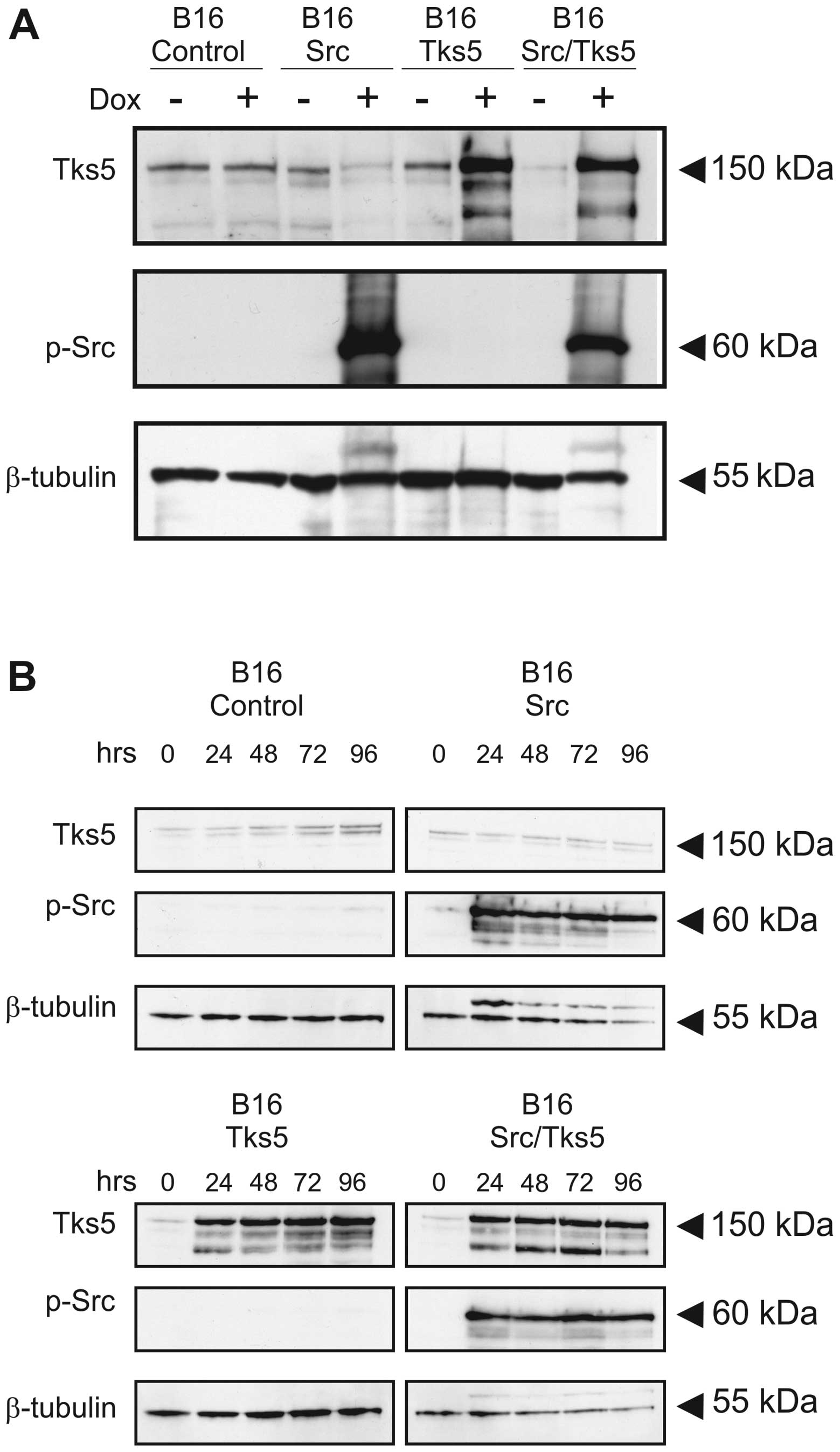
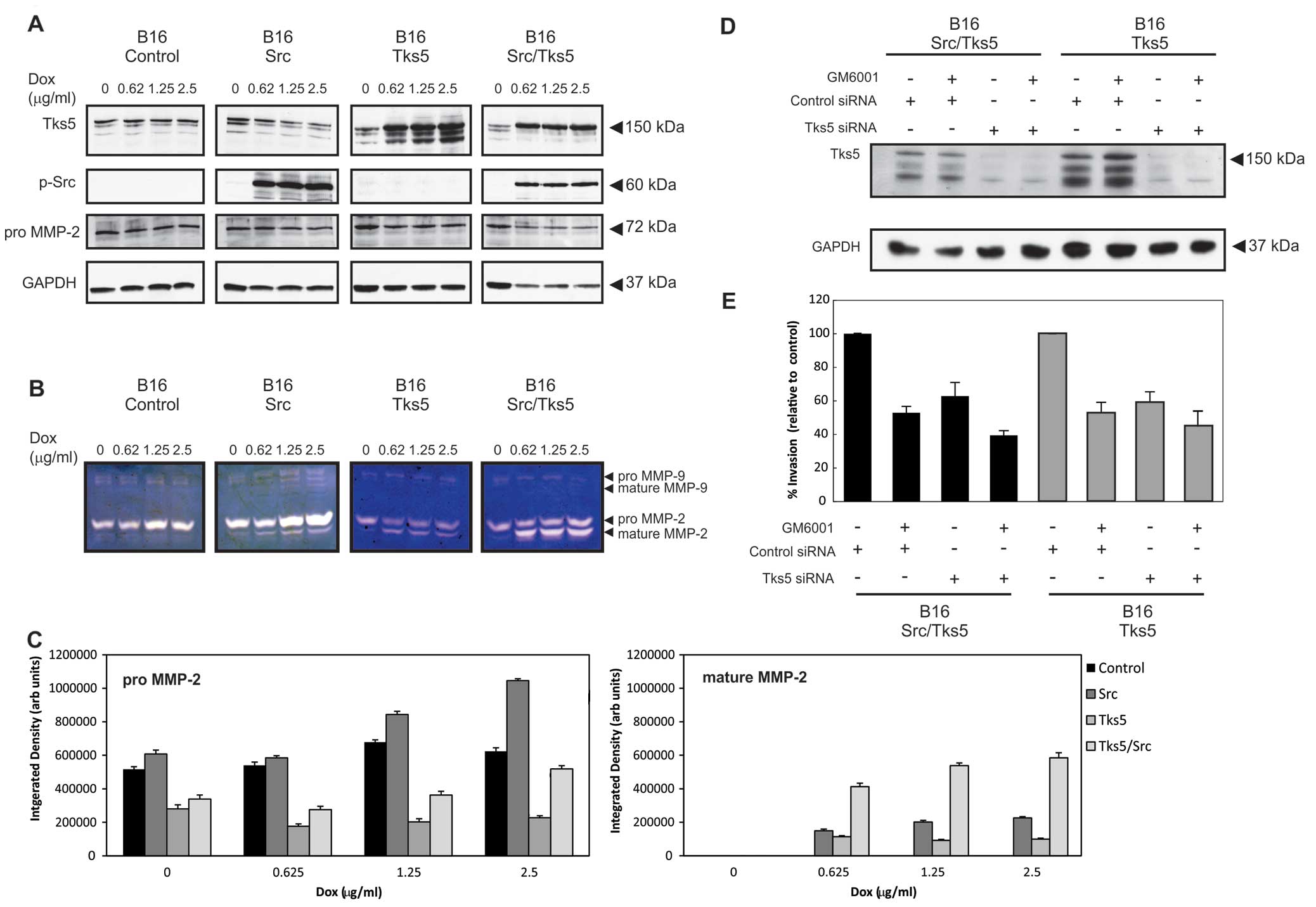
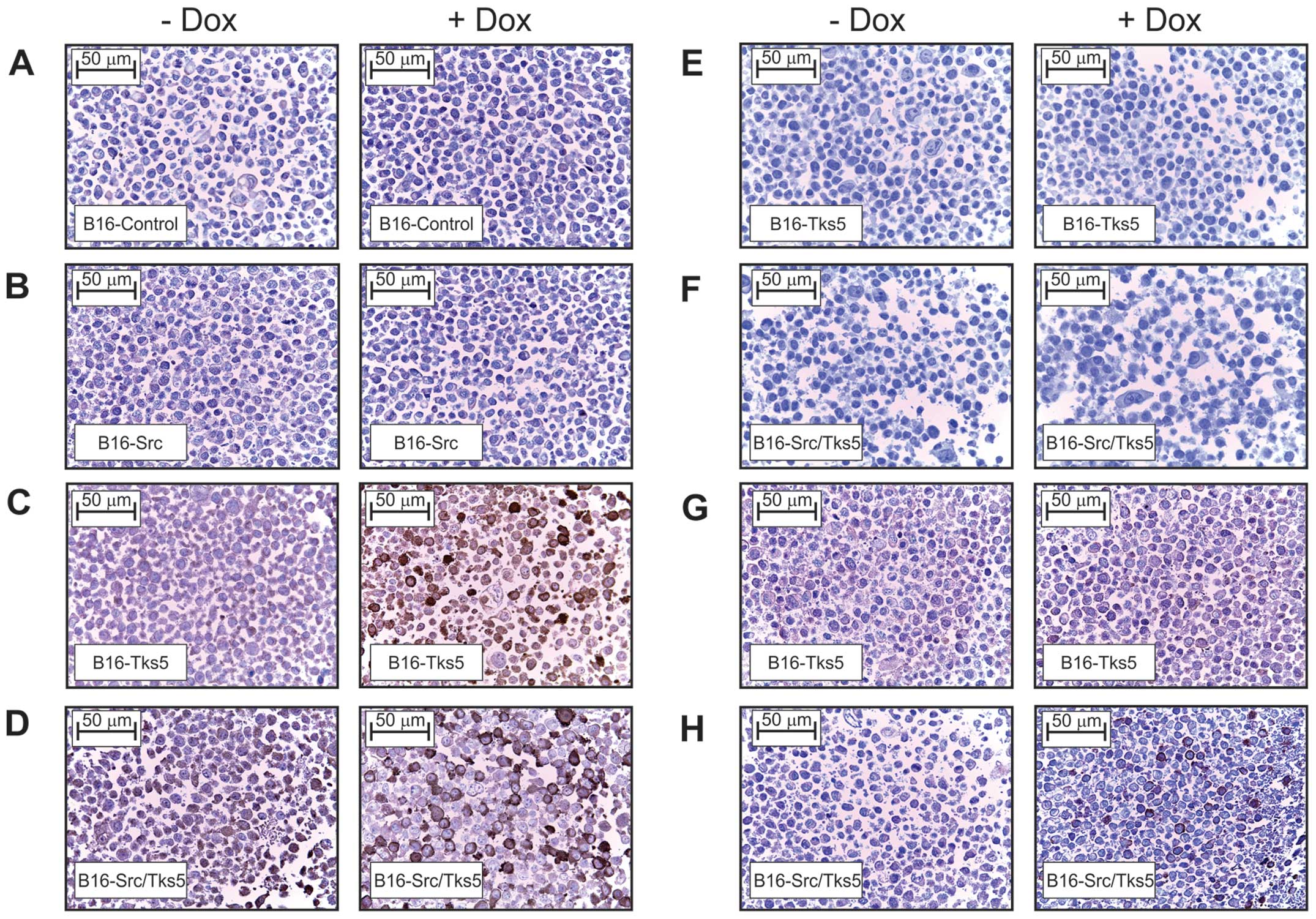
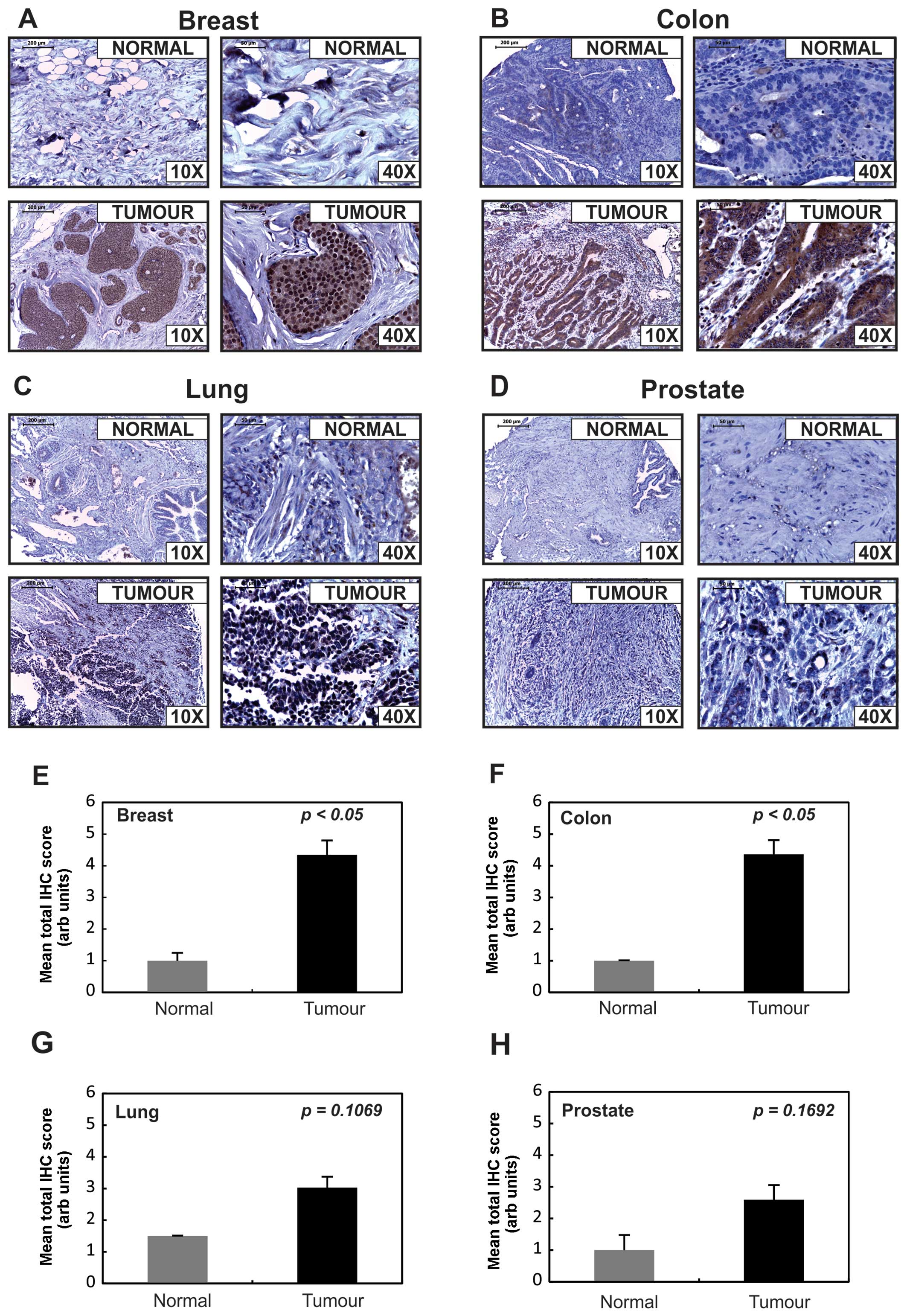
|
1
|
Sporn MB: The war on cancer. Lancet.
347:1377–1381. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abram CL, Seals DF, Pass I, et al: The
adaptor protein fish associates with members of the ADAMs family
and localizes to podosomes of Src-transformed cells. J Biol Chem.
278:16844–16851. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bowden ET, Barth M, Thomas D, Glazer RI
and Mueller SC: An invasion-related complex of cortactin, paxillin
and PKCμ associates with invadopodia at sites of extracellular
matrix degradation. Oncogene. 18:4440–4449. 1999.PubMed/NCBI
|
|
5
|
Buccione R, Caldieri G and Ayala I:
Invadopodia: specialized tumor cell structures for the focal
degradation of the extracellular matrix. Cancer Metastasis Rev.
28:137–149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clark ES and Weaver AM: A new role for
cortactin in invadopodia: regulation of protease secretion. Eur J
Cell Biol. 87:581–590. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Clark ES, Whigham AS, Yarbrough WG and
Weaver AM: Cortactin is an essential regulator of matrix
metalloproteinase secretion and extracellular matrix degradation in
invadopodia. Cancer Res. 67:4227–4235. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gimona M, Buccione R, Courtneidge SA and
Linder S: Assembly and biological role of podosomes and
invadopodia. Curr Opin Cell Biol. 20:235–241. 2008. View Article : Google Scholar
|
|
9
|
Linder S: The matrix corroded: podosomes
and invadopodia in extracellular matrix degradation. Trends Cell
Biol. 17:107–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Linder S and Aepfelbacher M: Podosomes:
adhesion hot-spots of invasive cells. Trends Cell Biol. 13:376–385.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weaver AM: Invadopodia: specialized cell
structures for cancer invasion. Clin Exp Metastasis. 23:97–105.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weaver AM: Cortactin in tumor
invasiveness. Cancer Lett. 265:157–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Webb BA, Jia L, Eves R and Mak AS:
Dissecting the functional domain requirements of cortactin in
invadopodia formation. Eur J Cell Biol. 86:189–206. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamaguchi H, Pixley F and Condeelis J:
Invadopodia and podosomes in tumor invasion. Eur J Cell Biol.
85:213–218. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Artym VV, Zhang Y, Seillier-Moiseiwitsch
F, Yamada KM and Mueller SC: Dynamic interactions of cortactin and
membrane type 1 matrix metalloproteinase at invadopodia: defining
the stages of invadopodia formation and function. Cancer Res.
66:3034–3043. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Daly RJ: Cortactin signalling and dynamic
actin networks. Biochem J. 382:13–25. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Tondravi M, Liu J, et al: Cortactin
potentiates bone metastasis of breast cancer cells. Cancer Res.
61:6906–6911. 2001.PubMed/NCBI
|
|
18
|
Yamaguchi H, Lorenz M, Kempiak S, et al:
Molecular mechanisms of invadopodium formation: the role of the
N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol.
168:441–452. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Staub E, Groene J, Heinze M, et al: An
expression module of WIPF1-coexpressed genes identifies patients
with favorable prognosis in three tumor types. J Mol Med.
87:633–644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
21
|
Pao W, Miller V, Zakowski M, et al: EGF
receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
|
|
22
|
Zeineldin R, Muller CY, Stack MS and
Hudson LG: Targeting the EGF receptor for ovarian cancer therapy. J
Oncol. 2010:4146762010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lock P, Abram CL, Gibson T and Courtneidge
SA: A new method for isolating tyrosine kinase substrates used to
identify fish, an SH3 and PX domain-containing protein, and Src
substrate. The EMBO J. 17:4346–4357. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stylli SS, Stacey TT, Verhagen AM, et al:
Nck adaptor proteins link Tks5 to invadopodia actin regulation and
ECM degradation. J Cell Sci. 122:2727–2740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fekete A, Bőgel G, Pesti S, Péterfi Z,
Geiszt M and Buday L: EGF regulates tyrosine phosphorylation and
membrane-translocation of the scaffold protein Tks5. J Mol Signal.
8:82013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Balzer EM, Whipple RA, Thompson K, et al:
c-Src differentially regulates the functions of microtentacles and
invadopodia. Oncogene. 29:6402–6408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oikawa T, Itoh T and Takenawa T:
Sequential signals toward podosome formation in NIH-src cells. J
Cell Biol. 182:157–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seals DF, Azucena EF Jr, Pass I, et al:
The adaptor protein Tks5/Fish is required for podosome formation
and function, and for the protease-driven invasion of cancer cells.
Cancer Cell. 7:155–165. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Murphy DA, Diaz B, Bromann PA, et al: A
Src-Tks5 pathway is required for neural crest cell migration during
embryonic development. PLoS One. 6:e224992011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stylli SS, I ST, Kaye AH and Lock P:
Prognostic significance of Tks5 expression in gliomas. J Clin
Neurosci. 19:436–442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu HJ, Iaria J and Sizeland AM: Smad7
differentially regulates transforming growth factor β-mediated
signaling pathways. J Biol Chem. 274:32258–32264. 1999.
|
|
32
|
Rhodes DR, Yu J, Shanker K, et al:
ONCOMINE: a cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ayala I, Baldassarre M, Caldieri G and
Buccione R: Invadopodia: a guided tour. Eur J Cell Biol.
85:159–164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baldassarre M, Pompeo A, Beznoussenko G,
et al: Dynamin participates in focal extracellular matrix
degradation by invasive cells. Mol Biol Cell. 14:1074–1084. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar
|
|
36
|
Deryugina EI, Ratnikov B, Monosov E, et
al: MT1-MMP initiates activation of pro-MMP-2 and integrin αvβ3
promotes maturation of MMP-2 in breast carcinoma cells. Exp Cell
Res. 263:209–223. 2001.
|
|
37
|
Finak G, Bertos N, Pepin F, et al: Stromal
gene expression predicts clinical outcome in breast cancer. Nat
Med. 14:518–527. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Curtis C, Shah SP, Chin SF, et al: The
genomic and transcriptomic architecture of 2,000 breast tumours
reveals novel subgroups. Nature. 486:346–352. 2012.PubMed/NCBI
|
|
39
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11:R72009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Buchholz M, Braun M, Heidenblut A, et al:
Transcriptome analysis of microdissected pancreatic intraepithelial
neoplastic lesions. Oncogene. 24:6626–6636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.
|
|
42
|
Iacobuzio-Donahue CA, Maitra A, Olsen M,
et al: Exploration of global gene expression patterns in pancreatic
adenocarcinoma using cDNA microarrays. Am J Pathol. 162:1151–1162.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen X, Leung SY, Yuen ST, et al:
Variation in gene expression patterns in human gastric cancers. Mol
Biol Cell. 14:3208–3215. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cho JY, Lim JY, Cheong JH, et al: Gene
expression signaturebased prognostic risk score in gastric cancer.
Clin Cancer Res. 17:1850–1857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
D’Errico M, de Rinaldis E, Blasi MF, et
al: Genome-wide expression profile of sporadic gastric cancers with
microsatellite instability. Eur J Cancer. 45:461–469.
2009.PubMed/NCBI
|
|
46
|
Korkola JE, Houldsworth J, Chadalavada RS,
et al: Down-regulation of stem cell genes, including those in a
200-kb gene cluster at 12p13.31, is associated with in vivo
differentiation of human male germ cell tumors. Cancer Res.
66:820–827. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Barretina J, Taylor BS, Banerji S, et al:
Subtype-specific genomic alterations define new targets for
soft-tissue sarcoma therapy. Nat Genet. 42:715–721. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Skotheim RI, Lind GE, Monni O, et al:
Differentiation of human embryonal carcinomas in vitro and in vivo
reveals expression profiles relevant to normal development. Cancer
Res. 65:5588–5598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yusenko MV, Kuiper RP, Boethe T, Ljungberg
B, van Kessel AG and Kovacs G: High-resolution DNA copy number and
gene expression analyses distinguish chromophobe renal cell
carcinomas and renal oncocytomas. BMC Cancer. 9:1522009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nindl I, Dang C, Forschner T, et al:
Identification of differentially expressed genes in cutaneous
squamous cell carcinoma by microarray expression profiling. Mol
Cancer. 5:302006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Talantov D, Mazumder A, Yu JX, et al:
Novel genes associated with malignant melanoma but not benign
melanocytic lesions. Clin Cancer Res. 11:7234–7242. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bredel M, Bredel C, Juric D, et al:
Functional network analysis reveals extended gliomagenesis pathway
maps and three novel MYC-interacting genes in human gliomas. Cancer
Res. 65:8679–8689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hou J, Aerts J, den Hamer B, et al: Gene
expression-based classification of non-small cell lung carcinomas
and survival prediction. PLoS One. 5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kaiser S, Park YK, Franklin JL, et al:
Transcriptional recapitulation and subversion of embryonic colon
development by mouse colon tumor models and human colon cancer.
Genome Biol. 8:R1312007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Aoyagi K, Tatsuta T, Nishigaki M, et al: A
faithful method for PCR-mediated global mRNA amplification and its
integration into microarray analysis on laser-captured cells.
Biochem Biophys Res Commun. 300:915–920. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
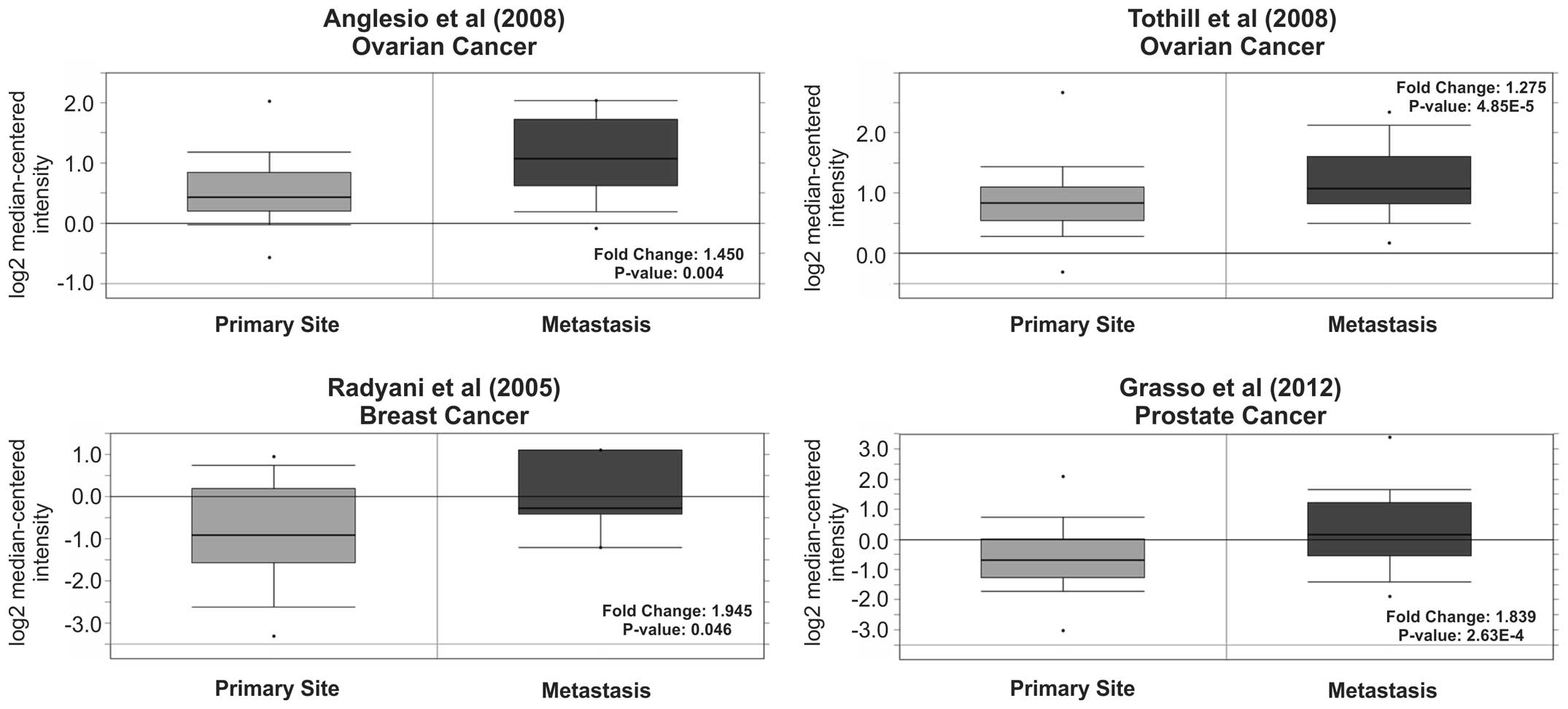
Anglesio MS, Arnold JM, George J, et al:
Mutation of ERBB2 provides a novel alternative mechanism for the
ubiquitous activation of RAS-MAPK in ovarian serous low malignant
potential tumors. Mol Cancer Res. 6:1678–1690. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tothill RW, Tinker AV, George J, et al:
Novel molecular subtypes of serous and endometrioid ovarian cancer
linked to clinical outcome. Clin Cancer Res. 14:5198–5208. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Radvanyi L, Singh-Sandhu D, Gallichan S,
et al: The gene associated with trichorhinophalangeal syndrome in
humans is overexpressed in breast cancer. Proc Natl Acad Sci USA.
102:11005–11010. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Grasso CS, Wu YM, Robinson DR, et al: The
mutational landscape of lethal castration-resistant prostate
cancer. Nature. 487:239–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
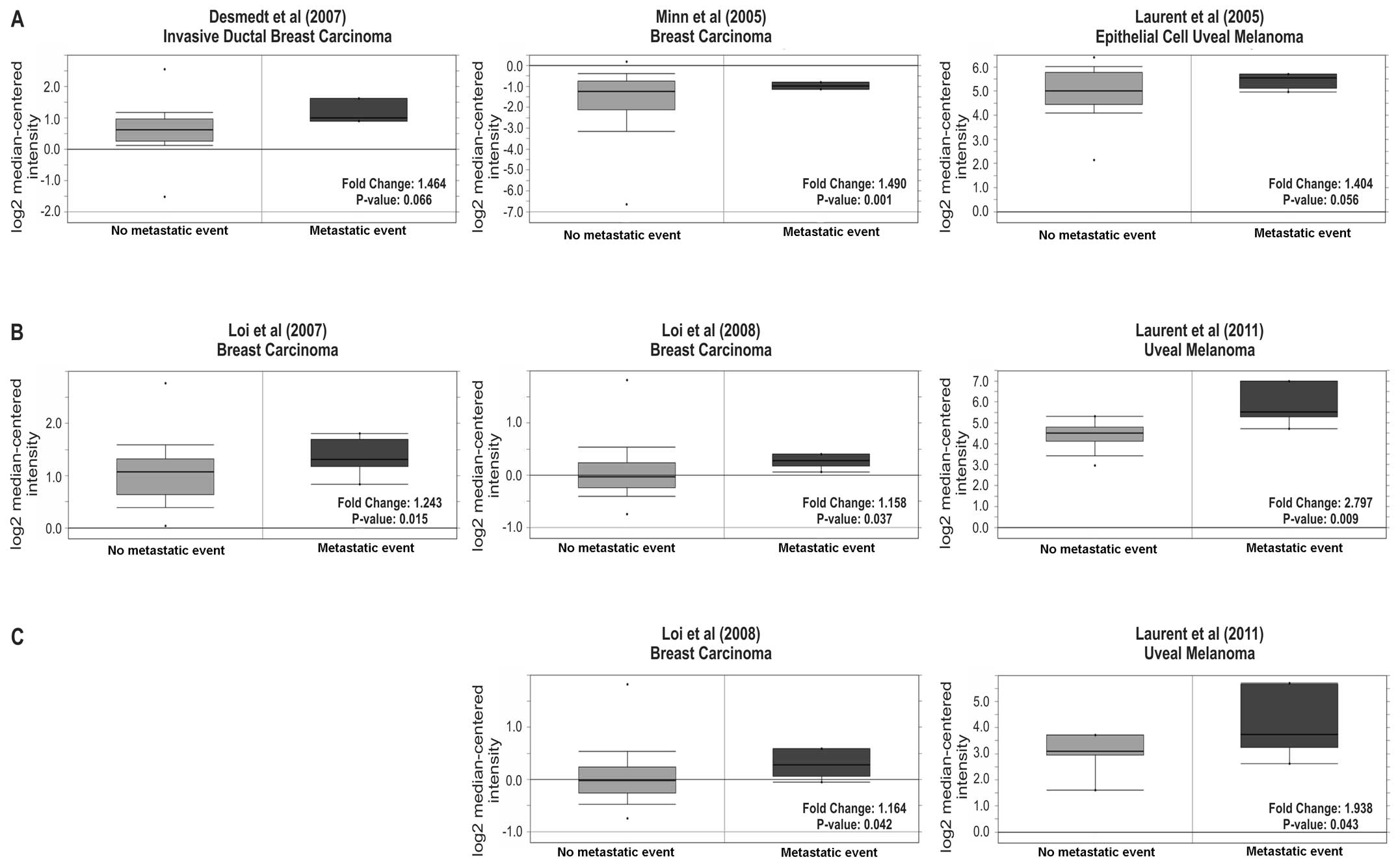
Desmedt C, Piette F, Loi S, et al: Strong
time dependence of the 76-gene prognostic signature for
node-negative breast cancer patients in the TRANSBIG multicenter
independent validation series. Clin Cancer Res. 13:3207–3214. 2007.
View Article : Google Scholar
|
|
61
|
Minn AJ, Gupta GP, Siegel PM, et al: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Laurent C, Valet F, Planque N, et al: High
PTP4A3 phosphatase expression correlates with metastatic risk in
uveal melanoma patients. Cancer Res. 71:666–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Loi S, Haibe-Kains B, Desmedt C, et al:
Predicting prognosis using molecular profiling in estrogen
receptor-positive breast cancer treated with tamoxifen. BMC
Genomics. 9:2392008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Loi S, Haibe-Kains B, Desmedt C, et al:
Definition of clinically distinct molecular subtypes in estrogen
receptor-positive breast carcinomas through genomic grade. J Clin
Oncol. 25:1239–1246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lambrechts A, Van Troys M and Ampe C: The
actin cytoskeleton in normal and pathological cell motility. Int J
Biochem Cell Biol. 36:1890–1909. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rohatgi R, Nollau P, Ho HY, Kirschner MW
and Mayer BJ: Nck and phosphatidylinositol 4,5-bisphosphate
synergistically activate actin polymerization through the
N-WASP-Arp2/3 pathway. J Biol Chem. 276:26448–26452. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rivera GM, Briceño CA, Takeshima F,
Snapper SB and Mayer BJ: Inducible clustering of membrane-targeted
SH3 domains of the adaptor protein Nck triggers localized actin
polymerization. Curr Biol. 14:11–22. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rowe RG and Weiss SJ: Navigating ECM
barriers at the invasive front: the cancer cell-stroma interface.
Annu Rev Cell Dev Biol. 25:567–595. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bowden ET, Onikoyi E, Slack R, et al:
Co-localization of cortactin and phosphotyrosine identifies active
invadopodia in human breast cancer cells. Exp Cell Res.
312:1240–1253. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kinley AW, Weed SA, Weaver AM, et al:
Cortactin interacts with WIP in regulating Arp2/3 activation and
membrane protrusion. Curr Biol. 13:384–393. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Oser M, Yamaguchi H, Mader CC, et al:
Cortactin regulates cofilin and N-WASp activities to control the
stages of invadopodium assembly and maturation. J Cell Biol.
186:571–587. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Webb BA, Zhou S, Eves R, Shen L, Jia L and
Mak AS: Phosphorylation of cortactin by p21-activated kinase. Arch
Biochem Biophys. 456:183–193. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gibcus JH, Mastik MF, Menkema L, et al:
Cortactin expression predicts poor survival in laryngeal carcinoma.
Br J Cancer. 98:950–955. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Hofman P, Butori C, Havet K, et al:
Prognostic significance of cortactin levels in head and neck
squamous cell carcinoma: comparison with epidermal growth factor
receptor status. Br J Cancer. 98:956–964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sheen-Chen SM, Huang CY, Liu YY, Huang CC
and Tang RP: Cortactin in breast cancer: analysis with tissue
microarray. Anticancer Res. 31:293–297. 2011.PubMed/NCBI
|
|
76
|
Bonome T, Levine DA, Shih J, et al: A gene
signature predicting for survival in suboptimally debulked patients
with ovarian cancer. Cancer Res. 68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Freije WA, Castro-Vargas FE, Fang Z, et
al: Gene expression profiling of gliomas strongly predicts
survival. Cancer Res. 64:6503–6510. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Giordano TJ, Kuick R, Else T, et al:
Molecular classification and prognostication of adrenocortical
tumors by transcriptome profiling. Clin Cancer Res. 15:668–676.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Smith JJ, Deane NG, Wu F, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nakayama R, Mitani S, Nakagawa T, et al:
Gene expression profiling of synovial sarcoma: distinct signature
of poorly differentiated type. Am J Surg Pathol. 34:1599–1607.
2010.PubMed/NCBI
|
|
81
|
Phillips HS, Kharbanda S, Chen R, et al:
Molecular subclasses of high-grade glioma predict prognosis,
delineate a pattern of disease progression, and resemble stages in
neurogenesis. Cancer Cell. 9:157–173. 2006. View Article : Google Scholar : PubMed/NCBI
|