Introduction
Nasopharyngeal carcinoma (NPC) is rare in most
populations, although its incidence is particularly high among
Asians, particularly in Southern China with an age standardized
incidence rate of ~25 cases per 100,000 individuals per year
(1). As a traditional treatment
modality, radiotherapy remains the mainstay of treatment for NPC
due to its radiosensitivity, even though it has been applied for
cancer therapy for more than a century. If diagnosed and treated at
an early stage, most NPC patients can be cured. However,
locoregional recurrence or distant metastasis may occur in some
patients after primary radiotherapy or chemo-irradiation due to the
radiation resistance of cancer cells (2). Due to the presence of tumor cell
heterogeneity, cancer cells may exhibit different degrees of
radiosensitivity even when they are of the same histological
differentiation status, which may contribute to the poor overall
survival of NPC patients after recurrence. It follows, therefore,
that the development of novel therapeutic strategies to overcome
radioresistance and enhance radiosensitivity of NPC are urgently
needed.
In regards to the primary mechanism of
radiation-induced cell death, apoptosis is not the predominant form
of cell death, accounting for only 20% of death (3). Another cell death pathway, namely
autophagy, has emerged as a crucial mechanism of tumor cell death
induced by radiation (4).
Autophagy, a multi-step process that involves degradation of
long-lived cellular proteins and organelles, is a genetically
programmed, highly conserved process that occurs in eukaryotes from
yeast to mammals. Recently, studies have shown that autophagy plays
a vital role in protecting cells against adverse conditions
(5–7), including irradiation (8). Inhibition of autophagy plays an active
role in radiosensitization in several cancer cell types (9) as well as in NPC cells (10). Although many studies have rigorously
attempted to elucidate the mechanism of autophagy in cancer
treatments, it remains unclear.
DNA double-strand breaks (DSBs) are the most
critical event in ionizing radiation (IR)-induced cell death and
can be efficiently repaired by DNA homologous recombination, which
is essential for maintaining genomic stability after IR, while
Rad51 is a key protein of homologous recombination in
resynthesizing the damaged region of the DNA (11). Overexpression of Rad51 may be a
possible mechanism with which to suppress recombination defects and
increase the resistance of mammalian cells to IR (12), suggesting that Rad51 expression may
play an essential role in radioresistance.
Previous research has demonstrated that homologous
recombination may mediate cellular resistance to radiation therapy
(13), and inhibition of autophagy
contributes to the radiosensitivity of CNE-2 cells (14). Therefore, we hypothesized that
inhibition of autophagy could enhance the radiosensitivity of NPC
cells by influencing the DNA homologous recombination system. To
test this hypothesis, we investigated the effect of autophagy
inhibition on the radiosensitivity of NPC cells and elucidated the
role of Rad51 in the regulation of radiosensitivity.
Materials and methods
Cell culture and irradiation
conditions
The human NPC cell lines, CNE-1 and CNE-2, were
obtained from the Chinese Academy of Sciences Cell Bank and were
cultured in Roswell Park Memorial Institute (RPMI)-1640 medium
supplemented with 10% fetal bovine serum, 100 U/ml of penicillin
and 100 μg/ml of streptomycin (all from Invitrogen, Carlsbad, CA,
USA). Cultures were maintained in a humidified atmosphere of 5%
CO2 at 37°C. 3-Methyladenine (3-MA) and chloroquine (CQ)
were obtained from Sigma-Aldrich (Shanghai, China). RI-1 was
purchased from ChemBridge Corp. (San Diego, CA, USA). BO2 was
purchased from Chief-East Tech Co., Ltd. (Beijing, China). All
irradiations were delivered using 6-MV X-rays with a linear
accelerator (Elekta, Sweden) with a dose rate of 220 cGy/min; SSD,
100 cm.
shRNA
A scrambled hairpin (SCR) was used as a negative
control (Invitrogen), and Stealth RNAi™ shRNA duplex
oligoribonucleotides targeting human Atg5 were obtained from
Invitrogen. The sequence of SCR-shRNA was CCT ACG CCA CCA ATT TCG
T; Atg5-1-shRNA was ATT GGC TCA ATT CCA TGA ATC and Atg5-2-shRNA
was AAG CAA ATA GTA TGG TTC TGC. The shRNA was transfected into
CNE1 and CNE2 cells using shRNA transfection reagent (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) according to the
manufacturer’s protocol.
Cell viability assay
The measurement of the viable cell mass was assessed
by the Cell Counting Kit-8 (Dojin Laboratories, Kumamoto, Japan),
as previously described (15).
Real-time PCR assay
Cells were collected to extract the total cellular
mRNA using TRIzol reagent (Invitrogen). cDNA was synthesized using
Moloney murine leukemia virus reverse transcriptase (Promega,
Madison, WI, USA) and 2 μg of total RNA and oligo(dT)18
primers. Two-microliter aliquots of cDNA were used for PCR
amplification. Real-time PCR was performed in triplicate using the
SYBR PrimeScript RT-PCR kit (Takara, Dalian, China). The sequences
of the Atg5 primers listed were TGG ATT TCG TTA TAT CC CCT TT AG
(sense) and CCT AGT GTG TGC AAC TGT CCA (antisense); the sequences
of the Rad51 primers listed were 5′-TGG CCC ACA ACC CAT TTC AC-3′
(sense) and 5′-TCA ATG TAC ATG GCC TTT CCT TCA C-3′ (antisense).
Total sample RNA was normalized to endogenous human
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Thermocycler
conditions included an initial hold at 50°C for 2 min and then 95°C
for 10 min; this was followed by a two-step PCR program of 95°C for
15 sec and 60°C for 60 sec repeated for 40 cycles on an Mx4000
system (Stratagene, La Jolla, CA, USA), on which data were
collected and quantitatively analyzed. The expression level of mRNA
was presented as fold-change relative to an untreated control.
Western blot analysis
Cells were washed in PBS solution, and the protein
was extracted according to an established protocol. Nuclear extract
proteins were quantified using the Bio-Rad protein assay. Proteins
were then mixed with Laemmli sample buffer, heated at 65°C for 10
min, loaded (20 μg/sample), separated by SDS-polyacrylamide gel
(7.5%) electrophoresis under denaturing conditions and
electroblotted on nitrocellulose membranes. The nitrocellulose
membranes were blocked by incubation in blocking buffer (1% BSA in
Tris-buffered saline, 0.1% Tween-20), incubated with the anti-Atg5
antibody (1:500 polyclonal; Bethyl), washed and incubated with the
anti-rabbit peroxidase-conjugated secondary antibody (1:10,000;
Sigma). Signals were visualized by chemiluminescent detection.
Blots were quantified using Quantity One software (Bio-Rad), and
Atg5 (Epitomics, Inc., USA) expression (peak intensity) was
normalized to values in the control group. Equal loading of samples
was verified by Coomassie blue staining of simultaneously run gels.
Gels were run four times, and the images shown are
representative.
Cell cycle analysis by flow
cytometry
Approximately 1×106 cells were collected
and fixed overnight in 70% ethanol at 4°C. Cells were then washed
in PBS and stained with PI (Sigma) in the presence of DNase-free
RNase. After a 30-min incubation at room temperature in the dark,
cells were filtered through a 40-μm diameter mesh to remove clumps
of nuclei, and cells within the cell cycle compartments (sub-G1, S
or G2-M) were determined via a flow cytometer using CellQuest
software (BD FACSAria, Becton-Dickinson), acquiring 10,000 events.
The percentages of sub-G1 nuclei in the population were determined
as apoptosis.
Cell apoptosis assay
Cells (2×105 per well) were cultured in
6-well plates to 70–80% confluency. Annexin V-FITC assay was used
to measure apoptotic cells by flow cytometry according to the
manufacturer’s instructions (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China; cat. KGA108). Briefly, cells were collected by
trypsinization, washed with ice-cold phosphate-buffered saline
(PBS) twice and resuspended in 300 μl 1X binding buffer containing
5 μl Annexin V and 5 μl PI for 30 min at room temperature in the
dark. After incubation, at least 10,000 cells were measured on a BD
FACSAria flow cytometer. Results are expressed as the percentage of
apoptotic cells at early stage (PI-negative and Annexin
V-positive). Meanwhile, apoptosis was detected by staining cells
with DAPI based on the nuclear morphology, identifying those cells
with condensed and fragmented nuclei as apoptotic.
Statistical analysis
All of the experiments were repeated at least three
times. The data are expressed as means ± SD. Statistical analysis
was performed using the Student’s t-test (two-tailed). The
criterion for statistical significance was determined to be
P<0.05.
Results
Suppression of Atg5 enhances the
radiosensitivity of human NPC cells
To investigate the function of the autophagy related
5 (Atg5) gene in NPC cells after radiation, specific
lentivirus-delivered shRNA to the Atg5 gene was used to induce Atg5
gene silencing by a mechanism that involves the RNA-induced
silencing complex (RISC). Transduced cells were expanded, and Atg5
mRNA expression was detected by RT-qPCR while the Atg5 protein
level was determined by western blot analysis 5 days after
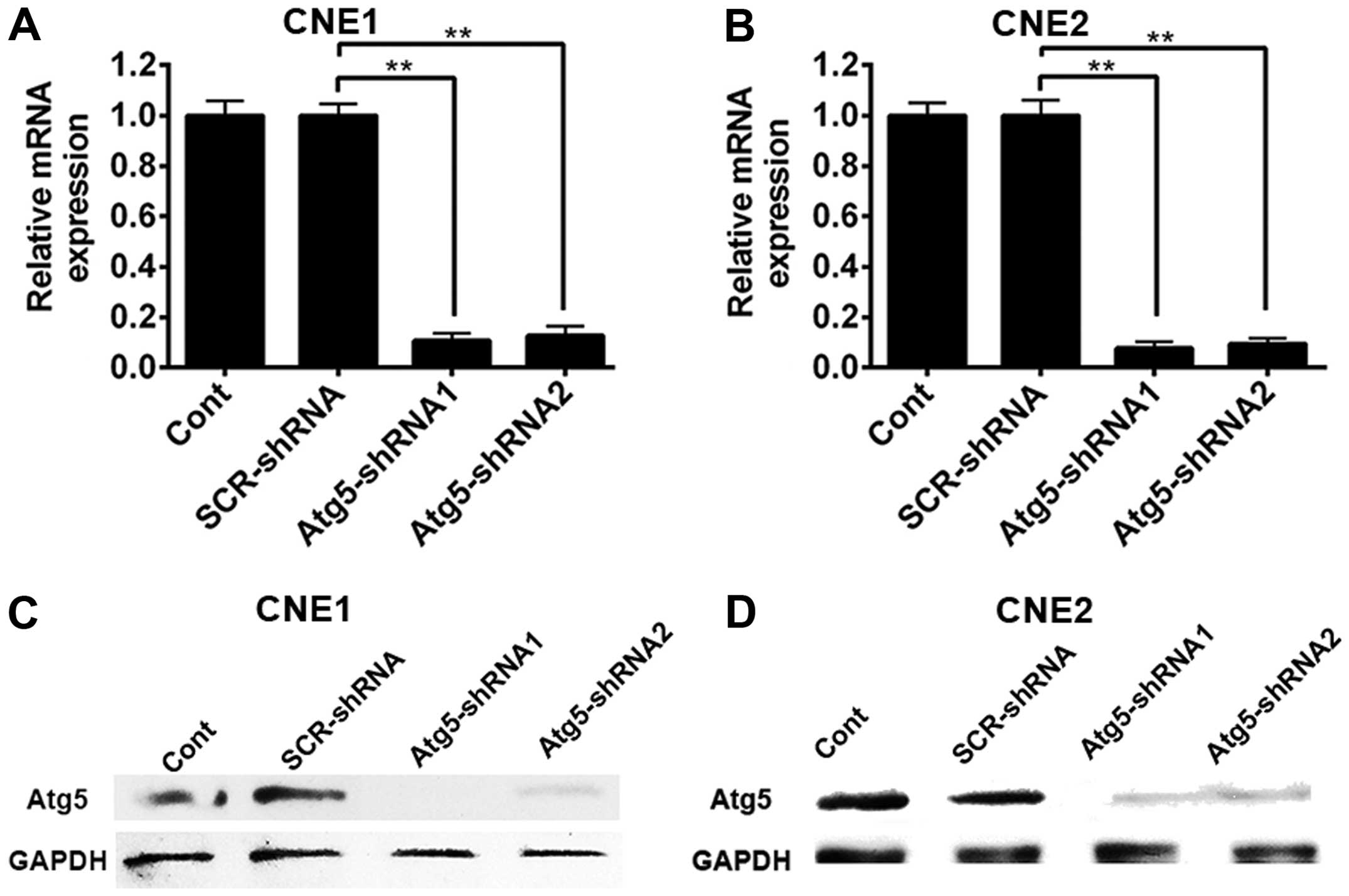
infection. As shown in Fig. 1A,
silencing of the Atg5 gene markedly decreased the expression of
Atg5 mRNA in the CNE-1 cells either transfected with Atg5-targeted
shRNA1 or Atg5-targeted shRNA2 compared with the non-targeted shRNA
CNE-1 cells and control cells. Similarly, transduction of CNE1
cells with the Atg5-shRNA1 and Atg5-shRNA2 construct virus resulted
in nearly complete elimination of Atg5 protein expression (Fig. 1C). In contrast, the levels of Atg5
protein in cells transduced with the lentiviral vector including
the non-specific construct shRNA sequence and control cells did not
exhibit an obvious change when compared with GAPDH. Similar results
were observed in the CNE-2 cells (Fig.
1B and D). The results above suggest that silencing of the Atg5
gene reduced the autophagy of NPC cells.
It has been demonstrated that cells die mainly via
two pathways, necrosis and apoptosis (16). Ionizing radiation (IR) kills cancer
cells mainly through the induction of DNA double-strand breaks
(DSBs), apoptosis as well as autophagy, whereas autophagy is widely
considered to be a protective mechanism against IR. Thus, we
hypothesized that autophagy may protect NPC cells from IR-induced
cell death by reducing apoptosis and DNA damage. To test this, we
further investigated the effect of Atg5 silencing on IR-induced
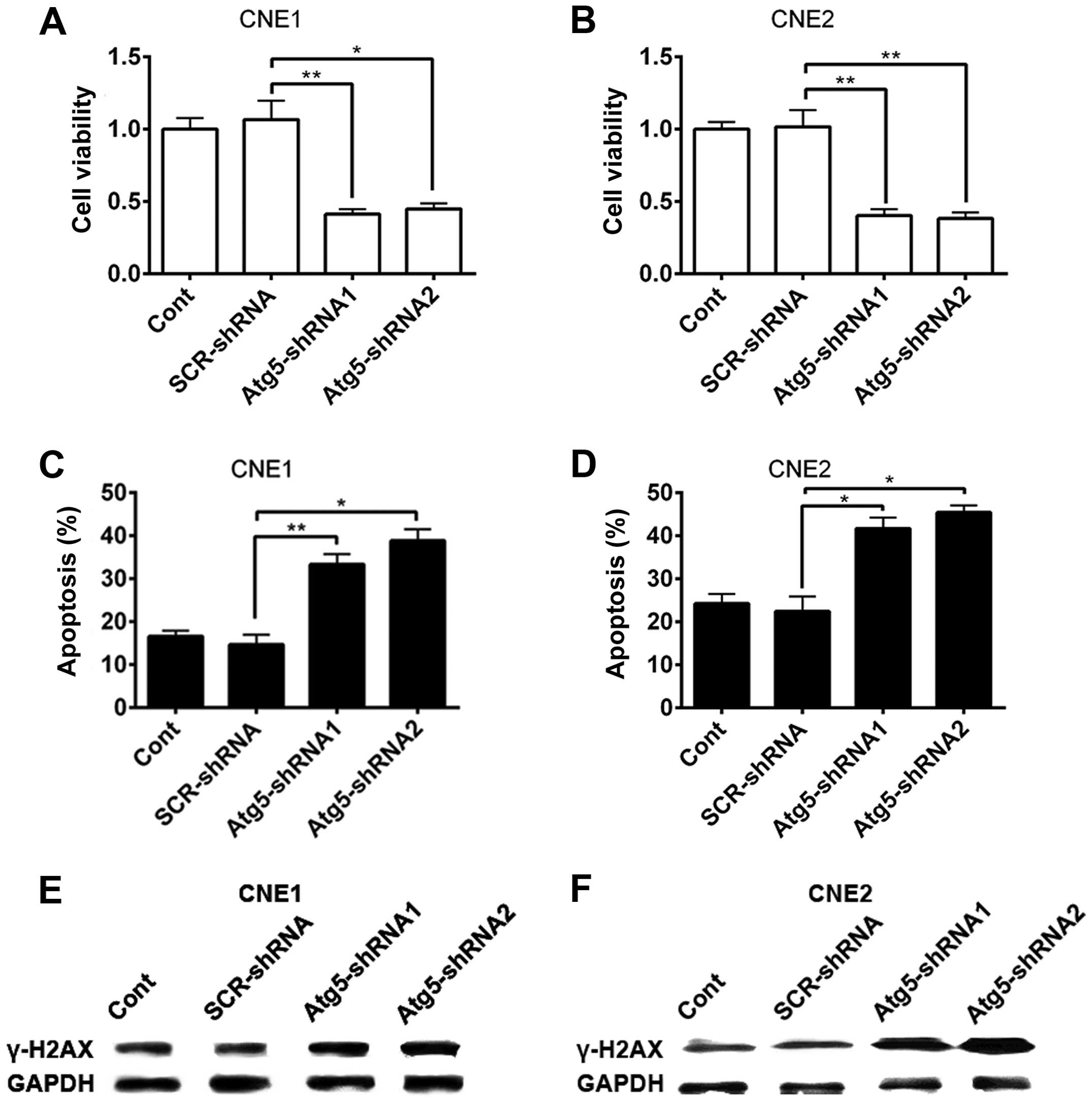
cell apoptosis and DNA damage. Forty-eight hours after 4-Gy X-ray
irradiation treatment, cell viability was evaluated by the CCK-8
assay. As shown in Fig. 2A and B,
following the silencing of the Atg5 gene, the cell viability of
CNE-1 and CNE-2 cells significantly decreased compared with that of
the scrambled and control cells.
The decrease in the viability of the cells
transfected with Atg5-targeted shRNA indicated an increase in cell
death of NPC cells after radiation. Apoptosis, known as type I
programmed cell death, could underlie the biological function of
radiation. We investigated whether there was an enhancement of cell
apoptosis induced by radiation after Atg5 silencing. To evaluate
apoptosis, the percentage of apoptotic cells was assessed by flow
cytometry with Annexin V/PI double staining analysis. Results
showed that CNE-1 cells transfected with the Atg5-targeted shRNA
had a higher proportion of apoptotic cells (Fig. 2C) compared to that of cells
transfected with the non-targeted shRNA and control cells. Similar
data were obtained using CNE-2 cells (Fig. 2D).
The formation of γ-H2AX, namely phosphorylation of a
histone protein H2AX, indicates the early response of cells to
IR-induced DNA damage (17). γ-H2AX
is commonly accepted as a biomarker that indicates the presence of
a DSB. Hence, we detected the protein level of γ-H2AX to confirm
the DNA damage after radiation in NPC cells. Western blot analysis
showed that the protein level of γ-H2AX in the CNE1 and CNE2 cells
was increased in all experimental groups after IR (Fig. 2E and F). However, a higher increase
in the protein level of γ-H2AX was observed as the cells were
pre-interfered by Atg5-targeted shRNA. Together, the results
suggest that suppression of Atg5, to some extent, enhanced the
susceptibility of NPC cells to radiation.
Inhibition of autophagy decreases the
expression of Rad51
The above data confirmed that suppression of Atg5
increased the susceptibility of NPC cells to radiation, while the
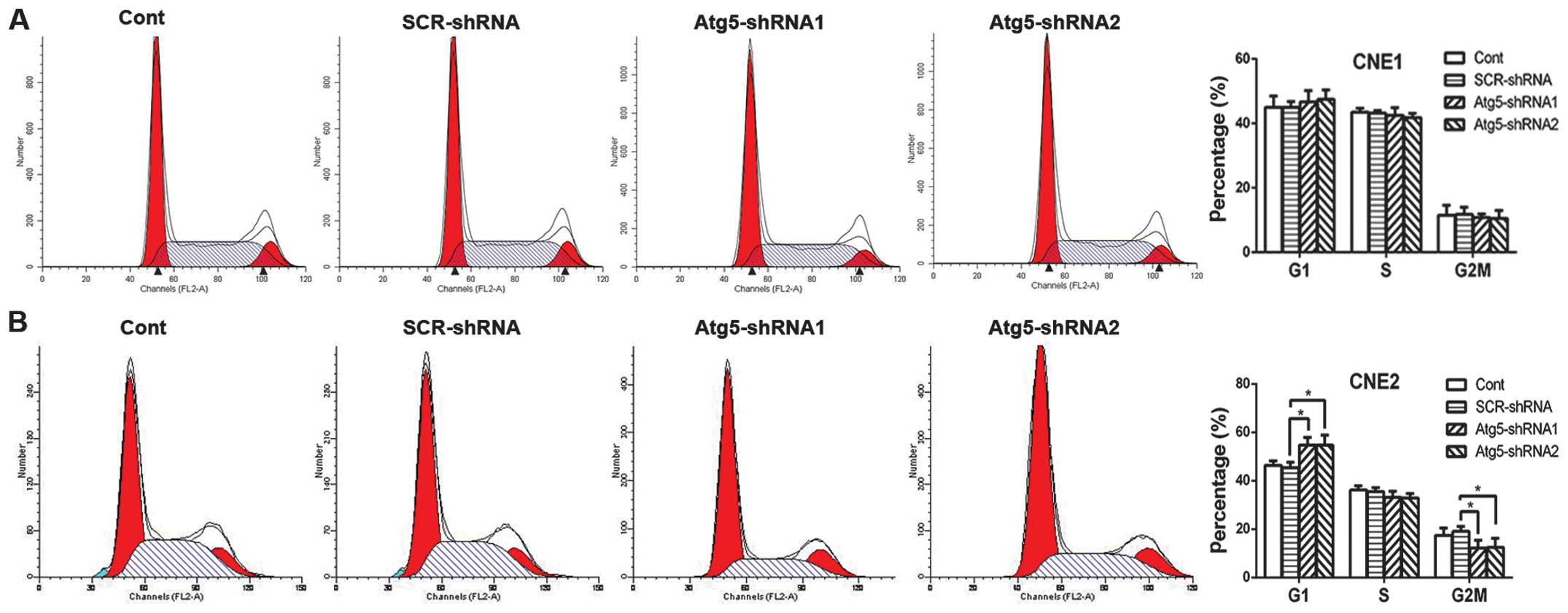
mechanism remains unclear. Radiation induces G2/M phase arrest in
various types of cancer cells (18,19).
This results in enhanced radiosensitivity of cancer cells for the
reason that cells are most radiosensitive in the M and G2 phases.
Therefore, we aimed to ascertain whether the radiosensitization
effect of Atg5 silencing was due to G2/M phase arrest. Contrary to
our expectation, cell cycle analysis by flow cytometric measurement
revealed that suppression of Atg5 led to cell cycle arrest in the
G1 phase where cells are less radiosensitive, while the percentage
of cells in the G2/M phase was decreased dramatically in the CNE-2
cells; both results achieving statistical significance (Fig. 3B). However, no difference in cell
cycle distribution was noted in the CNE-1 cells (Fig. 3A).
Radiation-induced cell damage mainly results in DNA
DSBs (20). Rad51, a key protein of
homologous recombination, has been demonstrated to play a critical
role in the repair of DNA DSBs. We aimed to ascertain whether Rad51
is related to the radiosensitization effect following silencing of
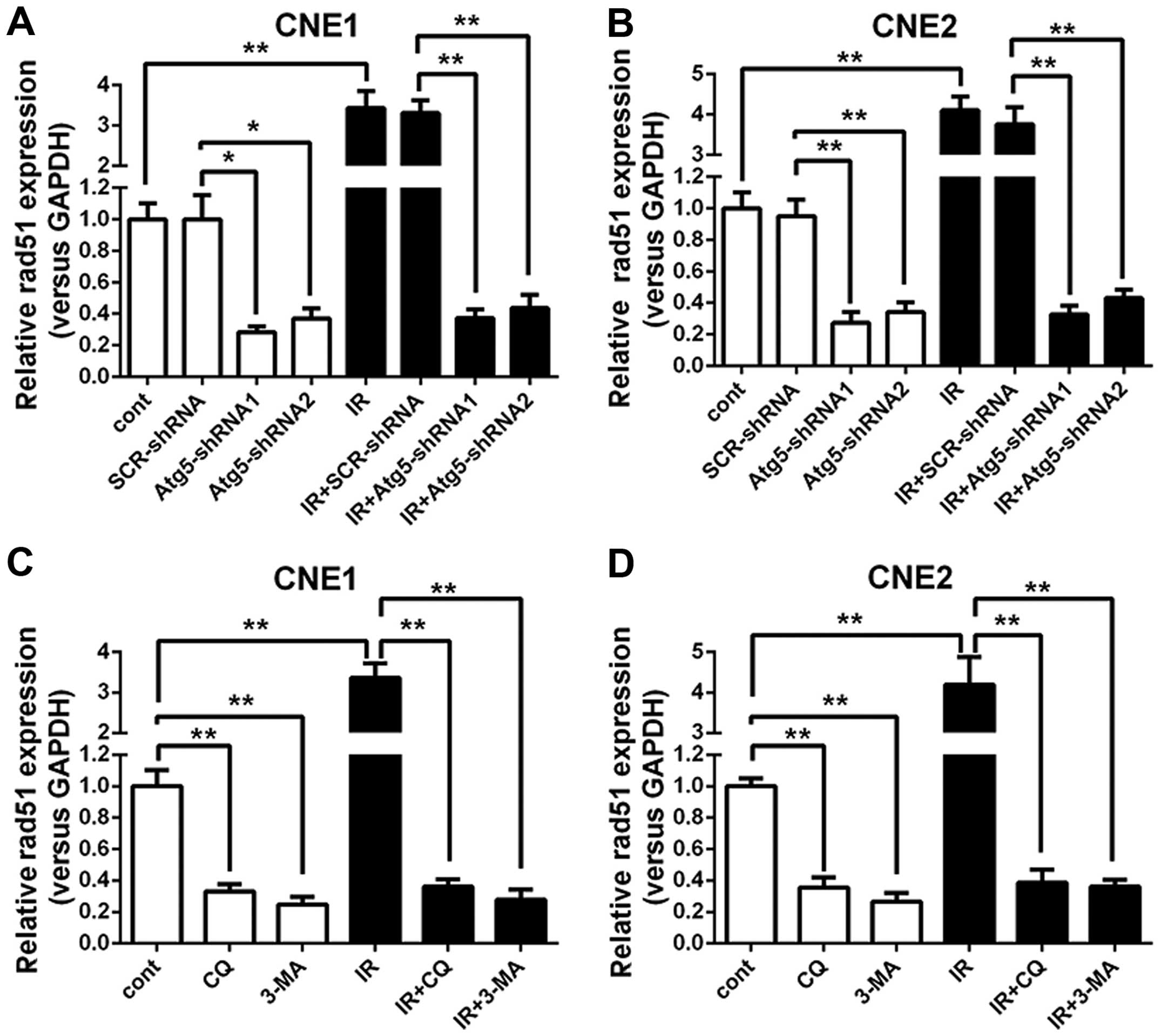
the Atg5 gene. Thus, expression of Rad51 mRNA in NPC cells was
analyzed. The RT-qPCR analysis revealed that the expression of
Rad51 mRNA demonstrated a notable rise in the negative control and
mock-transfected control cells when treated with radiation.
However, when cells were transfected with the Atg5-targeted shRNA,
which was proven to inhibit autophagy in the NPC cells, the Rad51
mRNA expression revealed a significant decline both in cells
treated with or without radiation when compared with that of the
negative control and mock-transfected control cells. Moreover, a
greater decrease in the expression of Rad51 mRNA was observed as
the cells were subjected to a combined treated with radiation
(Fig. 4A and B).
To further confirm the effect of autophagy
inhibition on Rad51 mRNA expression, the control cells were treated
with 3-MA or CQ alone or a combined treatment with irradiation
followed by Rad51 mRNA expression analysis with RT-qPCR. Fig. 4C and D shows that the Rad51 mRNA
expression levels exhibited a completely positive response which
were significantly upregulated when IR was administered to the
control cells; however, when the cells were treated with 3-MA or
CQ, the expression of Rad51 mRNA showed a marked decline compared
to that of the untreated cells. Moreover, the disparity became more
marked when cells received IR. Notably, IR barely altered the
expression of Rad51 mRNA as long as the cells were treated with
3-MA or CQ. As expected, the effect of autophagy inhibition on the
expression of Rad51 was ascertained by 3-MA and CQ, indicating that
autophagy inhibition may suppress the expression of Rad51.
Inhibition of autophagy enhances the
sensitivity of NPC cells to IR by reducing Rad51 expression
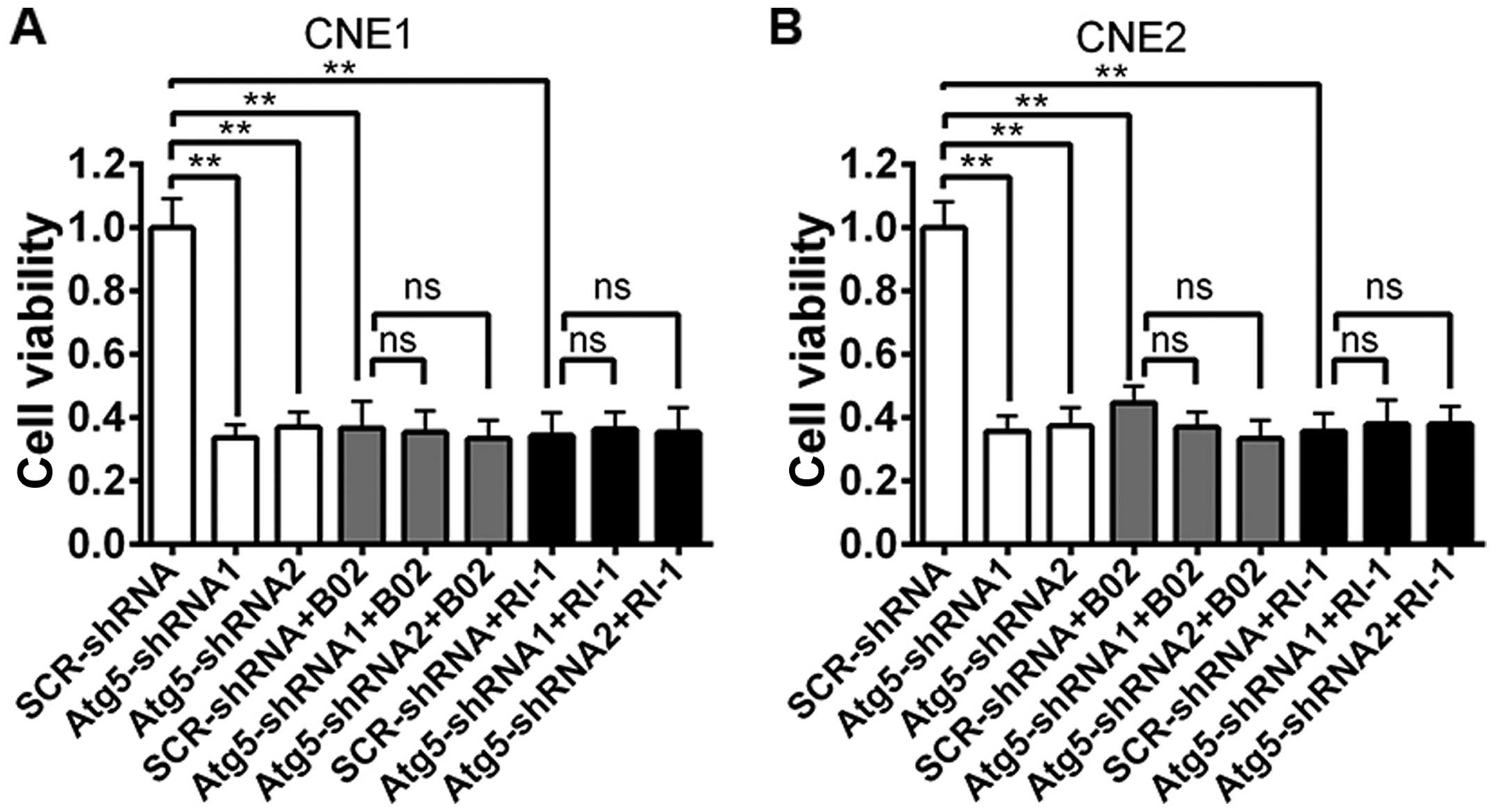
Prior studies have demonstrated that B02 is a
specific inhibitor of human Rad51 (21), while RI-1 acts as a chemical
inhibitor of Rad51 that disrupts homologous recombination in human
cells (22). Thus, to demonstrate
the function of Rad51 in the radiosensitivity of NPC cells, we
examined the influence of Rad51 suppression on radiation
sensitivity by treating NPC cells with B02 or RI-1 1 h before the
treatment of 4-Gy irradiation. After 48 h, cell viability was
evaluated using the CCK-8 assay. As shown in Fig. 5, a marked decrease in cell viability
was observed both in cells transfected with the Atg5-targeted shRNA
alone or scrambled cells treated with the Rad51 inhibitor when
compared to the cell viability of the scrambled cells. However, no
change was noted between the cells transfected with the
Atg5-targeted shRNA and the non-targeted shRNA when cells were
pre-treated with B02 or RI-1.
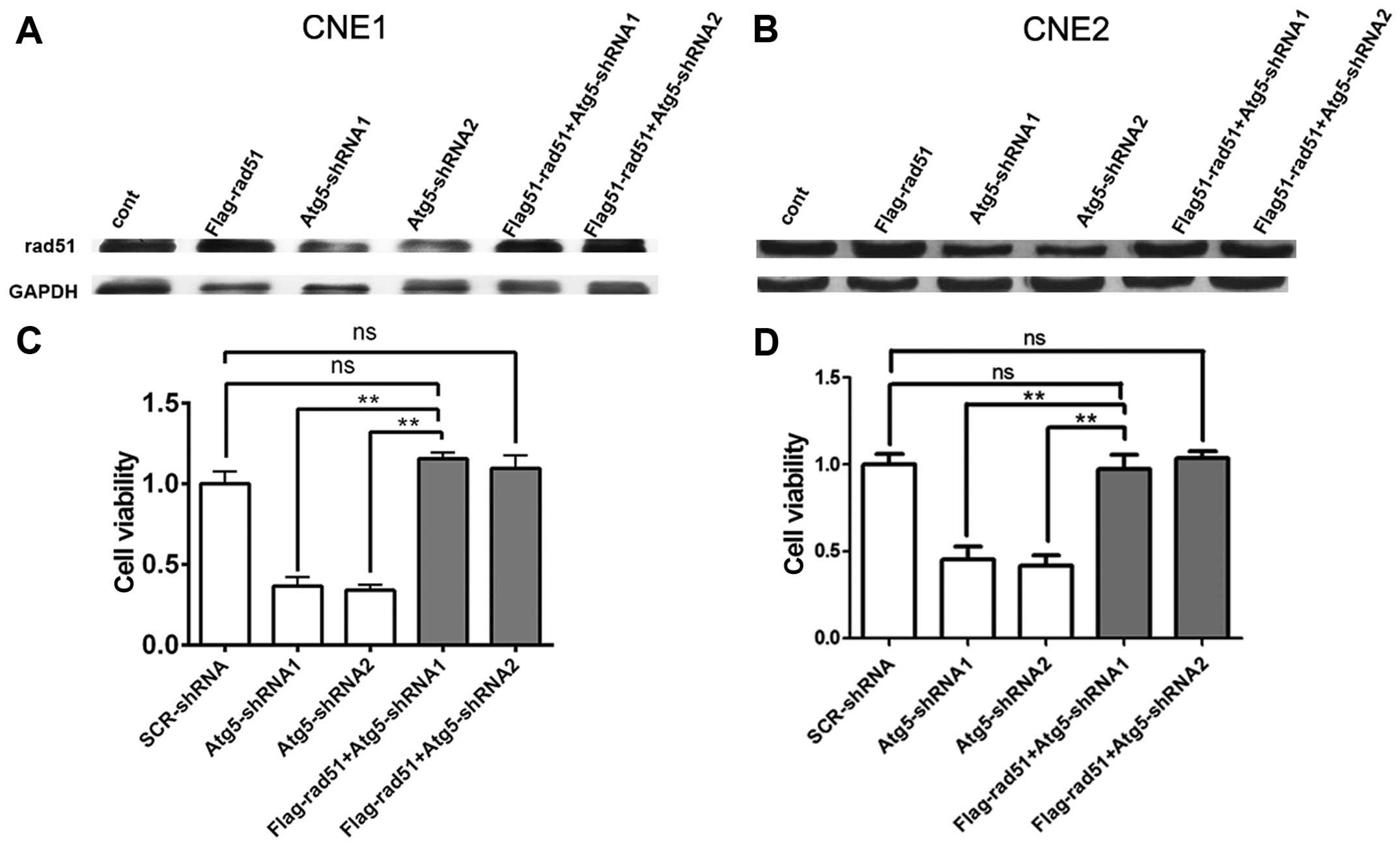
To further validate the relationship between Rad51
and the radioresistance of NPC cells, CNE1 and CNE2 cells were
transfected with the Flag-Rad51 vector. After 48 h, the total
protein level of Rad51 was determined by western blot analysis.
After the next 48 h when cells were treated by 4-Gy irradiation,
cell viability was evaluated by the CCK-8 assay. Western blot
analysis confirmed Rad51 overexpression in cells transfected with
the Flag-Rad51 vector and the control cells when compared to that
of cells transfected with the Atg5-targeted shRNA after
irradiation. However, the low expression of Rad51 in cells
transfected with the Atg5-targeted shRNA exhibited a reverse
upregulation when cells were transfected with the Flag-Rad51 vector
(Fig. 6A and B). Cell viability
assays revealed that the viability of the cells transfected with
the Atg5-targeted shRNA was significantly decreased compared to
that of the mock-transfected control cells, but showed an increase
when cells were transfected with the Flag-Rad51 vector before
irradiation, which reversed the enhanced radiosensitivity by Atg5
suppression in the CNE1 and CNE2 cells (Fig. 6C and D). Taken together, the above
data indicate a correlation between a reduced Rad51 protein level
and increased radiosensitivity to radiation. Thus, the level of
Rad51 protein is inversely correlated with sensitivity to radiation
in NPC cells.
Discussion
Autophagy, or ‘self-eating’, is a double-edged sword
that can both suppress cancer initiation and promote the growth of
established cancers. A large body of literature has strived to
elucidate the role of autophagy in cancer cells in response to
cancer therapy. However, whether autophagy contributes to cell
death or rather represents a survival mechanism remains
controversial (23,24). White revealed that suppression or
deficiency of autophagy genes are associated with diseases,
including cancer (25). Anticancer
agents, such as tamoxifen (26),
rapamycin (27), temozolomide
(28) and IR (29), have been reported to induce
autophagy. After exposure to IR, autophagy can frequently be
observed in cancer cells, and radiosensitivity can be increased by
inhibition of autophagy (30).
Consistent with this, in the present study, we showed that
inhibition of autophagy by knockdown of Atg5 (Fig. 1) enhanced the cytotoxicity of
radiotherapy in CNE-1 or CNE-2 cells, as determined from a markedly
decrease in cell viability and a higher proportion of apoptotic
cells (Fig. 2). Our findings were
further strengthened by a recent study suggesting that blockade of
autophagy by transduction of specific target siRNAs led to
downregulation of the autophagy-related genes, beclin 1, atg3,
atg4b, atg4c, atg5 and atg12 in cell lines, resulting in enhanced
cytotoxicity of radiotherapy in cancer cells. However, its
regulatory mechanisms remain elusive.
It has been demonstrated that the factors that
influence intrinsic radiosensitivity of cell subpopulations include
the level of hypoxia, DNA repair capacity and cell cycle phase.
Among these, regulation of the cell cycle may be the most important
determinant of IR sensitivity (31). Cells are most radiosensitive in the
M and G2 phases, less sensitive in the G1 phase and most
radioresistant in the S phase (31).
However, we found that the enhanced cytotoxicity of
radiotherapy in CNE-1 or CNE-2 cells did not involve cell cycle
arrest. Suppression of Atg5 only resulted in G1 phase arrest while
the percentage of cells in the G2/M phase was significantly reduced
in the CNE-2 cells following radiation, yet no difference in cell
cycle distribution in CNE-1 cells was detected (Fig. 3). Explanation of this may include
the short time of irradiation for only 48 h in our experiment. This
may not have been long enough to induce significant cell cycle
blockage and the cell type and the radiation dose used must be
taken into consideration. Therefore, in the present study, cell
cycle arrest was not the main mechanism of radiosensitivity in NPC
cells.
It has been reported that ionizing radiation mainly
results in the induction of DNA DSBs if left unrepaired (32), while Rad51 has been demonstrated to
be the central player in the initiation of homologous recombination
that plays a critical role in the repair of DNA DSBs. Rad51 is not
only involved in the progression of carcinogenesis but also in the
resistance to anticancer treatments (33). Enhanced Rad51 protein expression can
influence the chemoradiotherapy treatment outcome as well as
potentiate radioresistance in tumor cells (34). Further investigation by us indicated
that inhibition of autophagy with Atg5-targeted shRNA and an
autophagy inhibitor both resulted in decreased expression of Rad51,
yet exhibited no change even when cells were treated with IR
(Fig. 4). These data indicate that
inhibition of autophagy impairs the survival mechanism of NPC cells
after radiation by reducing Rad51 expression. Thus, we hypothesized
that suppression of Rad51 expression rather than cell cycle
blockage is a possible mechanism for autophagy-inhibited induced
radiosensitization in NPC cells.
For characterizing the role of Rad51, CNE1 and CNE2
cells were treated with RI-1or B02 prior to irradiation. Regardless
of whether cells were pre-treated with the Rad51 inhibitor or
subjected to knockdown of Atg5, the cell viability was markedly
decreased when compared to the scrambled cells without treatment by
the Rad51 inhibitor. Moreover, the extent of cell viability was
decreased between cells transfected with the Atg5-targeted shRNA
and the scrambled cells and was not meaningful by statistical
analysis when cells were pre-treated with B02 or RI-1 (Fig. 5A and B). These results indicate that
the target by which the inhibition of autophagy mediates the
radioresistance of NPC cells may be the same as that of B02 and
RI-1. Furthermore, the upregulation of Rad51 protein by
transfecting the Flag-Rad51 vector into CNE-1 and CNE-2 cells
reversed the enhanced radiosensitivity by Atg5 suppression with
increased cell viability to radiation (Fig. 6C and D). Corresponding to our
results, previous reseach concluded that overexpression of Rad51
contributes to the resistance of IR and other DNA-damaging agents
(12), while downregulation of
Rad51 protein may sensitize tumor cells to IR (35). These results indicate that Rad51 may
not only be involved in the resistance to radiation which results
in local residue but also in the locoregional recurrence or distant
metastasis of NPC. Nevertheless, the interaction between Rad51 and
autophagy in the radiation therapy of NPC is complex and further
studies are warranted to support the conclusion.
In summary, our study provides compelling data
supporting our hypothesis that inhibition of autophagy enhances the
radiosensitivity of NPC cells by reducing Rad51 expression. Thus,
the autophagic process of NPC cells is a self-protective mechanism
against radiation. Rad51 targeted therapy may be investigated as a
potential novel agent for the adjuvant treatment of traditional
radiation of NPC, particularly to ascertain whether their
combination and maintenance treatment of Rad51 targeted therapy
following radiation can enhance the therapeutic effect of
radiation, in order to minimize locoregional recurrence or distant
metastasis and maximize the outcome of NPC.
Acknowledgements
The study was supported by the Guangxi Medical
University Fund for Young Scientists (GXMUYSF11), the Guangxi
Natural Science Foundation (2013GXNSFAA019236) and the Guangxi
Natural Science Foundation for Young Scientists
(2014GXNSFBA118141).
References
|
1
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hou H, Li D, Cheng D, Li L, Liu Y and Zhou
Y: Cellular redox status regulates emodin-induced
radiosensitization of nasopharyngeal carcinoma cells in vitro and
in vivo. J Pharmaceutics. 2013.2013:
|
|
3
|
Verheij M and Bartelink H:
Radiation-induced apoptosis. Cell Tissue Res. 301:133–142. 2000.
View Article : Google Scholar
|
|
4
|
Zois CE and Koukourakis MI:
Radiation-induced autophagy in normal and cancer cells: towards
novel cytoprotection and radio-sensitization policies? Autophagy.
5:442–450. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu WM, Huang P, Kar N, et al: Lyn
facilitates glioblastoma cell survival under conditions of nutrient
deprivation by promoting autophagy. PloS One. 8:e708042013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oehme I, Linke JP, Böck BC, et al: Histone
deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl
Acad Sci USA. 110:E2592–E2601. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Selvakumaran M, Amaravadi RK, Vasilevskaya
IA and O’Dwyer PJ: Autophagy inhibition sensitizes colon cancer
cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res.
19:2995–3007. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lomonaco SL, Finniss S, Xiang C, et al:
The induction of autophagy by γ-radiation contributes to the
radioresistance of glioma stem cells. Int J Cancer. 125:717–722.
2009.
|
|
9
|
Apel A, Herr I, Schwarz H, Rodemann HP and
Mayer A: Blocked autophagy sensitizes resistant carcinoma cells to
radiation therapy. Cancer Res. 68:1485–1494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Yin W and Zhu X: Blocked autophagy
enhances radiosensitivity of nasopharyngeal carcinoma cell line
CNE-2 in vitro. Acta Otolaryngol. 134:105–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mazin AV and Mazina OM: RAD51 is a key
protein of DNA repair and homologous recombination in humans.
281–302. 2013.
|
|
12
|
Vispé S, Cazaux C, Lesca C and Defais M:
Overexpression of Rad51 protein stimulates homologous recombination
and increases resistance of mammalian cells to ionizing radiation.
Nucleic Acids Res. 26:2859–2864. 1998.
|
|
13
|
Somaiah N, Yarnold J, Lagerqvist A,
Rothkamm K and Helleday T: Homologous recombination mediates
cellular resistance and fraction size sensitivity to radiation
therapy. Radiother Oncol. 108:155–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou ZR, Zhu XD, Zhao W, et al:
Poly(ADP-ribose) polymerase-1 regulates the mechanism of
irradiation-induced CNE-2 human nasopharyngeal carcinoma cell
autophagy and inhibition of autophagy contributes to the radiation
sensitization of CNE-2 cells. Oncol Rep. 29:2498–2506. 2013.
|
|
15
|
Takasu H, Sugita A, Uchiyama Y, et al:
c-Fos protein as a target of anti-osteoclastogenic action of
vitamin D, and synthesis of new analogs. J Clin Invest.
116:528–535. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Menna-Barreto RF, Salomão K, Dantas AP, et
al: Different cell death pathways induced by drugs in
Trypanosoma cruzi: an ultrastructural study. Micron.
40:157–168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dickey JS, Redon CE, Nakamura AJ, Baird
BJ, Sedelnikova OA and Bonner WM: H2AX: functional roles and
potential applications. Chromosoma. 118:683–692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bae Y, Jung SH, Kim GY, Rhim H and Kang S:
Hip2 ubiquitin-conjugating enzyme overcomes radiation-induced G2/M
arrest. Biochim Biophys Acta. 1833:2911–2921. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyata H, Doki Y, Yamamoto H, et al:
Overexpression of CDC25B overrides radiation-induced G2-M arrest
and results in increased apoptosis in esophageal cancer cells.
Cancer Res. 61:3188–3193. 2001.PubMed/NCBI
|
|
20
|
Rodemann HP: Molecular radiation biology:
perspectives for radiation oncology. Radiother Oncol. 92:293–298.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang F, Motlekar NA, Burgwin CM, Napper
AD, Diamond SL and Mazin AV: Identification of specific inhibitors
of human RAD51 recombinase using high-throughput screening. ACS
Chem Biol. 6:628–635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Budke B, Logan HL, Kalin JH, et al: RI-1:
a chemical inhibitor of RAD51 that disrupts homologous
recombination in human cells. Nucleic Acids Res. 40:7347–7357.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eisenberg-Lerner A and Kimchi A: The
paradox of autophagy and its implication in cancer etiology and
therapy. Apoptosis. 14:376–391. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsuchihara K, Fujii S and Esumi H:
Autophagy and cancer: dynamism of the metabolism of tumor cells and
tissues. Cancer Lett. 278:130–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
White E: Deconvoluting the
context-dependent role for autophagy in cancer. Nat Rev Cancer.
12:401–410. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yenigun VB, Ozpolat B and Kose GT:
Response of CD44+/CD24−/low breast cancer
stem/progenitor cells to tamoxifen- and doxorubicin-induced
autophagy. Int J Mol Med. 31:1477–1483. 2013.
|
|
27
|
Tekirdag KA, Korkmaz G, Ozturk DG, Agami R
and Gozuacik D: MIR181A regulates starvation- and rapamycin-induced
autophagy through targeting of ATG5. Autophagy. 9:374–385. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y, Wang HD, Zhu L, et al: Knockdown
of Nrf2 enhances autophagy induced by temozolomide in U251 human
glioma cell line. Oncol Rep. 29:394–400. 2013.PubMed/NCBI
|
|
29
|
Liang N, Jia L, Liu Y, et al: ATM pathway
is essential for ionizing radiation-induced autophagy. Cell Signal.
25:2530–2539. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chaachouay H, Ohneseit P, Toulany M,
Kehlbach R, Multhoff G and Rodemann HP: Autophagy contributes to
resistance of tumor cells to ionizing radiation. Radiother Oncol.
99:287–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Corcelle E, Nebout M, Bekri S, et al:
Disruption of autophagy at the maturation step by the carcinogen
lindane is associated with the sustained mitogen-activated protein
kinase/extracellular signal-regulated kinase activity. Cancer Res.
66:6861–6870. 2006. View Article : Google Scholar
|
|
33
|
Takenaka T, Yoshino I, Kouso H, et al:
Combined evaluation of Rad51 and ERCC1 expressions for sensitivity
to platinum agents in non-small cell lung cancer. Int J Cancer.
121:895–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Du LQ, Wang Y, Wang H, Cao J, Liu Q and
Fan F-Y: Knockdown of Rad51 expression induces radiation- and
chemo-sensitivity in osteosarcoma cells. Med Oncol. 28:1481–1487.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamamori T, Meike S, Nagane M, Yasui H and
Inanami O: ER stress suppresses DNA double-strand break repair and
sensitizes tumor cells to ionizing radiation by stimulating
proteasomal degradation of Rad51. FEBS Lett. 587:3348–3353. 2013.
View Article : Google Scholar
|