Introduction
microRNAs (miRNAs) are an abundant class of small
non-coding RNAs (~22 nucleotides) with a critical regulative
function (1,2), that reduce mRNA stability and/or
suppress translation by perfect or imperfect sequence-complementary
binding to the 3′-untranslated region (3′-UTR) or the coding
sequences of target mRNAs (3,4).
miRNAs have been considered to contribute to the majority of basic
biological processes, such as development, differentiation,
apoptosis and cell proliferation (5). Their deregulation has also been linked
to cancer and other diseases (6,7).
Therefore, the in-depth validation of the biological roles of
miRNAs is important for understanding pathogenesis and raising new
curative therapies.
Several methods for gain- or loss-of-function have
been developed to modify the miRNA expression in vitro or
in vivo. However, the overexpression of exogenous miRNA
(gain-of-function) often leads to neomorphic phenotypes for a
strong concentration-dependent interaction between miRNAs and their
targets (8). For this reason, a
loss-of-function study is more likely to reveal the functions that
depend on physiological miRNA levels (9). The miRNA antisense oligonucleotides
inhibitors or antagomirs, genetic knockout and miRNA sponge
(miR-SPs) methods, have been used to block the activity of miRNA
(10–14). It has been reported that miR-SPs,
which contain multiple binding sites complementary to a mature
miRNA were introduced to sequester endogenous miRNAs and then their
functions were suppressed in the in vitro differentiation of
neurons (15), mesenchymal stem
cells (16), cancer cell xenografts
(17,18), bone marrow reconstitutions from
hematopoietic stem/progenitor cells (19) and animal models (20,21).
Due to the difficult operation of genetic knockouts and short-term
effects of antisense oligonucleotide inhibitors, miR-SPs are more
suitable to chronically block the activities of endogenous miRNA
in vivo.
microRNA-122 (miR-122), a highly abundant
liver-specific miRNA that accounts for 70% of total liver miRNA
expression (22,23), has been associated with hepatitis C
virus infection and replication (24,25),
the regulation of liver metabolism and hepatocellular carcinoma
(HCC) (26). miR-122 has been
identified to downregulate cyclin G1 (CCNG1) (27,28),
and thus leads to G1 arrest (29),
interrupts the interaction between CCNG1 and p53, and abrogates the
p53-mediated inhibition of hepatitis B virus replication (30). Oncogene Bcl-w is also the target of
miR-122, and the downregulation of Bcl-w by miR-122 subsequently
leads to the reduction of cell viability and activation of
caspase-3/7 (28,31). Overexpression of miR-122 affects HCC
intrahepatic metastasis by angiogenesis suppression, exerting some
of its actions via the regulation of its targets disintegrin and
metalloprotease 10 (ADAM10) and ADAM17, which are involved in
metastasis (29,32). The loss-of-function study of miR-122
is performed using miR-122 antisense oligonucleotides inhibitors or
genetic knockouts, while sponge-mediated miR-122 silencing has yet
to be studied in tumor cells.
In the present study, we generated a miR-sponge
based on CMV promoter to silence the activity of miR-122 on tumor
suppression using lentivirus. Our data showed that the miR-122
sponge (miR-122-SP) effectively blocked the functions of ectopic
liver-specific miR-122 by inhibiting cell proliferation, arresting
cell cycle, inducing apoptosis and suppressing metastasis, not only
in Huh7 hepatoma cells, but also in U2OS osteosarcoma cells. We
also found that miR-122-SP efficiently knocked down the expression
of endogenous miR-122 in Huh7 cells and promoted xenograft tumor
growth. Our studies provide a potential method in which to
investigate the miR-122 functions in liver development and
tumorigenesis in vitro or in vivo through the
application of miR-SPs.
Materials and methods
Construction of sponge plasmids and
reporters
The oligonucleotides with three repetitive
sequences, which are complementary to miR-122 with mismatches at
positions 9–12, were annealed, ligated, gel purified and cloned
into the XhoI/NotI site of a psiCHECK2 vector
(Promega, Madison, WI, USA), downstream of the Renilla
luciferase gene, to generate the vector psiCHECK2/miR-122-SP. We
constructed the GFP reporters using the same annealing method and
subcloned the sequences into the 3′-UTR of the EGFP in a pEGFP
vector with a CMV promoter.
Cell culture and stable cell line
production
The Huh7 cell line (American Type Culture
Collection, Manassas, VA, USA) was maintained at 37°C, 5%
CO2 and in DMEM/high glucose (Invitrogen Life
Technologies, Carlsbad, CA USA) with 10% fetal bovine serum (FBS).
The U2OS cell line (American Type Culture Collection) was
maintained at 37°C, 5% CO2 and in RPMI-1640 supplemented
with 10% FBS (both from Invitrogen Life Technologies) in 25-ml
culture flasks. For the miR-122-SP stable overexpression, the Huh7
or U2OS cells were infected with lentivirus that expressed
miR-122-SP (Genchem & Genpharm Co., Ltd., Jiangsu, China) or
its negative control at 30–50% confluence. The monoclonal
population of the stably infected cells was selected by a limiting
dilution assay using the GFP marker.
Luciferase and GFP reporter assay
For the luciferase reporter assay, 293T cells
(5×104 cells/well) were seeded in 24-well plates 24 h
prior to transfection. The cells were co-transfected with 0.5 μg of
the psiCHECK2/miR-122-SP plasmids together with 30 nM of miR-122
mimics or 30 nM of the negative control (both from GenePharma,
Shanghai, China) using Lipofectamine 2000 (Invitrogen Life
Technologies). Firefly and Renilla luciferase activities
were measured by using the dual-luciferase reporter assay (Promega)
48 h after transfection. The firefly luciferase activity was
normalized to the Renilla luciferase activity. For the GFP
reporter assay, U2OS cells (5×104 cells/well) seeded in
24-well plates were co-transfected with 0.5 μg of the
pEGFP/miR-122-SP plasmids together with 30 nM of miR-122 mimics or
30 nM of the negative control (both from GenePharma) using
Lipofectamine 2000 (Invitrogen Life Technologies). GFP expression
was examined by a microscope after 48 h of transfection.
RT-qPCR analysis
The total RNA from tissue samples and cell lines was
extracted using TRIzol reagent (Invitrogen Life Technologies)
following the manufacturer’s instructions. The RNA concentrations
and quality were determined with a spectrophotometer (Eppendorf AG,
Hamburg, Germany) and gel analysis. RT-qPCR was performed using
SYBR Premix Ex Taq (Takara Bio, Inc., Shiga, Japan) and Applied
Biosystems 7900 Real-Time PCR System (Applied Biosystems, Foster
City, CA, USA) supplied with its analytical software. The forward
and reverse primer pairs were designed for CCNG1, Bcl-w, and ADAM10
as described previously (28,29).
The primer pair for the 18SrRNA Q-PCR was as follows: forward,
5′-CCTGGATACCGCAGCTAGGA-3′ and reverse,
5′-GCGGCGCAATACGAATGCCCC-3′. The expression of mature miR-122 was
assayed using the miRCURY LNA™ Universal RT microRNA PCR Universal
cDNA synthesis kit with specific primers (both from Exiqon,
Vedbaek, Denmark) on cell lines. Reverse transcription reaction was
carried out starting from 1 μg of total RNA using real-time PCR was
performed according to the miRCURY LNA™ SYBR-Green master mix kit
(Exiqon) protocol. The ΔΔCt method for relative quantification was
used to determine the gene expression.
Western blot analysis
The cells transfected with the indicated vectors or
chemicals were subjected to western blotting with anti-CCNG1
(1:1,000), anti-Bcl-w (1:1,000) (both from Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), anti-ADAM10 (1:1,000;
eBioscience, Inc., San Diego, CA, USA) and actin (1:1,500; Santa
Cruz Biotechnology, Inc.) antibodies, respectively. The cells were
harvested and lysed in 0.5 ml of lysis buffer (10 mM Tris-HCl, pH
7.6, 5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM NaF,
0.1 mM Na3VO4, 1% Triton X-100, 1 mM PMSF and
protease inhibitors. Proteins (20 μg) were processed for SDS-PAGE,
which was performed on 10% gels. The proteins were
electrophoretically transferred to PVDF membranes (Millipore,
Billerica, MA, USA) and incubated with the primary antibodies
followed by incubation with an HRP-conjugated secondary antibody
(Santa Cruz Biotechnology, Inc.). After washing with TBS, the bound
antibodies were visualized by enhanced chemiluminescence (Pierce
Biotechnology, Inc., Rockford, IL, USA) and recorded on X-ray
film.
Cell proliferation assay
Ctrl-lentivirus (Ctrl-LV) and miR-122-SP-LV cells
(2.5×105 cells/well) were transfected with 30 nM miR-122
mimics or 30 nM negative control (GenePharma) using Lipofectamine
2000 (Invitrogen Life Technologies) as per the manufacturer’s
instructions in 6-well plates. The transfected cells
(1×103 cells/well) were then seeded in 96-well plates.
The MTT assay was performed on day 1, 3, 5 and 6. Absorbance of the
samples was measured with a spectrophotometer reader at 490 nm, and
each assay was performed in triplicate.
Colony formation assay
Each cell line transfected with synthesized
oligonucleotides or vectors was seeded in 12-well plates (200
cells/well), incubated for 7–10 days to allow colony growth, and
the colonies were stained with crystal violet. The colonies
containing ≥50 cells were counted. The plating efficiency was
calculated by dividing the average number of the colonies/dish by
the number of the cells plated. The survival fractions were
calculated by normalizing to the plating efficiency of the control
groups.
Wound-healing scratch assay
For the wound-healing scratch assay,
2.5×105 Ctrl-LV and miR-122-SP-LV cells transfected with
30 nM of miR-122 mimics or negative controls were seeded in 12-well
plates. After 24 h, a linear wound was made by scratching across
the bottom of the dish on the monolayer of the confluent cells
using a sterile p-200 pipette tip. The cells were rinsed gently
with phosphate-buffered saline (PBS) and cultivated in the
corresponding medium without serum supplemented with 0.5% FBS. The
same area of the gap was taken at ×100 magnifications using a
microscope equipped with a digital camera (Olympus, Centre Valley,
PA, USA) at 0 and 24 h after the scratching, and then quantified
using ImageJ software (http://rsbweb.nih.gov/ij/). The difference between the
initial and final areas was calculated.
Cell Matrigel invasion assay
For the invasion assay, 1×105 Ctrl-LV and
miR-122-SP-LV cells transfected with 30 nM of miR-122 mimics or
negative controls were seeded in a Matrigel-coated chamber with 8.0
μM pores (Corning Costar, Corning, NY, USA). The cells were seeded
in serum-free media in the upper chamber, and translocated towards
complete growth media for 36 h. The lower compartment of the
chamber was fixed in 1.5% glutaraldehyde for 1 h, stained with 0.1%
violet crystal dye for 15 min, and washed twice with distilled
water. The stained cells were viewed under a light microscope
(Olympus).
Cell cycle analysis
To examine the effect of miR-122-SP on the blocking
functions of miR-122 in cell cycle arrest, the Ctrl-LV and
miR-122-SP-LV cells were transfected with 30 nM of miR-122 mimics
or negative controls and then collected after 48 h and analyzed for
DNA content by flow cytometry.
Caspase-3/7 activity analysis
The Ctrl-LV and miR-122-SP-LV cells transfected with
30 nM of miR-122 mimics or negative controls were seeded in 96-well
plates (5×103 cells/well). After 24 h of incubation at
37°C, caspase-3/7 substrates (Promega) were added according to the
manufacturer’s instructions and incubated for 1 h at 37°C in the
dark. The relative light intensity was measured in each well using
a microplate luminometer.
Xenograft tumor model
Huh7-Ctrl-LV or Huh7-miR-122-SP-LV cells
(1×105) were inoculated subcutaneously into the right
flanks of female athymic nude nu/nu mice (3–4 weeks old),
respectively. The volume of the implanted tumor was measured every
4 days with a vernier caliper, using the formula: V = L ×
W2/2; where V, volume (mm3); L, biggest
diameter (mm); W, smallest diameter (mm). The mice were
subsequently sacrificed via cervical dislocation and the tumors
were weighed 4 weeks after inoculation.
Study approval
All animal procedures were undertaken according to
the United Kingdom Co-ordinating Committee on Cancer Research
(UKCCCR) Guidelines for the Welfare of Animals in Experimental
Neoplasia and were approved by the Central Laboratory, Shanghai
Tenth People’s Hospital of Tongji University (Shanghai, China).
Statistical analysis
Data were presented as means ± SD. The Student’s
t-test was applied to determine the differences between sample
means obtained from at least three experiments. Statistical
analyses were performed using Excel software. All tests of
statistical significance were two-sided. P<0.05 was considered
to indicate statistical significance.
Results
Construction of miR-122-SP
To generate a miR-122-SP, three repetitive
sequences, complementary to miR-122 with mismatches at positions
9–12 for enhanced stability, were introduced into the 3′-UTR of
Renilla luciferase in a psiCHECK2 vector (Fig. 1A and B). We first co-transfected
HEK293T cells with psiCHECK2/miR-122-SP plasmids and miR-122 mimics
and assayed the activity of the Renilla luciferase 48 h
after the transfection. The data showed that miR-122 decreased the
activity of luciferase by directly binding to the miR-122-SP sites
in the 3′-UTR region downstream of Renilla luciferase, while
the luciferase activities in the cells co-transfected with miR-122
mimics and psiCHECK2 vector, or microRNA negative control (NC) and
miR-122-SP, were not altered (Fig.
1D). To identify the effect of sponge, we sub-cloned miR-122-SP
sites into the 3′-UTR of the EGFP in a pEGFP vector with a CMV
promoter (Fig. 1C). We
co-transfected this pEGFP/miR-122-SP with miR-122 mimics into U2OS
cells, and found that the GFP was silenced in the U2OS cells with
miR-122 overexpression (Fig. 1E).
These results showed that the ectopic miR-122 directly bound to the
miR-122-SP, which subsequently resulted in the decrease of reporter
gene expression, suggesting that the construction of miR-122-SP was
successful.
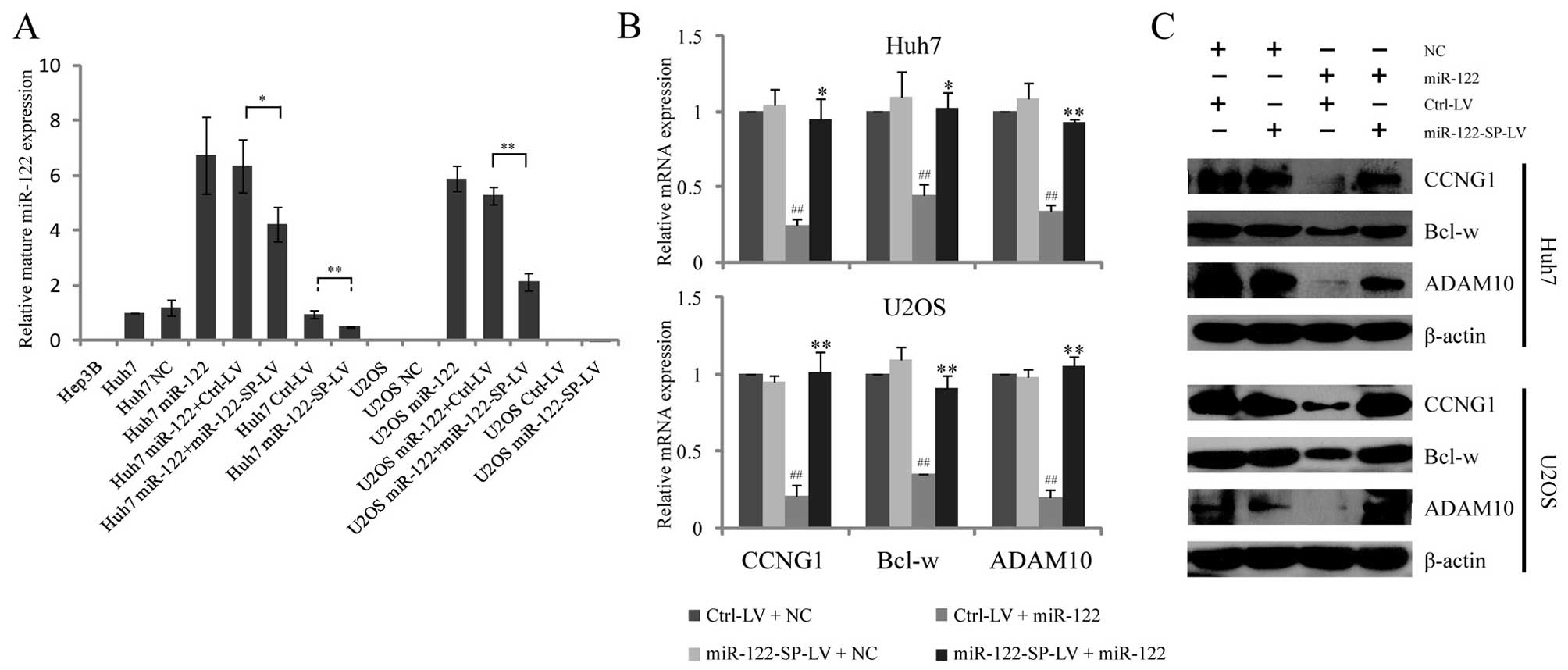
MiR-122-SP reduces miR-122 expression and
upregulates the target genes of miR-122
As a liver-specific microRNA, miR-122 expression was
confirmed to be downregulated in the HCC of rodent and human
(33). We detected miR-122
expression in different types of cell lines by RT-qPCR. No
endogenous expression of miR-122 was observed in Hep3B hepatoma
cells and U2OS osteosarcoma cells, but a low expression of miR-122
was identified in Huh7 cells (Fig.
2A). To study the miR-122-SP effects on blocking miR-122
functions, we chose Huh7 hepatoma cells with a low expression of
miR-122 and U2OS non-hepatoma cells without any expression of
miR-122 as cell models. Due to the low efficiency and short-term
effect of the transient transfection of plasmid, we used a
lentivirus-mediated expression system to express miR-122-SP in Huh7
and U2OS cells in the same manner as indicated in Fig. 1C. The RT-qPCR results showed that
stable overexpression of miR-122-SP led to a decrease of ectopic
miR-122 expression in Huh7 and U2OS cells, as well as the
downregulation of endogenous miR-122 expression in Huh7 cells
(Fig. 2A). These results indicated
that the lentivirus-mediated miR-122-SP expression was able to
efficiently reduce the endogenous or ectopic expression of
miR-122.
As a tumor suppressor, miR-122 has been identified
to downregulate the expression of CCNG1 (involved in the cell
cycle), Bcl-w (involved in apoptosis) and ADAM10 (involved in
migration). To investigate whether miR-122-SP decreased the
expression of these genes, we examined the mRNA and protein levels
in Huh7 and U2OS cells. As expected, the mRNA levels of the Huh7
and U2OS cells were inhibited by ectopic miR-122, while the cells
with miR-122-SP expression completely rescued the expression of
these three genes (Fig. 2B). The
western blot results revealed the same effects of miR-122-SP on
reducing the protein levels of CCNG1, Bcl-w and ADAM10 in the Huh7
and U2OS cells (Fig. 2C). However,
the mRNA and protein levels of CCNG1, Bcl-w and ADAM10 were not
significantly altered by miR-122-SP expression without ectopic
miR-122. This result may be due to the low expression of miR-122 in
Huh7 that could not obviously inhibit the expression of the three
genes. These results suggested that miR-122-SP would clearly be
able to restore the expression of CCNG1, Bcl-w and ADAM10 that was
downregulated by ectopic miR-122.
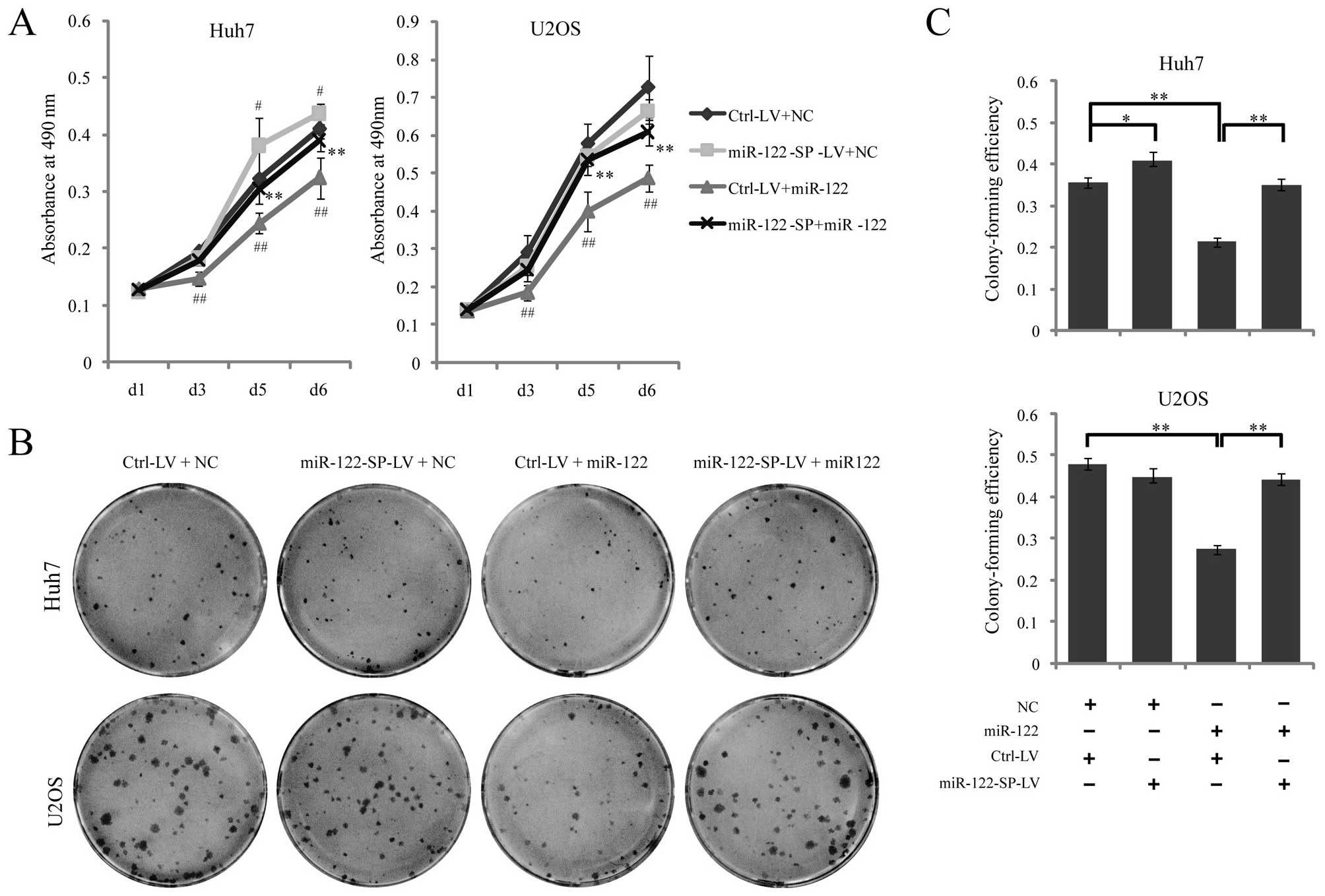
miR-122-SP blocks the functions of
miR-122 in cell proliferation
miR-122 has been identified to significantly inhibit
tumor cell proliferation. To identify whether miR-122-SP suppressed
the functions of miR-122 in cell proliferation, Ctrl-LV or
miR-122-SP-LV cells were transfected with miR-122 mimics or the
negative control. We found that Huh7 or U2OS Ctrl-LV cell
proliferation was significantly inhibited by the ectopic miR-122
expression through the MTT assay, while proliferations of the Huh7
or U2OS miR-122-SP-LV cells transfected with miR-122 were not
obviously altered (Fig. 3A).
Fig. 3C also showed that miR-122
could suppressed the colony-forming efficiency of Huh7 and U2OS
cells, while miR-122-SP with miR-122 mimics had no effect on colony
formation (Fig. 3B and C). In
accordance with the decrease of endogenous miR-122 expression by
miR-122-SP in Huh7, we also found that miR-122-SP promoted cell
proliferation and increased the cell colony formation ability of
Huh7 cells (Fig. 3A), which was
caused by the downregulation of endogenous miR-122 in Huh7-mediated
by miR-122-SP. These results indicated that miR-122-SP was able to
rescue the effects of liver-specific miR-122 on suppressing
proliferation, not only in Huh7 human hepatoma cells, but also in
U2OS osteosarcoma cells.
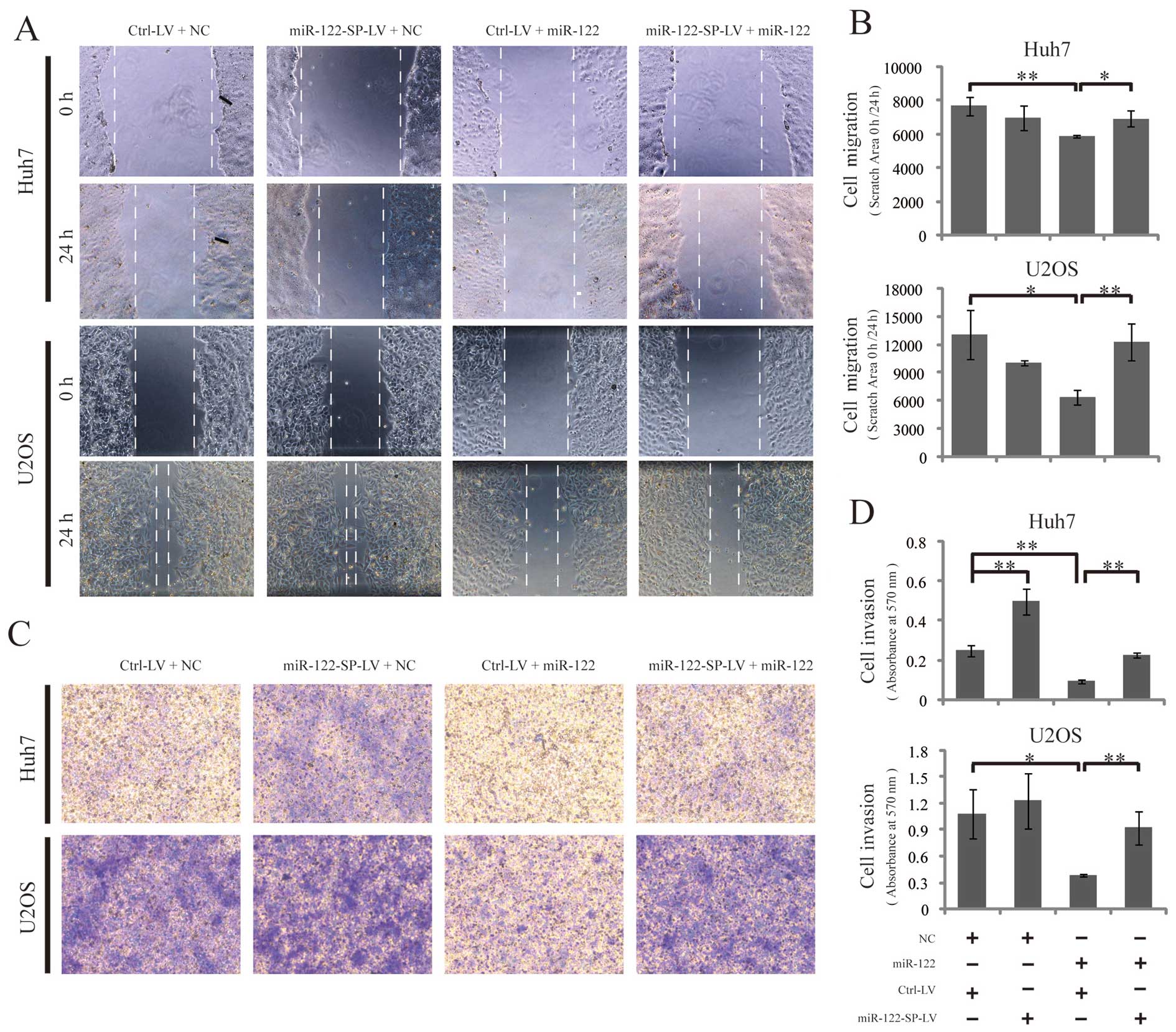
miR-122-SP blocks the functions of
miR-122 in cell migration and invasion
To study the effects of miR-122-SP on inhibiting the
functions of miR-122 in cell migration, we conducted wound-healing
scratch experiments to assess cell migration. Ctrl-LV or
miR-122-SP-LV cells were transfected with miR-122 mimics or
negative controls on 12-well plates. After culturing for 24 h in
medium with 10% FBS, the confluent culture monolayer of these cells
was scratched and cultured in medium with 0.5% FBS for another 24
h. The same wounded area/well was examined by phase-contrast
microscopy after scratching at 0 and 24 h. Transfection with
miR-122 mimics significantly inhibited the migration of U2OS and
Huh7 cells, while the miR-122-SP expression blocked the functions
of miR-122 by inhibiting cell migration (Fig. 4A and B). Subsequently, we subjected
a Matrigel invasion assay to define the effect of miR-122-SP on
blocking the functions of miR-122 in suppressing cell invasion. As
expected, the results showed that miR-122 decreased the number of
invading cells in Huh7 and U2OS, while this decrease induced by
miR-122 was significantly inhibited by miR-122-SP, suggesting a
suppressor role of miR-122-SP for miR-122 in cell migration and
invasion (Fig. 4C and D).
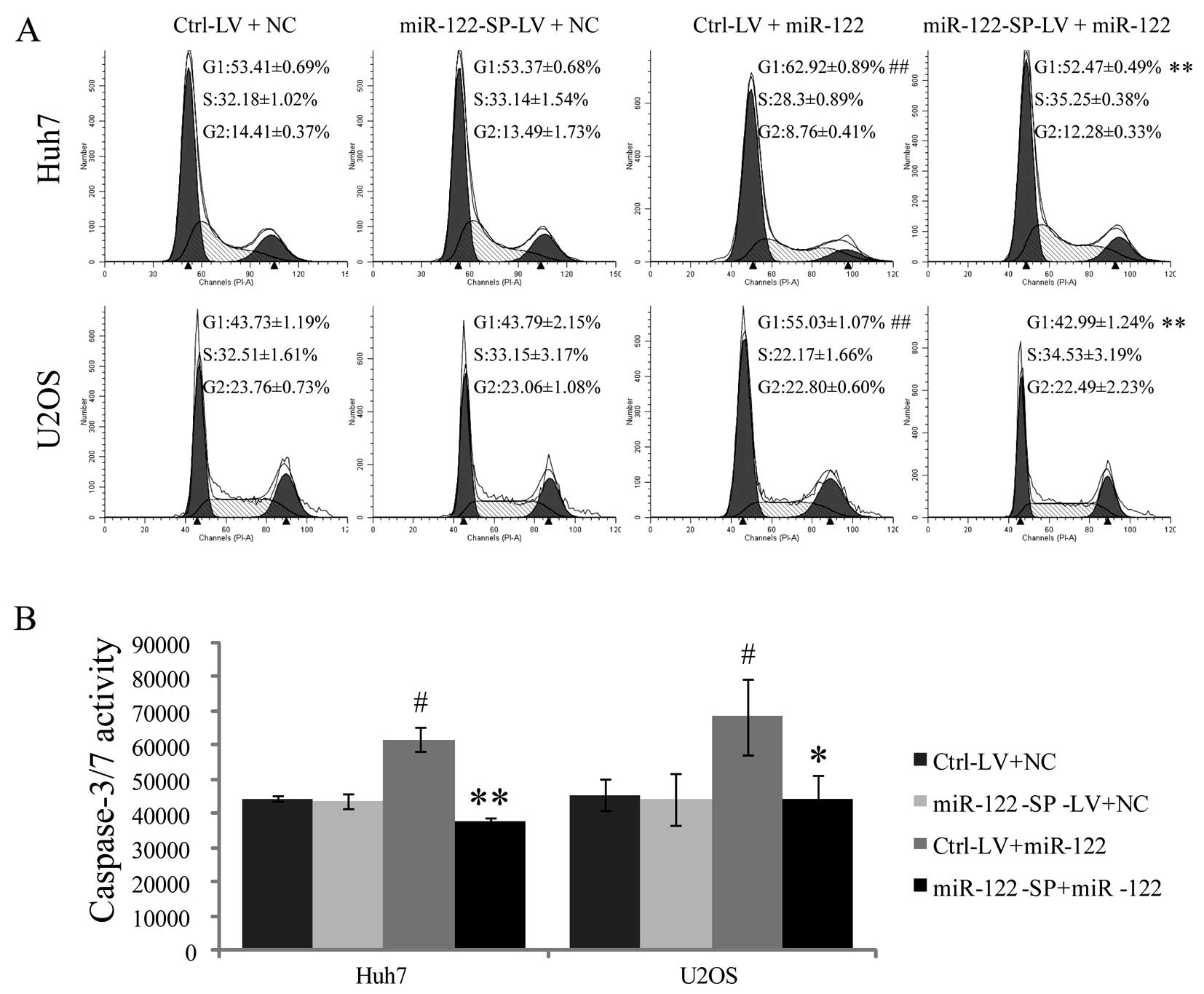
miR-122-SP blocks the functions of
miR-122 in the regulation of cell cycle and caspase-3/7
activity
CCNG1 and Bcl-w were identified as the targets of
miR-122, and ectopic miR-122 led to G1 arrest of cancer cells and
increased caspase-3/7 activity. Thus, we hypothesized that
miR-122-SP affected the cell cycle and the activity of caspase-3/7
through miR-122. To confirm this, we harvested Ctrl-LV and
miR-122-SP-LV cells 48 h after the cells were transfected with
miR-122 mimics or negative controls to analyze the cell cycle
distribution by FACS. Our results showed that there was a
significant increase in the population of cells at G1 phase between
the miR-122 expressing and the control cells (62.92 and 53.41% in
Huh7 cells, and 55.03 and 43.73% in U2OS cells). However,
miR-122-SP overexpression restored the distribution of G1 phase
from 62.92 to 52.47% in Huh7 cells and 55.03 to 42.99% in U2OS
(Fig. 5A). These results
demonstrated that miR-122-SP reduced the effects of miR-122 on
arresting cell cycle at G1 phase. Subsequently, we detected the
caspase-3/7 activity of the Ctrl-LV and miR-122-SP-LV cells that
were transfected with or without miR-122. The caspase-3/7 activity
was inhibited by miR-122; however, it was rescued by the
overexpression of miR-122-SP in Huh7 and U2OS cells (Fig. 5B). This suggested that miR-122-SP
was able to block the functions of miR-122 in promoting cell
apoptosis.
miR-122-SP blocks the functions of
miR-122 in tumor suppression
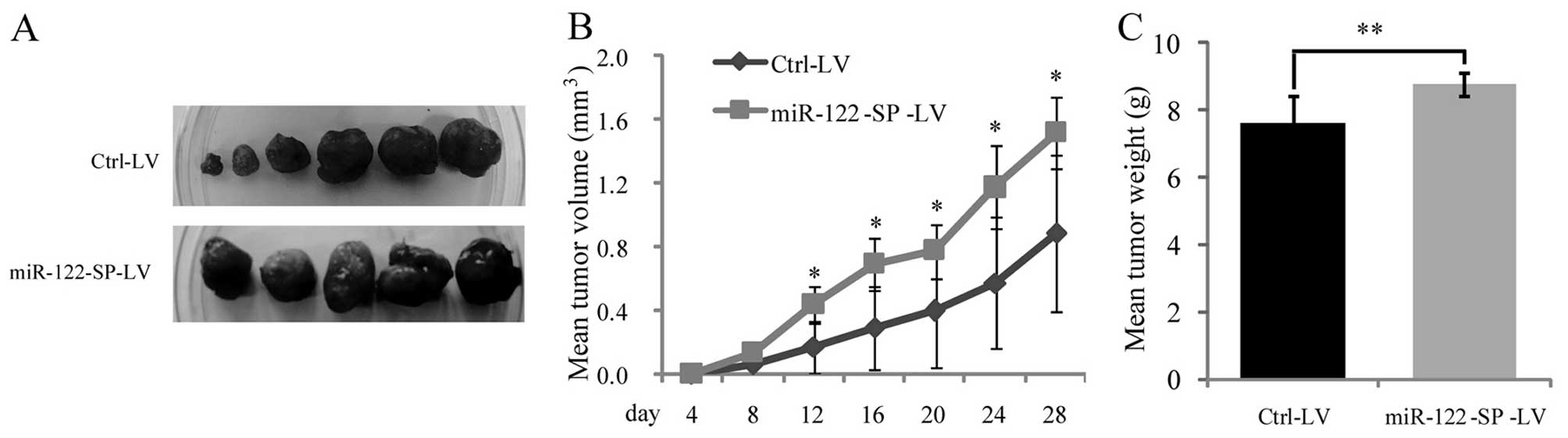
Although, a low expression of miR-122 was detected
in Huh7 cells, miR-122-SP was able to reduce the endogenous miR-122
expression and block the functions of miR-122 in inhibiting cell
proliferation. To determine whether miR-122-SP blocks activities of
miR-122 in suppressing tumorigenesis in vivo, Huh7 cells
infected with Ctrl-LV or miR-122-SP-LV were subcutaneously
inoculated into nude mice. The mean volume of xenograft tumors
formed in the presence of miR-122-SP was larger than that of
control (Fig. 6A and B). At the end
of the experimental period, the mean weight of tumors with
miR-122-SP overexpression was higher than that of the control
(Fig. 6C). Thus, these data
indicate that miR-122-SP blocks the functions of miR-122 in
inhibiting tumorigenesis in vivo.
Discussion
To investigate the functions of miRNAs, the
loss-of-function study is considered more reliable than that of
gain-of-function for its blocking endogenous miRNAs activity
(34). Several methods were
utilized to study miRNA loss-of-function, including genetic
knockouts, antisense oligonucleotide inhibitors (12,35,36)
and sponges (9). The use of
modified antisense oligonucleotide inhibitors, also designated as
antagomirs or locked nucleic acids (LNAs), was the first method to
block miRNA activity in animal cell cultures (12,36,37).
However, antagomirs, as well as LNAs did not reach some tissues,
such as the central nervous system, or the cells in a brain due to
the blood-brain barrier (12,38,39).
Moreover, they only provide short-term downregulation of the miRNA
targets and block single miRNA rather than whole miRNA families
(14), suggesting that antagomirs
and LNAs are not suitable for generating transgenic animal
models.
Compared to antisense oligonucleotide inhibitors,
miR-SPs offer more advantages for many in vivo research
applications. Firstly, sponges mediated by a lentiviral vector can
stably and chronically block the miRNA activity in
difficult-to-transfect cell lines or cells in vivo.
Secondly, a single miR-SP can block the activities of the whole
family of miRNAs sharing highly similar seed sequences while
locating in multiple distant loci (9). Furthermore, a reporter gene or a
selectable marker in sponge facilitates the introduction of sponge
for tracing, screening and sorting cells with miRNAs. A
tissue-specific or drug-inducible promoter in a sponge, such as the
tet-on and tet-off system, is more useful to study the functions of
many important miRNAs in development without leading to transgenic
model death.
Accumulating evidence has demonstrated that miR-122
is downregulated in chronic hepatitis B (CHB) and HCC of human
(30,33), suggesting the possible relationship
between hepatocarcinogenesis and liver-specific microRNA.
Currently, studies on miR-122 function in tumorigenesis is based on
the ectopic expression of miR-122 in hepatoma cells in
vitro. Although miR-122 has been confirmed to suppress
tumorigenic properties of Huh7 cells, which express miR-122 at a
lower level, by transfecion of anti-miR-122 oligonucleotides in
vitro (29), the roles of
endogenous of miR-122 in hepatoma cells in vivo have not
been reported. Thus, we designed lentivirus-mediated miR-122-SP,
which can be used to silence the activities of miR-122 in
vitro and in vivo, to confirm or identify the roles of
miR-122 in liver development and hepatocarcinogenesis. In this
study, we confirmed that ectopic miR-122 downregulated the
expression of CCNG1, Bcl-w and ADAM10, inhibited cell proliferation
and migration, arrested the cell cycle at G1 phase, and induced
cell apoptosis in Huh7 hepatoma cells in vitro. These
effects of miR-122 on suppressing tumorigenesis be efficiently
blocked by miR-122-SP. In addition, we may found similar results in
U2OS osteosarcoma cells, that miR-122-SP was able to silence the
functions of ectopic miR-122 in suppressing tumorigenic properties.
These results suggest that miR-122-SP is an effective tool for
loss-of-function research in vitro, and raise the potential
of miR-122 as small molecular medicine for other types of cancer
therapy.
In this study, we found that endogenous miR-122 in
Huh7 cells infected with miR-122-SP-LV was evidently downregulated
(Fig. 2A) compared to cells
infected with control lentivirus, and the effects of endogenous
miR-122 on cell proliferation (Fig.
3) and migration (Fig. 4C and
D) were efficiently blocked. However, the target expression of
miR-122 and the effects of miR-122 on cell cycle and apoptosis were
not significantly rescued by miR-122-SP, which we hypothesize are
due to a lower expression of miR-122 in Huh7 cells. An increased
expression level of miR-122 in Huh7, we identified a significant
differences in target expression, cell proliferation, migration and
apoptosis between the miR-122-SP-LV and control cells. Furthermore,
miR-122-SP was able to obviously promote xenograft tumor growth by
silencing the endogenous miR-122 in Huh7 (Fig. 6). These data suggest that
miR-122-SP-LV is a useful tool for in vivo research. Using
this system, we can examine more functions of miR-122 in liver
development and hepatocarcinogenesis.
In conclusion, our design of miR-122-SP can
successfully silence miR-122 effects on inhibiting tumorigenesis
in vitro and in vivo, suggesting that miR-122-SP has
potential roles for generating liver cancer models with a lower
expression of miR-122 to investigate the miR-122 functions in liver
development and diseases.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81301704 to J.M.; grant no.
81071524 to F.S.) (URL: http://www.nsfc.gov.cn/Portal0/default152.htm)
Abbreviations:
|
miRNAs
|
microRNAs
|
|
miR-122-SP
|
miR-122 sponge
|
|
3′-UTR
|
3′-untranslated region
|
|
miR-SPs
|
miRNA sponges
|
|
miR-122
|
microRNA-122
|
|
HCC
|
hepatocellular carcinoma
|
|
CCNG1
|
cyclin G1
|
|
ADAM10
|
disintegrin and metalloprotease 10
|
|
ADAM17
|
disintegrin and metalloprotease 17
|
References
|
1
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zeng Y: Principles of micro-RNA production
and maturation. Oncogene. 25:6156–6162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahmed FE: Role of miRNA in carcinogenesis
and biomarker selection: a methodological view. Expert Rev Mol
Diagn. 7:569–603. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Croce CM: Oncogenes and cancer. N Engl J
Med. 358:502–511. 2008. View Article : Google Scholar
|
|
8
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar
|
|
9
|
Ebert MS and Sharp PA: MicroRNA sponges:
progress and possibilities. RNA. 16:2043–2050. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown BD and Naldini L: Exploiting and
antagonizing microRNA regulation for therapeutic and experimental
applications. Nat Rev Genet. 10:578–585. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kloosterman WP, Lagendijk AK, Ketting RF,
Moulton JD and Plasterk RH: Targeted inhibition of miRNA maturation
with morpholinos reveals a role for miR-375 in pancreatic islet
development. PLoS Biol. 5:e2032007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Krützfeldt J, Rajewsky N, Braich R, et al:
Silencing of microRNAs in vivo with ‘antagomirs’. Nature.
438:685–689. 2005.
|
|
13
|
Park CY, Choi YS and McManus MT: Analysis
of microRNA knockouts in mice. Hum Mol Genet. 19:R169–R175. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barbato C, Ruberti F, Pieri M, et al:
MicroRNA-92 modulates K(+) Cl(−) co-transporter KCC2 expression in
cerebellar granule neurons. J Neurochem. 113:591–600.
2010.PubMed/NCBI
|
|
16
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem Cells. 28:357–364.
2010.PubMed/NCBI
|
|
17
|
Bonci D, Coppola V, Musumeci M, et al: The
miR-15a-miR-16-1 cluster controls prostate cancer by targeting
multiple oncogenic activities. Nat Med. 14:1271–1277. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Valastyan S, Reinhardt F, Benaich N, et
al: A pleiotropically acting microRNA, miR-31, inhibits breast
cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Papapetrou EP, Korkola JE and Sadelain M:
A genetic strategy for single and combinatorial analysis of miRNA
function in mammalian hematopoietic stem cells. Stem Cells.
28:287–296. 2010.PubMed/NCBI
|
|
20
|
Gentner B, Schira G, Giustacchini A, et
al: Stable knockdown of microRNA in vivo by lentiviral vectors. Nat
Methods. 6:63–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Loya CM, Lu CS, Van Vactor D and Fulga TA:
Transgenic microRNA inhibition with spatiotemporal specificity in
intact organisms. Nat Methods. 6:897–903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landgraf P, Rusu M, Sheridan R, et al: A
mammalian microRNA expression atlas based on small RNA library
sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jopling CL, Norman KL and Sarnow P:
Positive and negative modulation of viral and cellular mRNAs by
liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol.
71:369–376. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jopling CL: Regulation of hepatitis C
virus by microRNA-122. Biochem Soc Trans. 36:1220–1223. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Girard M, Jacquemin E, Munnich A, Lyonnet
S and Henrion-Caude A: miR-122, a paradigm for the role of
microRNAs in the liver. J Hepatol. 48:648–656. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
down-regulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma L, Liu J, Shen J, et al: Expression of
miR-122 mediated by adenoviral vector induces apoptosis and cell
cycle arrest of cancer cells. Cancer Biol Ther. 9:554–561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bai S, Nasser MW, Wang B, et al:
MicroRNA-122 inhibits tumorigenic properties of hepatocellular
carcinoma cells and sensitizes these cells to sorafenib. J Biol
Chem. 284:32015–32027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang S, Qiu L, Yan X, et al: Loss of
microRNA 122 expression in patients with hepatitis B enhances
hepatitis B virus replication through cyclin G(1)-modulated P53
activity. Hepatology. 55:730–741. 2012. View Article : Google Scholar
|
|
31
|
Lin CJ, Gong HY, Tseng HC, Wang WL and Wu
JL: miR-122 targets an anti-apoptotic gene, Bcl-w, in human
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
375:315–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsai WC, Hsu PW, Lai TC, et al:
MicroRNA-122, a tumor suppressor microRNA that regulates
intrahepatic metastasis of hepatocellular carcinoma. Hepatology.
49:1571–1582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kutay H, Bai S, Datta J, et al:
Downregulation of miR-122 in the rodent and human hepatocellular
carcinomas. J Cell Biochem. 99:671–678. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sahu SC, O’Donnell MW Jr and Sprando RL:
Interactive toxicity of usnic acid and lipopolysaccharides in human
liver HepG2 cells. J Appl Toxicol. 32:739–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meister G, Landthaler M, Dorsett Y and
Tuschl T: Sequence-specific inhibition of microRNA- and
siRNA-induced RNA silencing. RNA. 10:544–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ørom UA, Kauppinen S and Lund AH:
LNA-modified oligonucleotides mediate specific inhibition of
microRNA function. Gene. 372:137–141. 2006.PubMed/NCBI
|
|
37
|
Davis S, Lollo B, Freier S and Esau C:
Improved targeting of miRNA with antisense oligonucleotides.
Nucleic Acids Res. 34:2294–2304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Obad S, dos Santos CO, Petri A, et al:
Silencing of microRNA families by seed-targeting tiny LNAs. Nat
Genet. 43:371–378. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Otaegi G, Pollock A and Sun T: An
optimized sponge for microrna miR-9 affects spinal motor neuron
development in vivo. Front Neurosci. 5:1462012. View Article : Google Scholar : PubMed/NCBI
|