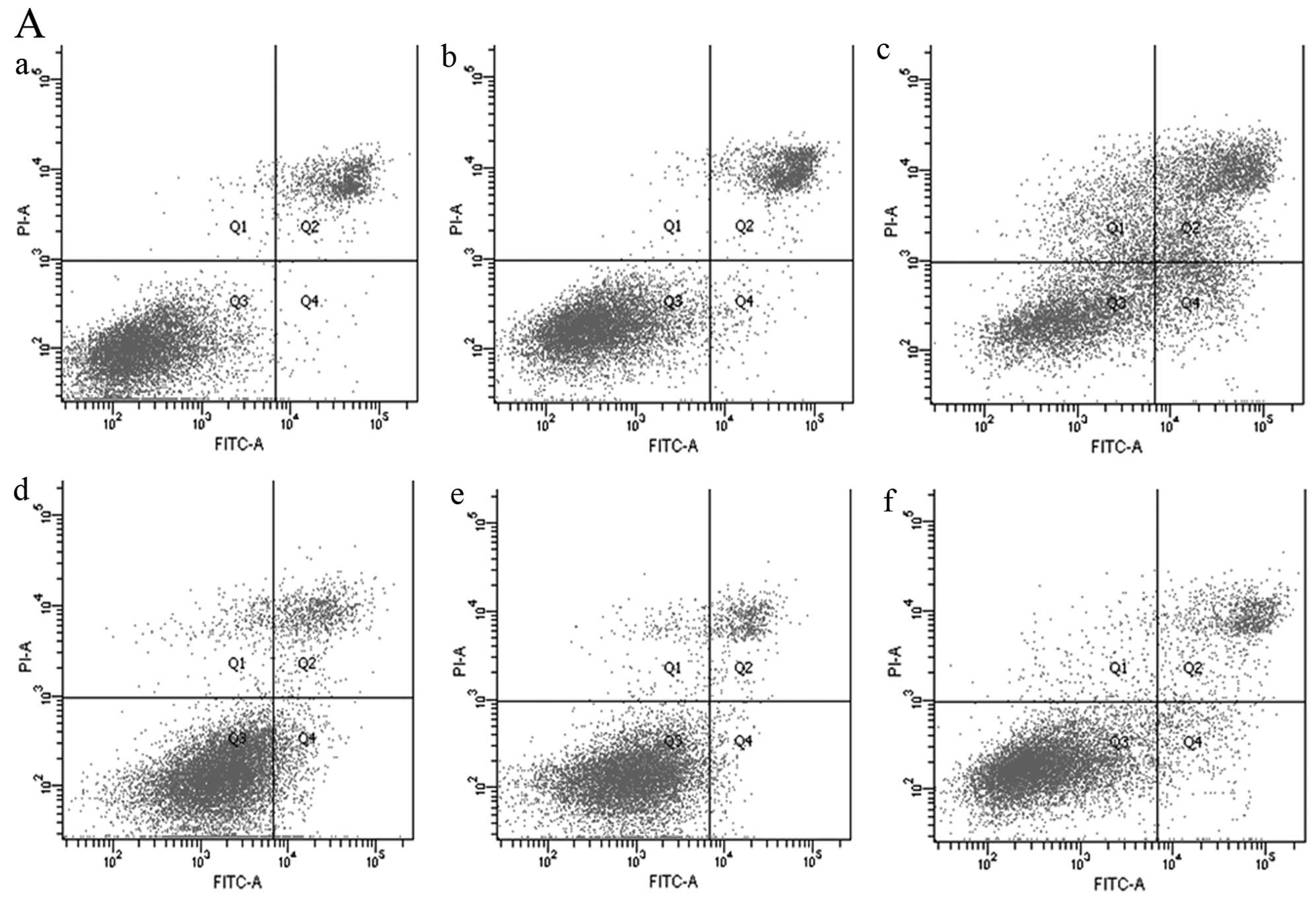 | Figure 2Apoptosis of gastric cancer cells
induced by glucose starvation. (A) Apoptosis of MGC803 cells
induced by glucose starvation. (B) Apoptosis of AGS cells induced
by glucose starvation. (C) Apoptosis of SGC7901 cells induced by
glucose starvation. FITC-labeled Annexin V and PI were both used as
indices for apoptosis induced by medium containing 2,000, 1,000 or
500 mg/ml glucose (a–c) with or without NEA (d–f). (a, 2,000 DG
group; b, 1,000 DG group; c, 500 DG group; d, 2,000 DG-AA group; e,
1,000 DG-AA group; and f, 500 DG-AA group). |
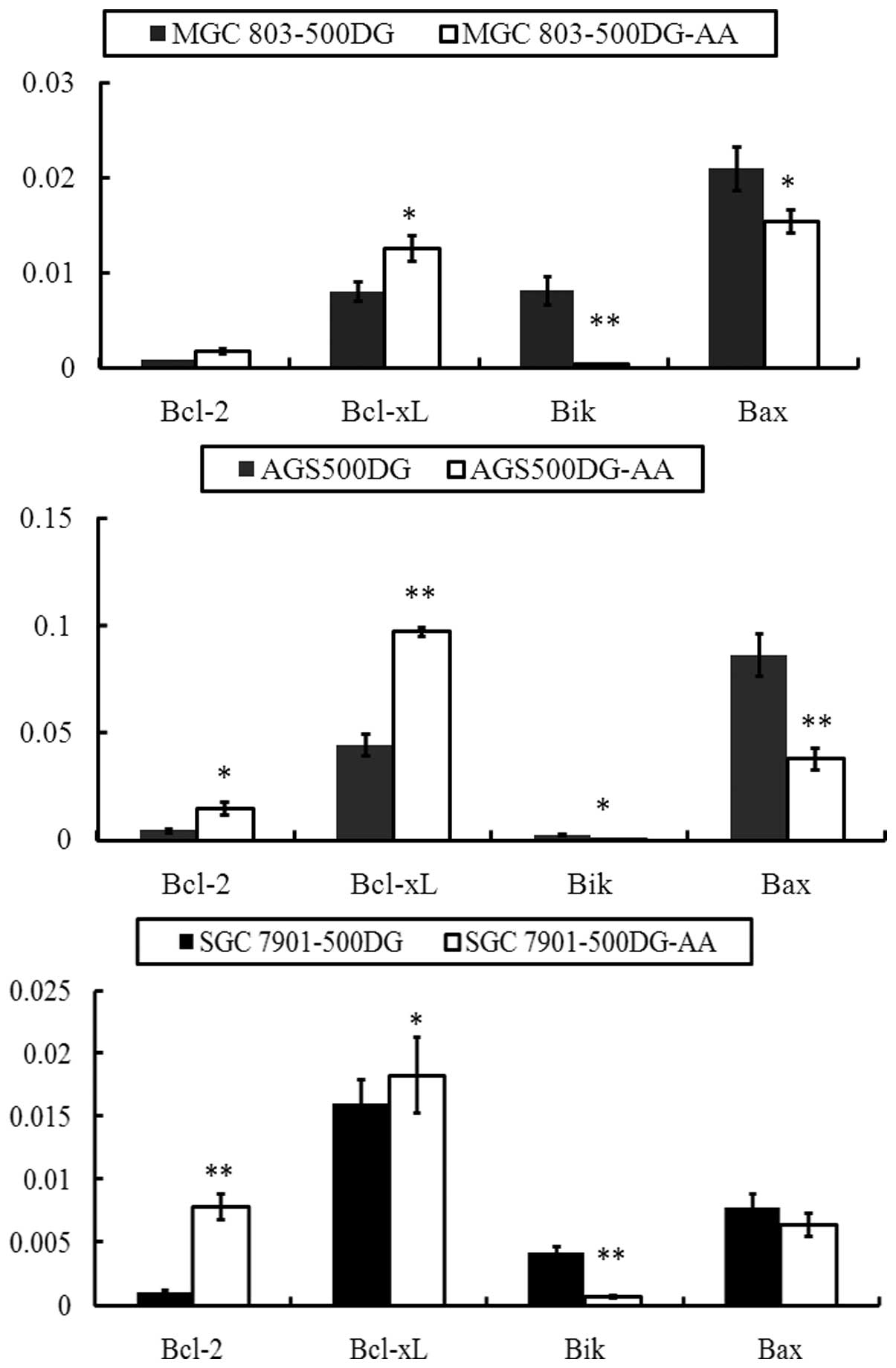
Altered expression of the Bcl-2 gene
family
Apoptosis is a programmed procedure involving
various genes. The Bcl-2 gene family is a member of the
apoptosis-associated genes and plays an important role in promoting
or inhibiting apoptosis (23,24).
The family contains Bcl-2, Bcl-xL, Bak, Bik and other genes
(24). The general function of
Bcl-2 and Bcl-xL is anti-apoptotic and that of Bax and Bik is
pro-apoptotic (24,25). Our data showed that the groups
treated with 500 DG and 500 DG-AA medium had significantly
different rates of proliferation and apoptosis. Thus, we evaluated
the expression of Bcl-2, Bcl-xL, Bak and Bik in gastric cancer
cells treated with 500 DG or 500 DG-AA medium. As shown in Fig. 3, Bcl-2 and Bcl-xL genes demonstrated
decreased expression when gastric cancer cells were exposed to 500
DG medium compared with 500 DG-AA medium. Conversely, the
expression of the Bik and Bax genes increased, which suggested that
NEA sustained gastric cancer cells by inducing the expression of
anti-apoptotic members of the Bcl-2 gene family and preventing the
expression of pro-apoptotic members.
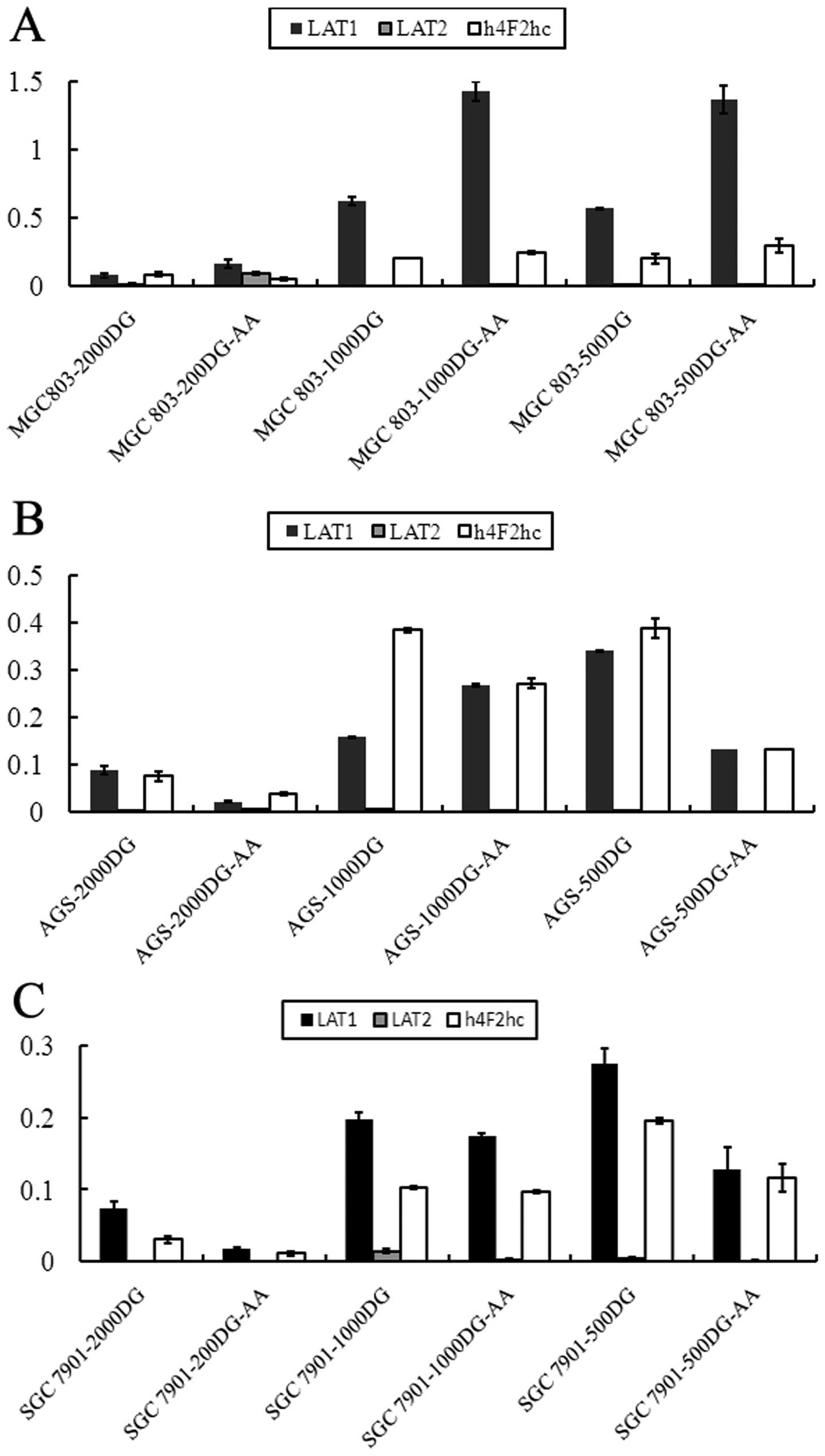
Overexpression of amino acid transporter
genes induced by glucose starvation
The data presented above demonstrate that amino
acids inhibit apoptosis. We hypothesized that the expression of
amino acid transporters in the cell membrane may increase to
fulfill the energy requirements of the cell. Thus, we examined the
relative expression levels of amino acid transporter genes L-type
amino acid transporter 1 (LAT1), LAT2 and h4F2hc. LAT1, as a
transporter gene, is reported to be highly expressed in various
human cancers (26–32), and its expression is strongly
correlated with that of another amino acid transporter gene, h4F2hc
(26). We performed qRT-PCR to
determine transporter gene expression after 48 h of low-glucose
culture and found that the decrease in glucose concentration led to
the significant upregulation of amino acid transporters (LAT1 and
h4Fhc). As shown in Fig. 4, LAT1
expression in MGC803 cells was upregulated under low-glucose
conditions by 8.17- and 7.48-fold (1,000 and 500 DG, respectively)
compared with the 2,000 DG group. LAT2 expression was decreased
under low-glucose conditions, and the expression of h4F2hc was
upregulated by 2.48- and 2.40-fold (1,000 and 500 DG, respectively;
Fig. 4A) compared with the 2,000 DG
group. Additionally, the ratios of LAT1, LAT2 and h4F2hc in the
1,000 and 500 DG groups compared with the 2,000 DG group were
changed from 1.79 to 3.87, 2.42 to 0.69 and 5.09 to 5.14,
respectively, in AGS cells (Fig.
4B). The corresponding ratios were 2.68 and 3.72, 20.23 and
6.20, and 3.41 and 4.50 in SGC7901 cells (Fig. 4C). Thus, glucose starvation was
capable of upregulating amino acid transporter genes.
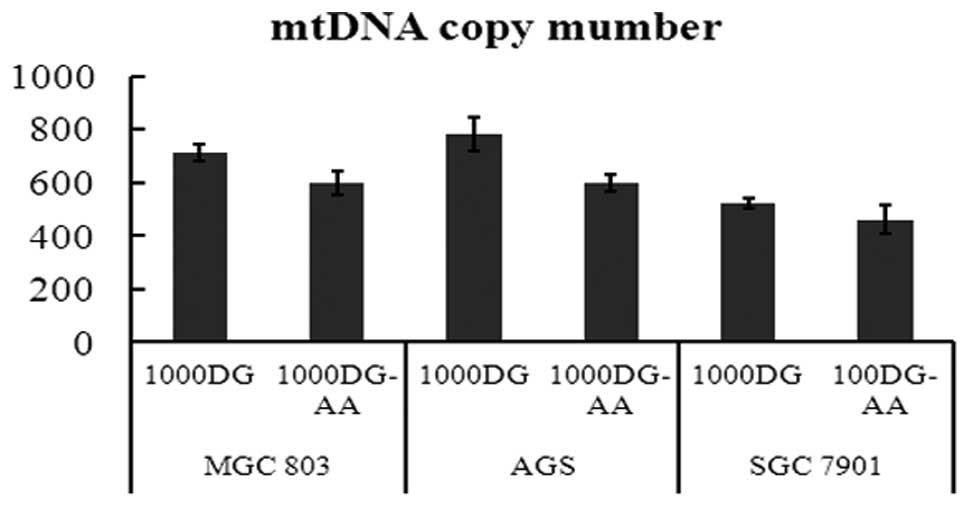
Mitochondrial content determination: DNA
copy number
Mitochondrial disorders are complicated
heterogeneous diseases that may be caused by molecular or cellular
defects (33,34). It is clear that a constant number of
mtDNA copies is essential for maintaining cell homeostasis
(35–37). Similarly, we investigated whether
the copy number of mtDNA varied in gastric cancer cells during
glucose starvation. We extracted the total genomic DNA of gastric
cancer cells stimulated by 1,000 DG and 1,000 DG with NEA for 48 h
and investigated the relative copy number of mtDNA compared to nDNA
with real-time PCR. With nDNA as an internal control, the relative
mtDNA copy number increased from 599 to 712, 597 to 783, and 461 to
523 in MGC803, AGS and SGC7901 cells, respectively, in the medium
with NEA (p<0.05) (Fig. 5).
NEAs prevent the loss of the
mitochondrial membrane potential of gastric cancer cells during
glucose starvation
Depolarization of the mitochondrial inner membrane
resulting from ion channel opening is regarded as one of the signs
of cell death. To examine whether low glucose and amino acids
affected mitochondrial inner membrane potential, we used the
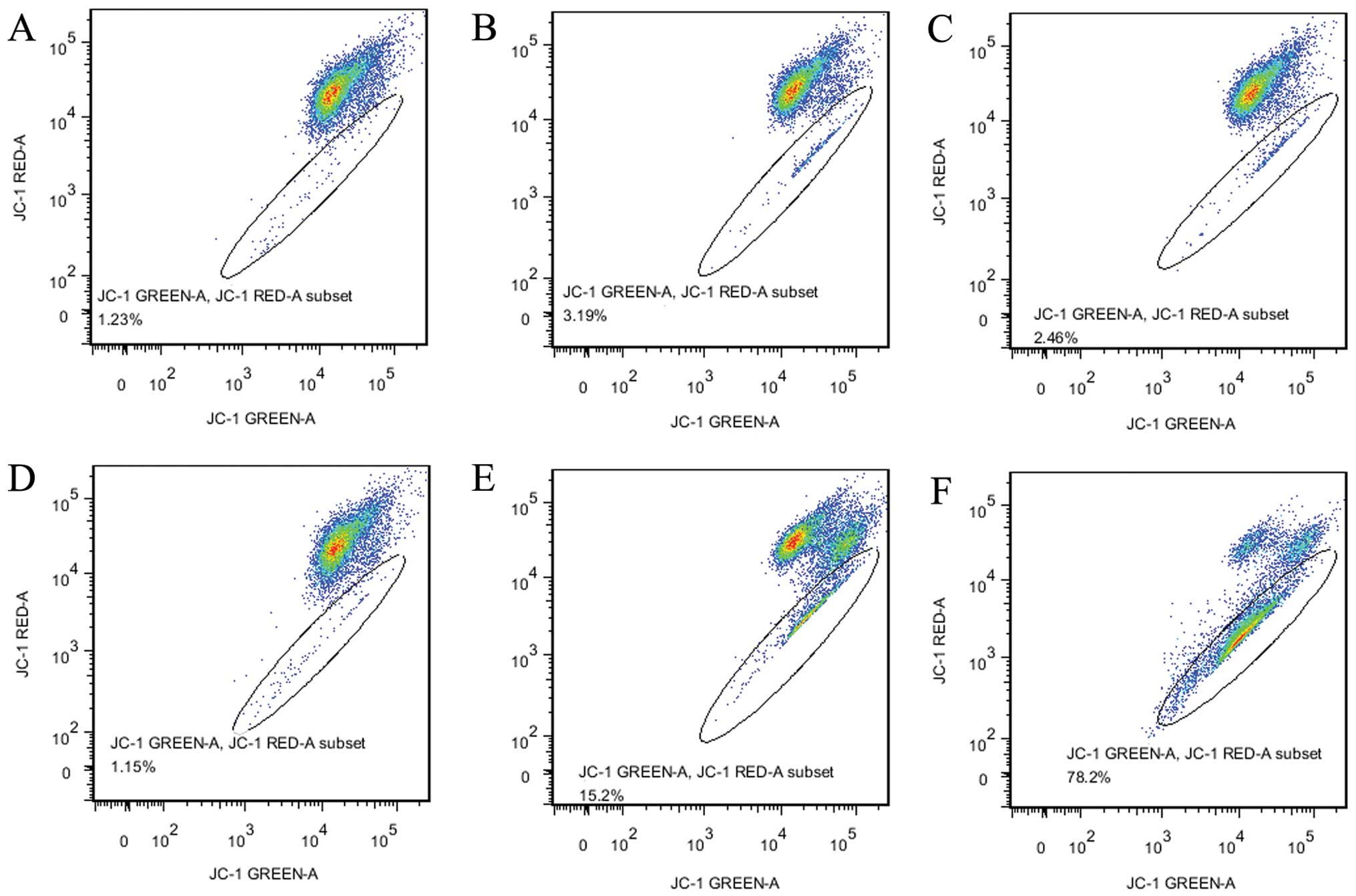
voltage-sensitive fluorescent dye JC-1 to stain gastric cancer AGS
cells that were treated for 24 h with low-glucose medium or medium
with NEA. The ratio of JC-1 red (595 nm)/green (535 nm) represents
the percentage of cells undergoing apoptosis. The ratio of JC-1
red/green in AGS cells is shown in Fig.
5. We determined that there were no significant differences in
mitochondrial membrane potential between the groups cultured with
or without NEA at 2,000 or 1,000 DG (2.45–1.23 and 1.15–3.19%,
respectively), but a significant difference was found between the
500 DG groups with and without NEA (78% of the 500 DG group and
15.2% of the 500 DG-AA group) (Fig. 6C
and D).
Discussion
Gastric cancer, similar to other types of solid
cancer, requires angiogenesis when cancer cells undergo rapid
growth and proliferation (38,39).
Cancer cells require more energy support compared to normal cells,
and glucose is a major source of cellular energy (40). Otherwise, glucose is consumed not
for ATP production but for cell growth and the production of new
cells. Glycolysis is the major mechanism of glucose metabolism in
cancer cells, which is called the Warburg effect, as it was
observed by Otto Warburg for the first time in 1956 (3,41,42).
Without sufficient angiogenesis, glucose starvation or deprivation
is common in fast-growing cancer cells, and autophagy may be an
alternative energy source for cancer cells (6,9). Amino
acids, as the products of autophagy, could serve as a back-up
source of nutrients for rapidly growing malignant cells (13,14).
Based on the above-referenced studies, we inferred that amino acids
could prevent apoptosis of gastric cancer cells undergoing glucose
deprivation.
Here, we presented evidence for the first time
supporting the hypothesis that non-essential amino acids can
attenuate glucose starvation-induced apoptosis of gastric cancer
cells. Survival, apoptosis, the potential of mitochondria,
mitochondria DNA copy number, and amino acid transporter proteins
are all directly or indirectly regulated by non-essential amino
acids added to low-glucose media. These data demonstrated that
non-essential amino acids may be an alternative energy source for
gastric cancer cells under glucose-starved conditions.
Energy deprivation of somatic or cancer cells could
trigger marked reprogramming of cell homeostasis to facilitate the
adjustment of cellular metabolism (43,44).
To elucidate the bio-function of amino acids in gastric cancer
cells undergoing glucose starvation, we adopted a strategy of
treating cancer cells with culture medium containing a glucose
concentration gradient and investigated whether amino acids
performed important effects to attenuate glucose starvation. The
results of a series of experiments measuring proliferation,
apoptosis, and the expression of related genes showed that low
glucose concentration could induce gastric cancer cells to undergo
apoptosis, but non-essential amino acids could attenuate that
effect. Several studies have mentioned that glucose starvation
could result in apoptosis of cells though signaling pathways
(43,45).
The amino acid transporter gene LAT1 has been
reported to be upregulated in the membranes of many cancer cells
(27,28,46–48).
This protein transports amino acids into cells and is different
from two other amino acid transporter genes, namely LAT2 and h4F2hc
(49). Energy deprivation can
trigger response systems to adjust the state of cells, and
increased amino acid intake may be one of those responses.
Several recent studies have shown that amino acids
are associated with proliferation in cancer (50–54).
Jain et al found that 219 metabolites were released from
rapidly proliferating cancer cells, and glycine consumption and the
glycine biosynthetic pathway were strongly correlated with the rate
of proliferation of almost 60 cell lines (50). Zhang et al reported that
glycine may be a critical factor associated with tumor-initiating
cells and tumorigenesis of non-small-cell lung cancer (54). In Arabidopsis thaliana, low
energy status induced the upregulation of the basic leucine zipper
transcriptional factors bZIP1 and bZIP53, which are crucial factors
in proline, asparagine, and branched amino acid metabolism in
response to energy starvation (55). Proline and asparagine are included
in non-essential amino acids. Increasingly more evidence shows that
amino acids may have a critical functional role in proliferation,
resistance to anticancer drugs, invasion or metastasis of cancer
cells. Amino acids are constructed units of proteins and
additionally may serve as an energy resource such as glucose. As
shown in the present study, non-essential amino acids were able to
mitigate the stress caused by glucose starvation.
All cells and tissues have an amino acid sensor
system that can help monitor the state of intercellular amino acids
(56,57). In higher eukaryotic cells, mTOR1 is
believed to regulate protein synthesis in the amino acid signaling
pathway (57). The protein mTOR is
a central cell growth regulator and promotes cell growth and cell
size by controlling protein synthesis (56). At the same time, mTORC can be
stimulated by or control amino acid metabolism through several key
components (such as Rag GTPase), and the signaling is described in
detail in a review (56).
Mitochondria have important roles in various
cellular activities, including apoptosis and energy metabolism
(58). The content of mitochondria
(mitochondrial DNA copy number) changes with the changed status of
cells and may increase or decrease (59–62).
In the present study, we found that non-essential amino acids
maintained a constant mitochondria content in gastric cancer cells
undergoing glucose starvation, which indicates that non-essential
amino acids may reduce the decreasing effect on the content of
mitochondria that are induced by glucose starvation and thus
sustain cell viability (63). The
potential of mitochondria is associated with cell apoptosis and
mitochondria DNA copy number and is also a marker of cell activity.
All the effects that were observed in this study reflected the
finding that non-essential amino acids could increase the activity
of gastric cancer cells facing glucose starvation.
The results of the present study confirmed the
hypothesis that non-essential amino acids and not total amino acid
or glutamine levels were capable of promoting gastric cancer
survival in cells under glucose starvation. The promoting effects
of the non-essential amino acids may be achieved through the
increased expression of amino acid transporter proteins,
upregulation of the expression of anti-apoptotic genes, and
mitochondrial content. Future studies should focus on the signaling
pathways by which amino acids are regulated in response to low
glucose conditions.
Acknowledgements
The present study was supported by grants from the
National Health Key Special Fund (No. 200802112), the Health
Department Fund (No. 2007A093), the Traditional Chinese Medicine
Bureau Fund (No. 2007ZA019), the Natural Science Foundation of
Zhejiang Province (Nos. Y2080001, Y12H160121 and Z2080514), the Key
Project of Zhejiang Province (No. 2009C03012-5 and No.
2013C03044-5), and the National Natural Science Foundation of China
(No. 81372302).
References
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
3
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Denko NC: Hypoxia, HIF1 and glucose
metabolism in the solid tumour. Nat Rev Cancer. 8:705–713. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar
|
|
7
|
Levine B: Cell biology: autophagy and
cancer. Nature. 446:745–747. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mizushima N: Autophagy: process and
function. Genes Dev. 21:2861–2873. 2007. View Article : Google Scholar
|
|
9
|
Degenhardt K, Mathew R, Beaudoin B, et al:
Autophagy promotes tumor cell survival and restricts necrosis,
inflammation, and tumorigenesis. Cancer Cell. 10:51–64. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boya P, González-Polo RA, Casares N, et
al: Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol.
25:1025–1040. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Colell A, Ricci JE, Tait S, et al: GAPDH
and autophagy preserve survival after apoptotic cytochrome c
release in the absence of caspase activation. Cell. 129:983–997.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karantza-Wadsworth V, Patel S, Kravchuk O,
et al: Autophagy mitigates metabolic stress and genome damage in
mammary tumorigenesis. Genes Dev. 21:1621–1635. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuma A, Hatano M, Matsui M, et al: The
role of autophagy during the early neonatal starvation period.
Nature. 432:1032–1036. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Onodera J and Ohsumi Y: Autophagy is
required for maintenance of amino acid levels and protein synthesis
under nitrogen starvation. J Biol Chem. 280:31582–31586. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Yang Y, Yang T, et al:
microRNA-22, downregulated in hepatocellular carcinoma and
correlated with prognosis, suppresses cell proliferation and
tumourigenicity. Br J Cancer. 103:1215–1220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gasz B, Rácz B, Roth E, et al: Pituitary
adenylate cyclase activating polypeptide protects cardiomyocytes
against oxidative stress-induced apoptosis. Peptides. 27:87–94.
2006. View Article : Google Scholar
|
|
17
|
Gartlon J, Kinsner A, Bal-Price A, Coecke
S and Clothier RH: Evaluation of a proposed in vitro test strategy
using neuronal and non-neuronal cell systems for detecting
neurotoxicity. Toxicol In Vitro. 20:1569–1581. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coleman MD, O’Neil JD, Woehrling EK, et
al: A preliminary investigation into the impact of a pesticide
combination on human neuronal and glial cell lines in vitro. PLoS
One. 7:e427682012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cappello C, Zwergal A, Kanclerski S, et
al: C/EBPβ enhances NF-κB-associated signalling by reducing the
level of IκB-α. Cell Signal. 21:1918–1924. 2009.
|
|
20
|
Venegas V, Wang J, Dimmock D and Wong LJ:
Real-time quantitative PCR analysis of mitochondrial DNA content.
Curr Protoc Hum Genet. Unit 19.17:2011.PubMed/NCBI
|
|
21
|
Pola C: Cancer microenvironment: p53 acts
in the hood. Nat Med. 19:5462013. View
Article : Google Scholar
|
|
22
|
Yermolaieva O, Xu R, Schinstock C, et al:
Methionine sulfoxide reductase A protects neuronal cells against
brief hypoxia/reoxygenation. Proc Natl Acad Sci USA. 101:1159–1164.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Finnegan NM, Curtin JF, Prevost G, Morgan
B and Cotter TG: Induction of apoptosis in prostate carcinoma cells
by BH3 peptides which inhibit Bak/Bcl-2 interactions. Br J Cancer.
85:115–121. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hardwick JM and Soane L: Multiple
functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol.
5:a0087222013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou F, Yang Y and Xing D: Bcl-2 and
Bcl-xL play important roles in the crosstalk between autophagy and
apoptosis. FEBS J. 278:403–413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kaira K, Oriuchi N, Takahashi T, et al:
LAT1 expression is closely associated with hypoxic markers and mTOR
in resected non-small cell lung cancer. Am J Transl Res. 3:468–478.
2011.PubMed/NCBI
|
|
27
|
Fukumoto S, Hanazono K, Komatsu T, Iwano
H, Kadosawa T and Uchide T: L-type amino acid transporter 1 (LAT1)
expression in canine mammary gland tumors. J Vet Med Sci.
75:431–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kaira K, Oriuchi N, Takahashi T, et al:
L-type amino acid transporter 1 (LAT1) expression in malignant
pleural mesothelioma. Anticancer Res. 31:4075–4082. 2011.PubMed/NCBI
|
|
29
|
Kaira K, Oriuchi N, Imai H, et al: L-type
amino acid transporter 1 (LAT1) is frequently expressed in thymic
carcinomas but is absent in thymomas. J Surg Oncol. 99:433–438.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ohkawa M, Ohno Y, Masuko K, et al:
Oncogenicity of L-type amino-acid transporter 1 (LAT1) revealed by
targeted gene disruption in chicken DT40 cells: LAT1 is a promising
molecular target for human cancer therapy. Biochem Biophys Res
Commun. 406:649–655. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
del Amo EM, Urtti A and Yliperttula M:
Pharmacokinetic role of L-type amino acid transporters LAT1 and
LAT2. Eur J Pharm Sci. 35:161–174. 2008.PubMed/NCBI
|
|
32
|
Dickens D, Webb SD, Antonyuk S, et al:
Transport of gabapentin by LAT1 (SLC7A5). Biochem Pharmacol.
85:1672–1683. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parr RL, Mills J, Harbottle A, et al:
Mitochondria, prostate cancer, and biopsy sampling error. Discov
Med. 15:213–220. 2013.PubMed/NCBI
|
|
34
|
Bratic A and Larsson NG: The role of
mitochondria in aging. J Clin Invest. 123:951–957. 2013. View Article : Google Scholar
|
|
35
|
Purdue MP, Hofmann JN, Colt JS, et al: A
case-control study of peripheral blood mitochondrial DNA copy
number and risk of renal cell carcinoma. PLoS One. 7:e431492012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qian W and Van Houten B: Alterations in
bioenergetics due to changes in mitochondrial DNA copy number.
Methods. 51:452–457. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang BC and Hays L: Mitochondrial DNA
copy number changes in human gliomas. Cancer Lett. 105:167–173.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kitadai Y: Angiogenesis and
lymphangiogenesis of gastric cancer. J Oncol. 4687252010.PubMed/NCBI
|
|
39
|
Maehara Y, Hasuda S, Abe T, et al: Tumor
angiogenesis and micrometastasis in bone marrow of patients with
early gastric cancer. Clin Cancer Res. 4:2129–2134. 1998.PubMed/NCBI
|
|
40
|
Lunt SY and Vander Heiden MG: Aerobic
glycolysis: meeting the metabolic requirements of cell
proliferation. Annu Rev Cell Dev Biol. 27:441–464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ferguson EC and Rathmell JC: New roles for
pyruvate kinase M2: working out the Warburg effect. Trends Biochem
Sci. 33:359–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: the metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009.PubMed/NCBI
|
|
43
|
Mendivil-Perez M, Jimenez-Del-Rio M and
Velez-Pardo C: Glucose starvation induces apoptosis in a model of
acute T leukemia dependent on caspase-3 and apoptosis-inducing
factor: a therapeutic strategy. Nutr Cancer. 65:99–109. 2013.
View Article : Google Scholar
|
|
44
|
Liu K, Tang Q, Fu C, et al: Influence of
glucose starvation on the pathway of death in insect cell line Sl:
apoptosis follows autophagy. Cytotechnology. 54:97–105. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moruno-Manchón JF1, Pérez-Jiménez E and
Knecht E: Glucose induces autophagy under starvation conditions by
a p38 MAPK-dependent pathway. Biochem J. 449:497–506.
2013.PubMed/NCBI
|
|
46
|
Hayashi K, Jutabha P, Endou H and Anzai N:
c-Myc is crucial for the expression of LAT1 in MIA Paca-2 human
pancreatic cancer cells. Oncol Rep. 28:862–866. 2012.PubMed/NCBI
|
|
47
|
Haining Z, Kawai N, Miyake K, et al:
Relation of LAT1/4F2hc expression with pathological grade,
proliferation and angiogenesis in human gliomas. BMC Clin Pathol.
12:42012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ichinoe M, Mikami T, Yoshida T, et al:
High expression of L-type amino-acid transporter 1 (LAT1) in
gastric carcinomas: comparison with non-cancerous lesions. Pathol
Int. 61:281–289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Khunweeraphong N, Nagamori S,
Wiriyasermkul P, et al: Establishment of stable cell lines with
high expression of heterodimers of human 4F2hc and human amino acid
transporter LAT1 or LAT2 and delineation of their differential
interaction with α-alkyl moieties. J Pharmacol Sci. 119:368–380.
2012.PubMed/NCBI
|
|
50
|
Jain M, Nilsson R, Sharma S, et al:
Metabolite profiling identifies a key role for glycine in rapid
cancer cell proliferation. Science. 336:1040–1044. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Maddocks OD, Berkers CR, Mason SM, et al:
Serine starvation induces stress and p53-dependent metabolic
remodelling in cancer cells. Nature. 493:542–546. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Filipp FV, Ratnikov B, De Ingeniis J,
Smith JW, Osterman AL and Scott DA: Glutamine-fueled mitochondrial
metabolism is decoupled from glycolysis in melanoma. Pigment Cell
Melanoma Res. 25:732–739. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shyh-Chang N, Locasale JW, Lyssiotis CA,
et al: Influence of threonine metabolism on
S-adenosylmethionine and histone methylation. Science.
339:222–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang WC, Shyh-Chang N, Yang H, et al:
Glycine decarboxylase activity drives non-small cell lung cancer
tumor-initiating cells and tumorigenesis. Cell. 148:259–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dietrich K, Weltmeier F, Ehlert A, et al:
Heterodimers of the Arabidopsis transcription factors bZIP1
and bZIP53 reprogram amino acid metabolism during low energy
stress. Plant Cell. 23:381–395. 2011.
|
|
56
|
Kim J and Guan KL: Amino acid signaling in
TOR activation. Annu Rev Biochem. 80:1001–1032. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lamb RF: Amino acid sensing mechanisms: an
Achilles heel in cancer? FEBS J. 279:2624–2631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Crimi M and Rigolio R: The mitochondrial
genome, a growing interest inside an organelle. Int J Nanomed.
3:51–57. 2008.PubMed/NCBI
|
|
59
|
Thyagarajan B, Wang R, Nelson H, Barcelo
H, Koh WP and Yuan JM: Mitochondrial DNA copy number is associated
with breast cancer risk. PLoS One. 8:e659682013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mondal R, Ghosh SK, Choudhury JH, et al:
Mitochondrial DNA copy number and risk of oral cancer: a report
from Northeast India. PLoS One. 8:e577712013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Monnot S, Samuels DC, Hesters L, et al:
Mutation dependance of the mitochondrial DNA copy number in the
first stages of human embryogenesis. Hum Mol Genet. 22:1867–1872.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yu M, Wan Y and Zou Q: Reduced
mitochondrial DNA copy number in Chinese patients with
osteosarcoma. Transl Res. 161:165–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gomes LC, Di Benedetto G and Scorrano L:
During autophagy mitochondria elongate, are spared from degradation
and sustain cell viability. Nat Cell Biol. 13:589–598. 2011.
View Article : Google Scholar : PubMed/NCBI
|