Introduction
Hepatocellular carcinoma (HCC) is the most common
type of liver cancer and is the third leading cause of
cancer-related deaths worldwide. Understanding the molecular
biology and exploring the mechanisms involved in progression of HCC
may facilitate the development of new therapeutic strategies.
Recent studies have confirmed that the process of
epithelial-mesenchymal transition (EMT) is an absolute requirement
for tumor invasion and metastasis (1,2). EMT
may be regulated by a group of transcriptional factors (3,4), and
signaling pathways activated by intrinsic or extrinsic stimuli
converge on these transcriptional factors and regulate the
phenotypic changes of cancer cells (5). Developmental genetics research has
identified many acting transcription factors that play crucial
roles in embryogenesis by orchestrating EMT (6–9). In
recent years, these embryonic transcription factors were found to
have a close relationship with the malignant traits of cancer
cells, such as motility, invasiveness, and resistance to apoptosis.
Slug (SNAI2), a member of the Snail family of zinc-finger
transcription factors, plays a crucial role in the regulation of
EMT during embryogenesis (10,11).
Researchers have found that Slug is involved in cancer cell
invasion, resistance to apoptosis and stem cell features (11–15).
To date, the mechanism involved in the promotion of
HCC progression by Slag is currently unknown. Therefore, in the
present study, we aimed to demonstrate the critical role of Slug in
HCC progression and thus provide novel therapeutic strategies for
HCC.
Materials and methods
Patient samples
HCC tissue specimens were obtained from 113 patients
who underwent hepatectomy for HCC between 2001 and 2010 at the
Tianjin Cancer Hospital, Tianjin Medical University. The diagnoses
of the HCC samples were reviewed by senior pathologists. Detailed
pathological and clinical data were collected.
Immunohistochemical methods
The immunohistochemical assay was performed as
previously described (17–20).
Cell culture, stable cell lines and
expression plasmids
As described in our previous study (21), human liver cancer cell lines (HepG2
and SMMC7221) were obtained from the American Type Culture
Collection (ATCC, USA), and the Cell Bank of the Chinese Academy of
Medical Sciences (Beijing, China). Transfection of HepG2 cells was
performed with Lipofectamine 2000 reagent, and the clones were
selected by G418. For the expression plasmids, full-length Slug
complementary DNA (cDNA) was generated by normal human embryo total
cDNA, and digested with XhoI/EcoRI and subcloned into
pcDNA3.1 vectors. The resulting constructs were confirmed by DNA
sequencing.
Retrovirus vectors and infections
For siRNA-mediated inhibition, the siRNA sequences
against human Slug (5′-CAGACCCATTCTGATGTAAAG-3′) were cloned into
the psiHIV-nH1 lentiviral vector system (GeneCopoeia, FulenGen Co.,
Ltd., Guangzhou, China). Lentiviruses were produced by transient
transfection of 293T cells with the plasmids, and lentiviral
supernatants were collected 48 h post transfection and centrifuged
at 500 × g for 10 min to get rid of the cell debris. Following
centrifugation, the supernatant was filtered through 0.45-μm
polyethersulfone low protein-binding filters. Then the virus
suspension diluted in complete medium with Polybrene
(Sigma-Aldrich, China) at a final concentration of 8 μg/ml was used
to infect the target cells.
Western blot analysis
The whole cell lysates were resolved by way of
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred onto polyvinylidene difluoride membranes (Millipore,
Billerica, MA, USA). Blots were blocked and incubated with the
primary antibody Slug (Cell Signaling Technology, Boston, MA, USA),
CD133 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA), sox2
(GeneTex, San Antonio, TX, USA), nanog (Novus Biologicals,
Littleton, CO, USA), oct4, E-cadherin (both from Santa Cruz
Biotechnology Inc.) and vimentin (Epitomics, Burlingame, CA, USA)
followed by incubation with a secondary antibody (Santa Cruz
Biotechnology, Inc.). Blots were developed using an enhanced
chemiluminescence detection kit (Amersham Pharmacia Biotech,
Piscataway, NJ, USA). For protein loading analyses, a monoclonal
β-actin antibody (Santa Cruz Biotechnology, Inc.) was used.
Apoptosis measurements
The cells were pelleted by centrifugation and
resuspended for apoptosis analysis using the FITC-Annexin V and PI
detection kit (Sigma-Aldrich) according to the manufacturer’s
instructions.
ChIP-on-chip analysis (GEO accession
number: GSE41028)
Samples were harvested from three groups
(HepG2-control cells in regular culture, HepG2-Slug cells in
regular culture and HepG2-Slug cells on Matrigel) and sent to
CapitalBio Corporation (Beijing, China) for further analysis.
Cells (1×108) were fixed with 1%
formaldehyde in culture medium for 10 min at room temperature
followed by quenching with 0.125 M glycine for 5 min. The cells
were washed twice with ice-cold PBS and washed in 10 ml of lysis
buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2,
0.5% IGEPAL, 1 mM PMSF) three times at 4°C. The crosslinked
chromatin was sheared to an average size of 500 bp by ten 30-sec
pulses using a sonicator. The chromatin solution was then incubated
overnight with an anti-Slug antibody at 4°C. After incubation with
protein A beads for 2 h at 4°C, the immune complexes were collected
by centrifugation and washed with the following buffers each for 10
min at 4°C: RIPA buffer (150 mM NaCl, 50 mM Tris pH 8.0, 0.1% SDS),
high-salt buffer (500 mM NaCl, 50 mM Tris pH 8.0, 0.1% SDS, 1%
NP-40), LiCl buffer (250 mM LiCl, 50 mM Tris pH 8.0, 0.5% Na
deoxycholate, 1% NP-40) and 2X TE (20 mM Tris pH 8.0, 2 mM EDTA).
The protein-DNA complexes were eluted from the beads in 450 μl
elution buffer (1% SDS, 100 mM NaHCO3) at 55°C for 2 h
followed by the addition of proteinase K to 500 μg/ml and overnight
incubation at 65°C. Genomic DNA was isolated from the precipitated
material as well as from the sheared chromatin input (1/100 of the
material used for ChIP) by phenol extraction and ethanol
precipitation. One microgram ChIP DNA was directly labeled by DSL
technology at CapitalBio Corporation. The labeled ChIP DNA was
precipitated with 0.1 volume 5 M NaCl and 1 volume isopropanol, and
hybridized in 80 μl of hybridization buffer (3X SSC, 0.2% SDS, 5X
Denhart’s, 25% formamide). Arrays were hybridized in CapitalBio
hybridization stations for 16–18 h at 42°C, and then washed at 42°C
in 0.2% SDS/0.2X SSC, at room temperature in 0.2X SSC, and in 0.05X
SSC. Data of arrays were analyzed by the technicians at CapitalBio
Corporation.
Microarray analysis (GEO accession
number: GSE41028)
Total RNA was extracted from three samples
(HepG2-control cells in regular culture, HepG2-Slug cells in
regular culture and HepG2-Slug cells on Matrigel) using TRIzol
(Invitrogen Life Technologies, Carlsbad, CA, USA) following the
manufacturer’s instructions. Three samples of total RNA were sent
to CapitalBio Corporation for microarray analysis.
Sulforhodamine B (SRB) assay
HepG2 and HepG2-Slug cells were cultured in a
96-well plate at a concentration of 104/100 μl.
Hydroxyurea (Sigma-Aldrich) was used to induce DNA damage (final
concentration, 2 mM). The cells were treated with 10%
trichloroacetic acid for 5 min after 48 h, and then stained by
sulforhodamine B for 30 min at 37°C. A microplate reader (BioTek,
Winooski, VT, USA) was used to measure the absorbance value at 490
nm.
Xenografts
Male BALB/c nude mice, 5 weeks of age, were
purchased from Beijing, China. Viable cells (5×106) were
injected under the skin of 20 nude mice with a 26-gauge needle. The
nude mice with xenografts were monitored for 28 days before
sacrifice.
Ethics statement
Human HCC tissue collection and analysis in this
study were approved by the Ethics Committee of Tianjin Medical
University, China. All animal research was approved by the Animal
Ethics Committee of Tianjin Medical University, China.
Availability of supporting data
section
The microarray data has been deposited in NCBI. The
following is the link (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE41028).
Results
Expression of Slug is correlation with
metastasis and shorter survival time in HCC patients
Slug expression in 113 cases of primary HCC was
examined by IHC. Slug expression was identified in the cytoplasm as
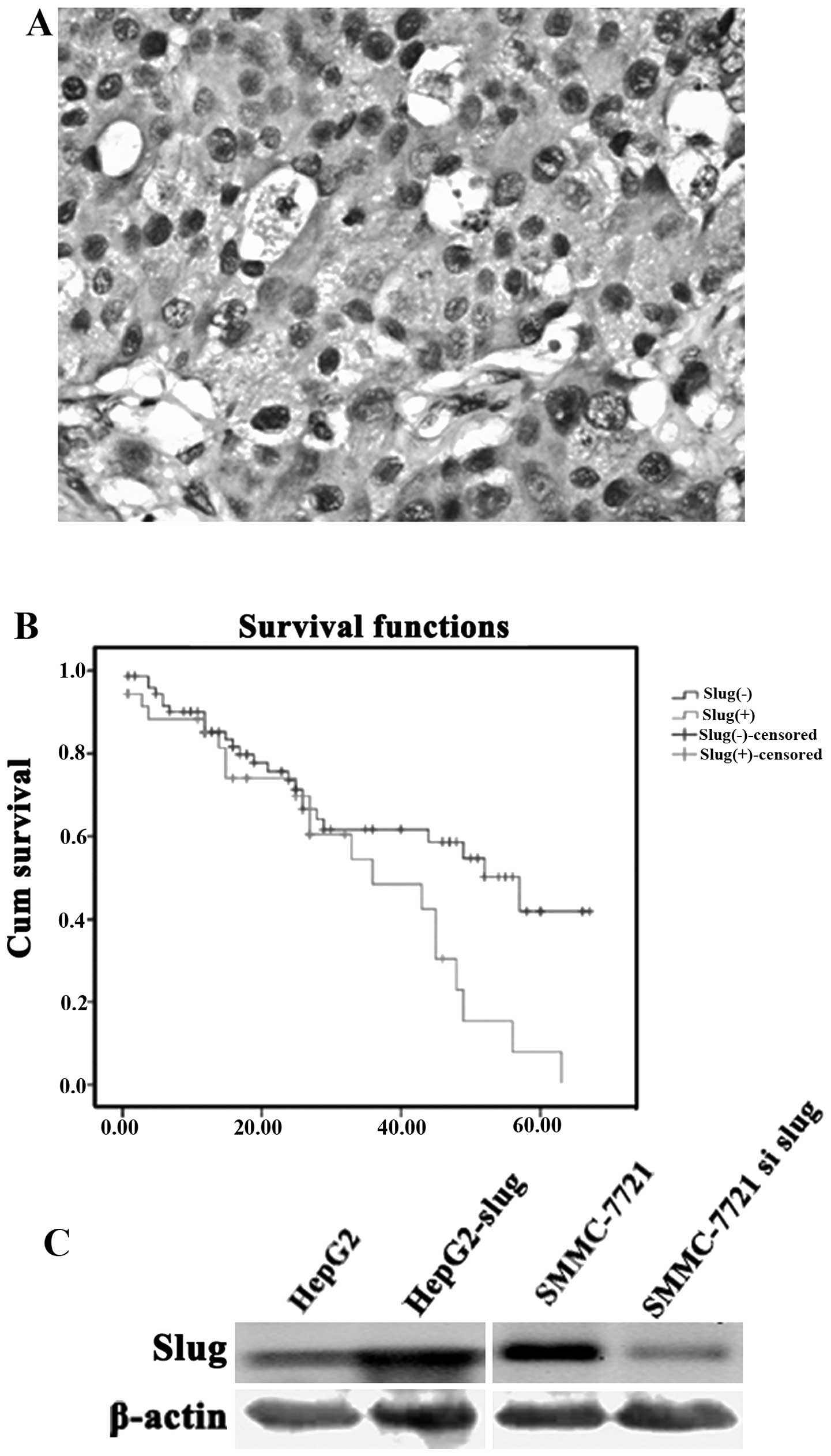
well as in the nucleus of the cancer cells (Fig. 1A). Cases with a percentage of
positive cells ≥10% were considered as Slug-positive. Correlations
between Slug expression and clinicopathological characteristics of
the patients are shown in Table I.
Among all factors compared, the metastasis status was significantly
different between the Slug-positive group and the Slug-negative
group (P=0.020). The positive rate of Slug was 46.8% in the
metastasis group, higher than that of the nonmetastasis group
(25.8%).
 | Table ICorrelation between Slug expression
and clinicopathological characteristics of the patients with
hepatocellular carcinoma. |
Table I
Correlation between Slug expression
and clinicopathological characteristics of the patients with
hepatocellular carcinoma.
| Slug expression | |
|---|
|
| |
|---|
| Clinicopathological
parameters | Positive (n=39) | Negative (n=74) | P-value |
|---|
| Age (years) | 54.6±1.6 | 53.5±1.4 | 0.603 |
| Gender | | | 0.445 |
| Male | 31 | 63 | |
| Female | 8 | 11 | |
| Differentiation
grade | | | 0.246 |
| I/II | 11 | 29 | |
| III/IV | 28 | 45 | |
| Stage | | | 0.508 |
| I/II | 18 | 39 | |
| III/IV | 21 | 35 | |
| Metastasis | | | 0.020 |
| Yes | 22 | 25 | |
| No | 17 | 49 | |
Survival analysis indicated that patients with Slug
positive expression in HCC tissue were significantly associated
with poor overall survival (Fig.
1B). The mean (95% CI) overall survival time was 34.311
(27.084–41.538) and 44.721 (38.192–51.251) months respectively for
patients with and without Slug positive expression in HCC tissue
(P=0.025).
Molecular profiling of Slug downstream
targets in HCC cells cultured on Matrigel
Slug expression was not similarly expressed in the
different HCC cell lines as detected by western blotting. We found
that there was a lower level of Slug expression in the HepG2 cells
compared with the SMMC-7721 cells which showed a higher level
(Fig. 1C).
HepG2 cells were then transfected with Slug cDNA and
showed an increased Slug protein expression (Fig. 1C). Matrigel induces cells to
migrate, and this migratory behavior can be referred to as a model
of tumor cell metastasis in vitro. Thus, we cultured HepG2
and HepG2-Slug cells on Matrigel in order to delineate the Slug
downstream targets during HCC cell migration. On Matrigel,
HepG2-Slug cells showed a more aggressive behavior by forming
tubular structure, suggesting that Slug has the potential to
promote cell migration in vitro.
Slug acts as a transcriptional repressor that binds
to E-box motifs, and the binding site of Slug is known as E-box
(5′-CANNTG-3′) (16). Next, we
examined the promoters that bind to Slug using combined ChIP and
Affymetrix Gene Chip (ChIP-on-chip) for HepG2-control cells in
regular culture (HCR), HepG2-Slug cells in regular culture (HSR)
and HepG2-Slug cells on Matrigel (HSM). The results showed that the
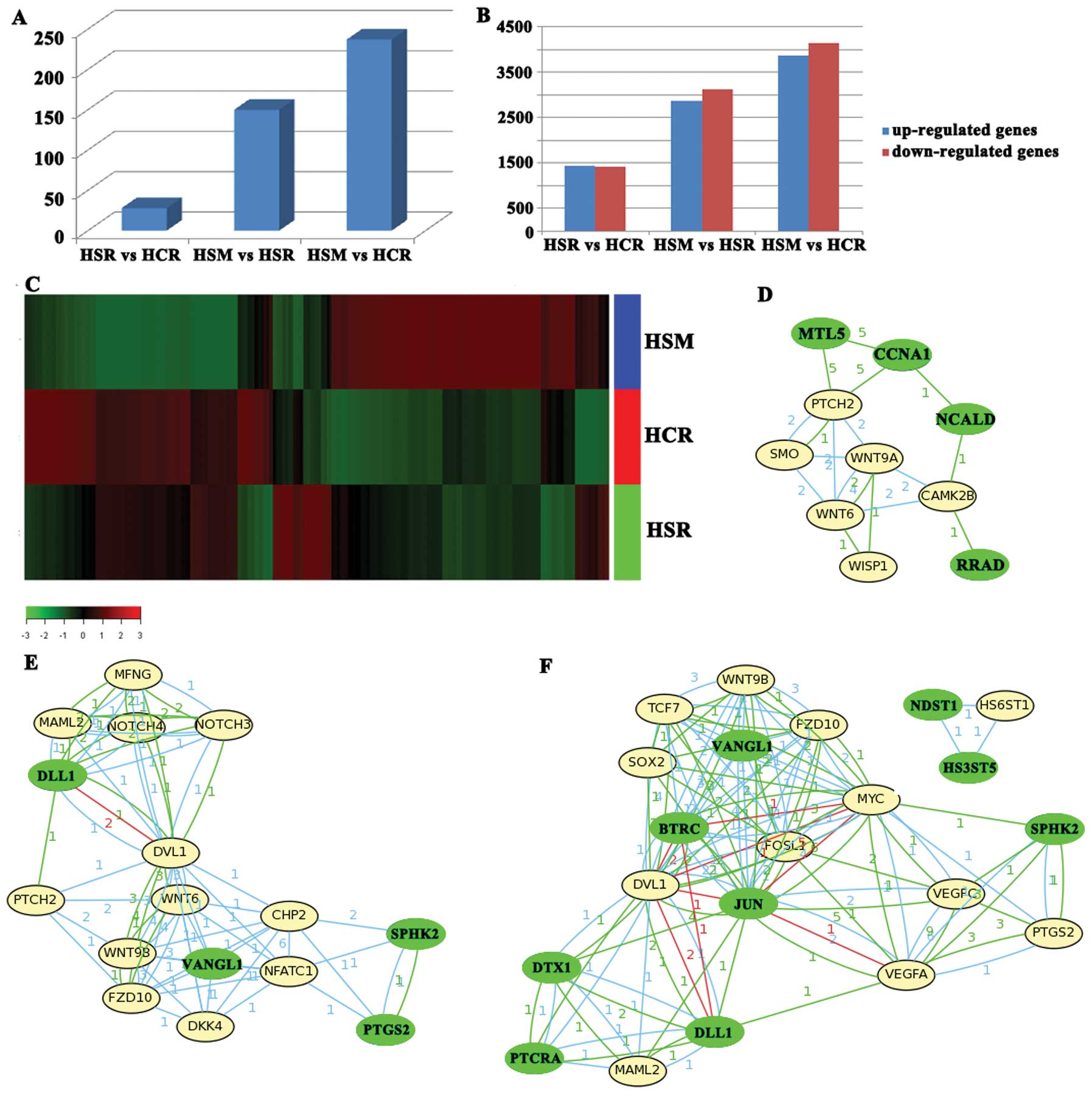
number of gene promoters that bound to Slug only increased to 28 in
the HSR vs. HCR; however, on Matrigel, the number of gene promoters
that bound to Slug in HSM increased significantly and the increased
promoter number reached 150 for HSM vs. HSR, and 237 for HSM vs.
HCR (Fig. 2A). Our study
demonstrated that the peak binding of the promoter by Slug occurred
in HepG2-Slug cells on Matrigel.
Roche NimbleGen microarray analysis was employed to
assess global genome expression in the HCR, HSR and HSM. Our
analysis identified 2,873 genes that were differentially expressed
for HSR vs. HCR; however, there were 6,023 and 8018 genes that were
differentially expressed for HSM vs. HSR and HSM vs. HCR (Fig. 2B and C). The results suggest that
during the process of HCC cell migration when cells were cultured
on Matrigel such as HSM, Slug could bind more genes and provoked
more genes to be upregulated or downregulated thus contributing to
HCC progression.
By Molecule Annotation System (MAS) analysis, many
pathways were identified in HSR vs. HCR, HSM vs. HSR as well as HSM
vs. HCR, such as ECM-receptor interaction pathway, systemic lupus
erythematosus pathway and focal adhesion pathway. Since our results
showed that Slug expression contributed to HCC progression, we
identified the cancer-related pathway as the major signaling
pathway. Between HSR vs. HCR, the involved pathways were Wnt and
Hedgehog pathways initiated by Slug downregulated genes containing
E-boxes (MTL5, RRAD, NCALD and CCNA1). The downregulation of MTL5,
RRAD, NCALD and CCNA1 activated genes of the Wnt and Hedgehog
pathways (WNT9A, WNT6, CAMK2B, WISP1, SMO and PTCH2) (Fig. 2D).
Similarly, the Wnt, Notch and Hedgehog pathways and
the VEGF pathway were identified as major pathways in HSM vs. HSR
and in HSM vs. HCR (Fig. 2E and F).
Furthermore, more genes were involved in HSM compared with HSR and
HCR, suggesting that Wnt, Notch and Hedgehog pathway genes promoted
by Slug overexpression play an important role in the process of
cancer cell invasion. Importantly, the activation of the Wnt and
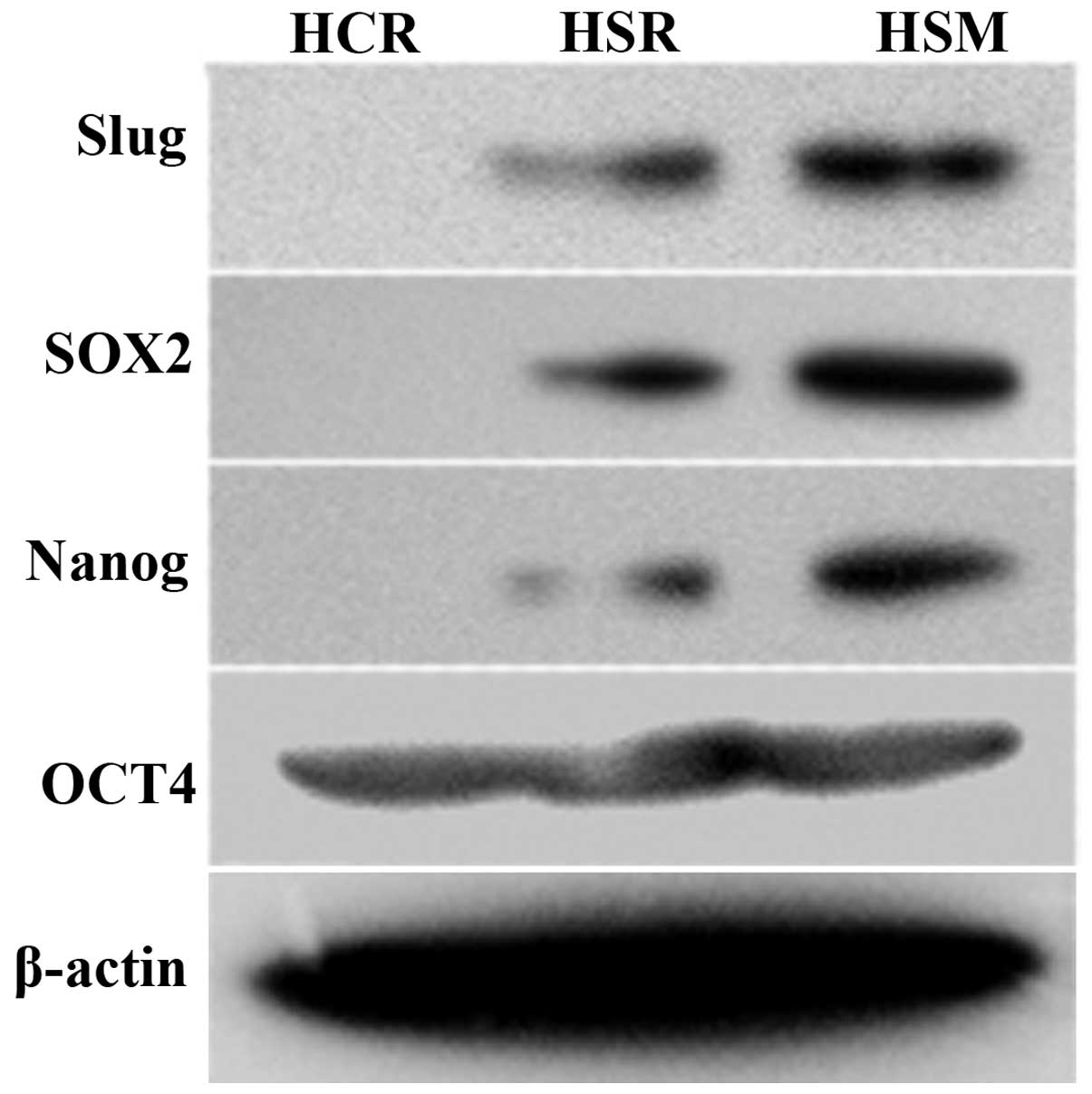
Notch pathways promoted sox2 expression by microarray analysis and
the upregulation was evident in HSM. Further western blotting
validated the elevated sox2 and nanog expression present in HSM
(Fig. 3). Our study suggests that
the reprogramming factors sox2 and nanog contribute to tumor
progression in HCC. In addition, the VEGF pathway was also
activated in HSM, which may be induced by SPHK2 downregulation
initiated by Slug (Fig. 2E and
F).
Slug overexpression has a close
relationship with increased sox2 and nanog expression in HCC
patients
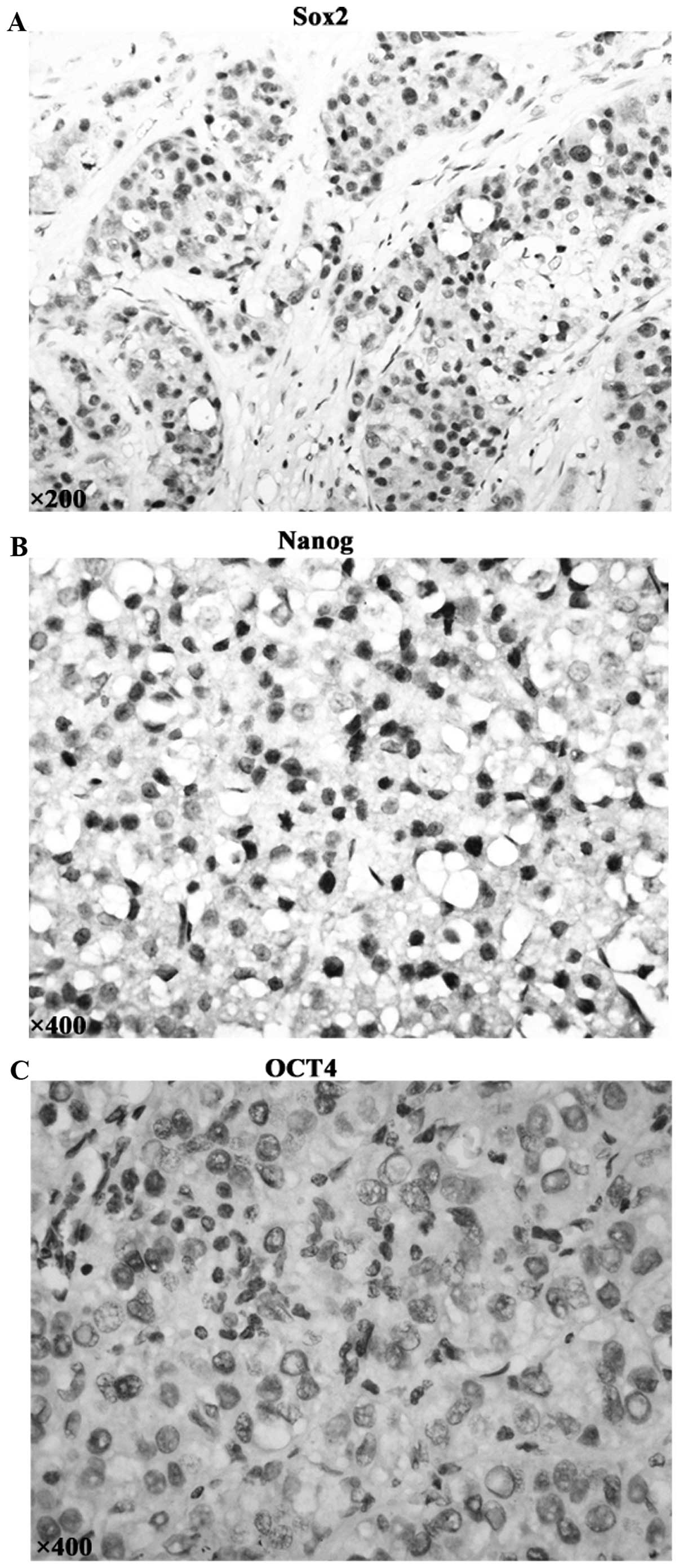
The expression of pluripotency maintaining factors
(sox2, nanog, oct4) which are involved in specification and
maintenance of cancer stem cells were examined by
immunohistochemistry in HCC specimen. Positive cells were indicated
by the presence of brown staining in the nucleus (Fig. 4A and B). The percentage of positive
cells ≥10% was considered as positive. Sox2 and Nanog were detected
in 29.2 and 13.3% of hepatocellular cancer tissues; whereas there
was lack of Oct4 expression in all the 113 HCC cases (Fig. 4C). Importantly, there was a
significant correlation between Slug and sox2 expression (r=0.230,
P=0.014) as well as Slug and nanog expression (r=0.210,
P=0.026).
Slug silencing induces apoptosis and
inhibits cell migration in HCC cells in vitro
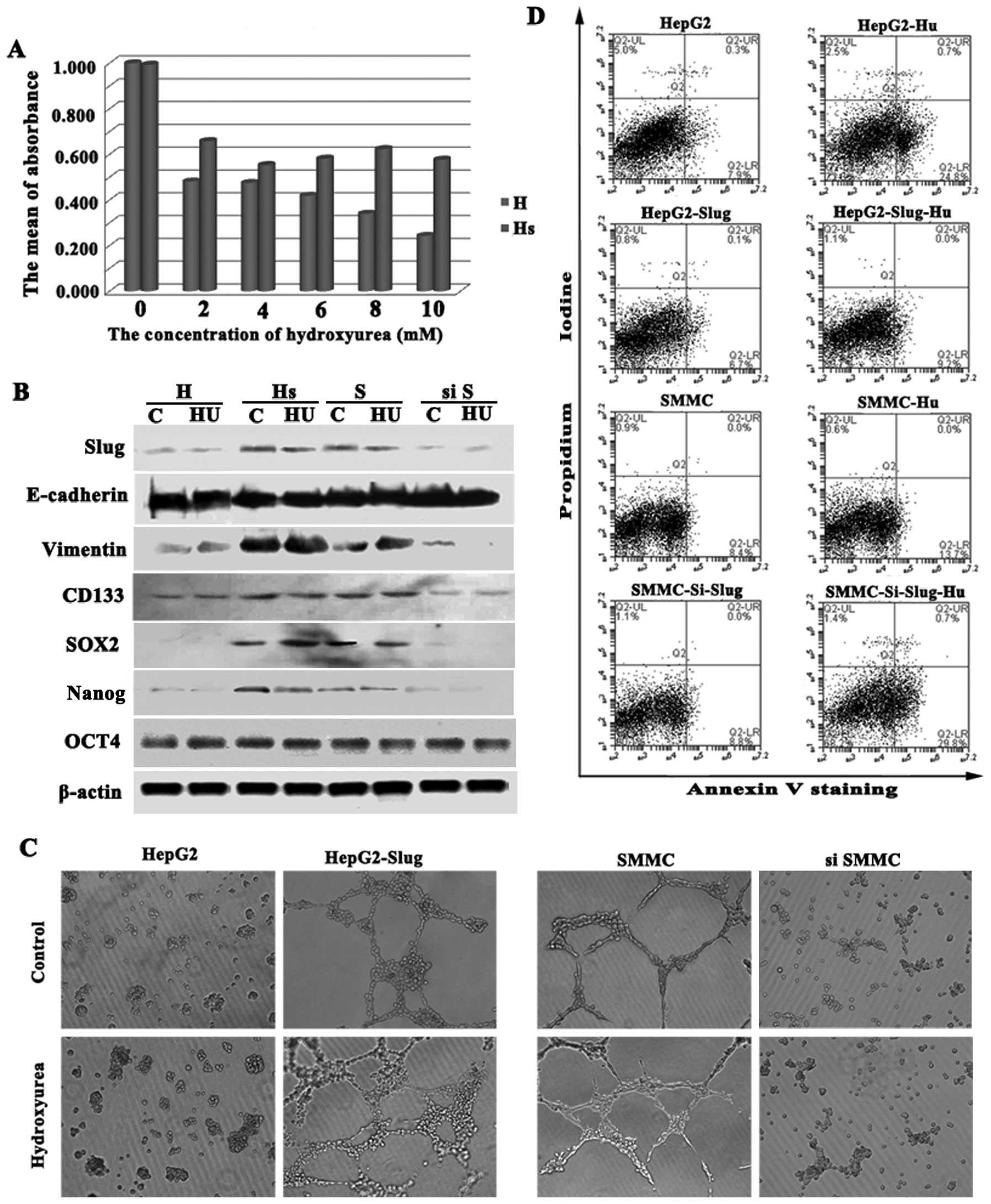
HepG2-Slug and HepG2-control cells were treated with
2–10 mM hydroxyurea (HU) for 48 h and were then assessed for cell
proliferation employing the SRB assay. Cell proliferation in the
HepG2-control cells was significantly inhibited by HU to different
extents depending on the dose. However, HepG2-Slug cells showed
increased resistance to the cytotoxic effects of HU compared to the
cultured HepG2-control cells. Cell proliferation in the HepG2-Slug
cells was inhibited to a lesser extent and the inhibition was
independent of the dose (Fig.
5A).
To evaluate whether endogenous Slug plays any role
in HCC cells with high Slug expression, we knocked down Slug
expression in SMMC-7721 cells using Slug siRNA. The concomitant
decrease in the Slug protein level in the Slug siRNA-treated cells
was evident from the western blot data (Fig. 1C). Since Slug overexpression
conferred more resistance to HU, we next observed whether or not HU
treatment had an effect on Slug expression. HepG2-control,
HepG2-Slug, SMMC-7721-control and SMMC-7721-siRNASlug cells were
treated with 2 mM HU for 48 h and western blotting showed that the
expression level of Slug was not reduced by HU in the HepG2-Slug
and SMMC-7721 cells with higher Slug expression (Fig. 5B). In addition, western blot
analysis demonstrated the maintenance of mesenchymal marker
vimentin expression and the CD133+ CSC phenotype when
HepG2-Slug and SMMC-7721 cells were treated with HU. Importantly,
neither sox2 nor nanog expression was reduced by HU treatment
(Fig. 5B). Additionally, cell
migration was not inhibited when HepG2-Slug and SMMC-7721 cells
were treated with HU (Fig. 5C).
Analysis of Annexin V+ cells showed that
the fraction of Annexin V+ cells in the HepG2-Slug and
SMMC-7721 cells with high Slug expression did not increase
significantly when exposed to HU compared with the control cells.
However, SMMC-7721 cells depleted of Slug, similar to the HepG2
cells with low Slug expression treated with HU, showed a
significant increase in the proportion of Annexin V+
cells (Fig. 5D). Therefore Slug
silencing played a major role in the commitment to apoptosis.
Moreover, SMMC-7721 cells with Slug silencing showed reduced
mesenchymal marker vimentin expression and the CD133−
non-CSC phenotype (Fig. 5B).
Remarkably, Slug silencing inhibited SMMC-7721 cell migration on
Matrigel with or without HU treatment (Fig. 5C).
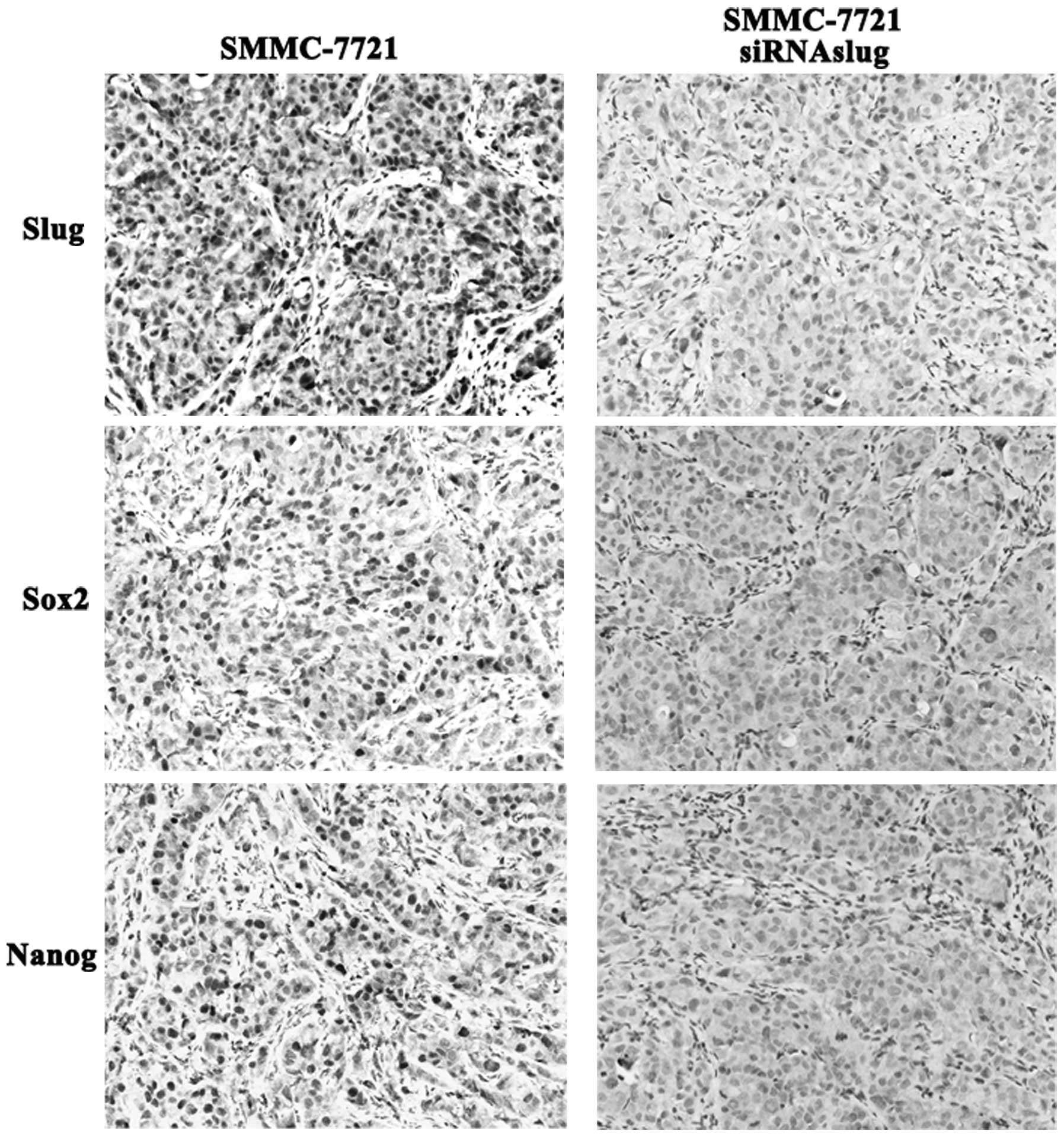
Slug silencing inhibits sox2 and nanog
expression in vivo
To further confirm the relationship of Slug with HCC
progression in vivo, a xenograft model of human HCC
progression employing SMMC-7721 cells and SMMC-7721-siRNASlug cells
was established. Although neither SMMC-7721 nor SMMC-7721-siRNASlug
xenografts displayed propensity to metastasize to distant organs
such as liver and lung, the SMMC-7721-siRNASlug cells failed to
grow vigorously in the nude mice as compared to the parental
SMMC-7721 cells. Remarkably, after in vivo growth,
SMMC-7721-siRNASlug xenografts displayed lower sox2 and nanog
expression while higher levels were noted in the SMMC-7721
xenografts (Fig. 6), suggesting
that Slug has a close relationship with sox2 and nanog
expression.
Discussion
Although the specific role of Slug in the
downregulation of E-cadherin is not completely clear, Slug is
critical to induce the EMT phenotype, cancer stem-like properties
and mediate radioresistance and chemoresistance (22–25).
In the present study, we detected Slug expression in 113 cases of
HCC tissue samples to characterize the linkage between the
activation of Slug and metastasis. Statistical analysis showed that
expression of Slug was correlated with metastasis and a shorter
survival time in HCC patients. Therefore, our study suggests that
Slug overexpression could serve as a poor prognosis marker.
ChIP-on-chip analysis showed that the greatest
number of binding peaks of Slug occurred in the HepG2-Slug cells on
Matrigel. We also identified the novel non-canonical pathway, Wnt
and Notch pathway, leading to sox2 and nanog overexpression in
vitro. Recent research showed that Sox2, Nanog and Oct4, can
directly reprogram somatic cells to a pluripotent stem cell state
(26–29). In our study, HCC progression may be
induced through the activation of a reprogramming-like mechanism
promoted by Sox2 and nanog.
Our data also showed that a correlation between
expression levels of Slug and increased sox2 and nanog expression
was obvious in the human HCC tissue specimens and HCC xenografts
in vivo, thus indicating that Slug is sufficient to induce
sox2 and nanog overexpression. Interestingly, Slug overexpression
potentiated the chemoresistance properties of HepG2 cells to DNA
damage reagent HU. Moreover, HU treatment could not affect EMT, the
CSC phenotype and cell migration in HCC cells with Slug
overexpression. Notably, our study showed that Slug silencing
inhibited cell migration on Matrigel in vitro, suggesting
that Slug plays a crucial role in HCC progression.
In conclusion, our findings reveal a previously
unidentified role of Slug to promote HCC progression by activation
of reprogramming-related genes sox2 and nanog.
Acknowledgements
This study was partly supported by a grant from the
Key Project of the National Natural Science Foundation of China
(no. 81230050), the National Natural Science Foundation of China
(nos. 81172046 and 81173091), the Key Project of the Tianjin
Natural Science Foundation (no. 12JCZDJC23600) and the Tianjin
Natural Science Foundation (no. 12JCYBJC15500).
References
|
1
|
Książkiewicz M, Markiewicz A and Zaczek
AJ: Epithelial-mesenchymal transition: a hallmark in metastasis
formation linking circulating tumor cells and cancer stem cells.
Pathobiology. 79:195–208. 2012. View Article : Google Scholar
|
|
2
|
Tiwari N, Gheldof A, Tatari M and
Christofori G: EMT as the ultimate survival mechanism of cancer
cells. Semin Cancer Biol. 22:194–207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sánchez-Tilló E, Liu Y, de Barrios O,
Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A
and Postigo A: EMT-activating transcription factors in cancer:
beyond EMT and tumor invasiveness. Cell Mol Life Sci. 69:3429–3456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Balli D, Ustiyan V, Zhang Y, Wang IC,
Masino AJ, Ren X, Whitsett JA, Kalinichenko VV and Kalin TV: Foxm1
transcription factor is required for lung fibrosis and
epithelial-to-mesenchymal transition. EMBO J. 32:231–244. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Craene BD and Berx G: Regulatory networks
defining EMT during cancer initiation and progression. Nat Rev
Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wehbe M, Soudja SM, Mas A, Chasson L,
Guinamard R, de Tenbossche CP, Verdeil G, Van den Eynde B and
Schmitt-Verhulst AM: Epithelial-mesenchymal-transition-like and
TGFβ pathways associated with autochthonous inflammatory melanoma
development in mice. PLoS One. 7:e494192012. View Article : Google Scholar
|
|
7
|
Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS,
Hwang SG, An S, Yoon G, Gye MC, Yi JM, Kim MJ and Lee SJ: Claudin-1
induces epithelial-mesenchymal transition through activation of the
c-Abl-ERK signaling pathway in human liver cells. Oncogene.
32:4873–4882. 2013. View Article : Google Scholar
|
|
8
|
Tanaka Y, Terai Y, Kawaguchi H, Fujiwara
S, Yoo S, Tsunetoh S, Takai M, Kanemura M, Tanabe A and Ohmichi M:
Prognostic impact of EMT
(epithelial-mesenchymal-transition)-related protein expression in
endometrial cancer. Cancer Biol Ther. 14:13–19. 2013. View Article : Google Scholar :
|
|
9
|
Zheng X, Vittar NB, Gai X,
Fernandez-Barrena MG, Moser CD, Hu C, Almada LL, McCleary-Wheeler
AL, Elsawa SF, Vrabel AM, Shire AM, Comba A, Thorgeirsson SS, Kim
Y, Liu Q, Fernandez-Zapico ME and Roberts LR: The transcription
factor GLI1 mediates TGFβ1 driven EMT in hepatocellular carcinoma
via a SNAI1-dependent mechanism. PLoS One. 7:e495812012. View Article : Google Scholar
|
|
10
|
Kim NH, Kim HS, Li XY, Lee I, Choi HS,
Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher
CA, Weiss SJ and Yook JI: A p53/miRNA-34 axis regulates
Snail1-dependent cancer cell epithelial-mesenchymal transition. J
Cell Biol. 195:417–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mathsyaraja H and Ostrowski MC: Setting
Snail2’s pace during EMT. Nat Cell Biol. 14:1122–1123. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith BN and Odero-Marah VA: The role of
Snail in prostate cancer. Cell Adh Migr. 6:433–441. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimokawa M, Haraguchi M, Kobayashi W,
Higashi Y, Matsushita S, Kawai K, Kanekura T and Ozawa M: The
transcription factor Snail expressed in cutaneous squamous cell
carcinoma induces epithelial-mesenchymal transition and
down-regulates COX-2. Biochem Biophys Res Commun. 430:1078–1082.
2013. View Article : Google Scholar
|
|
14
|
Kume K, Haraguchi M, Hijioka H, Ishida T,
Miyawaki A, Nakamura N and Ozawa M: The transcription factor Snail
enhanced the degradation of E-cadherin and desmoglein 2 in oral
squamous cell carcinoma cells. Biochem Biophys Res Commun.
430:889–894. 2013. View Article : Google Scholar
|
|
15
|
Lemma S, Karihtala P, Haapasaari KM,
Jantunen E, Soini Y, Bloigu R, Pasanen AK, Turpeenniemi-Hujanen T
and Kuittinen O: Biological roles and prognostic values of the
epithelial-mesenchymal transition-mediating transcription factors
Twist, ZEB1 and Slug in diffuse large B-cell lymphoma.
Histopathology. 62:326–333. 2013. View Article : Google Scholar
|
|
16
|
Bhat-Nakshatri P, Appaiah H, Ballas C,
Pick-Franke P, Goulet R Jr, Badve S, Srour EF and Nakshatri H:
SLUG/SNAI2 and tumor necrosis factor generate breast cells with
CD44+/CD24− phenotype. BMC Cancer.
10:4112010. View Article : Google Scholar
|
|
17
|
Sun T, Sun BC, Zhao XL, Zhao N, Dong XY,
Che N, Yao Z, Ma YM, Gu Q, Zong WK and Liu ZY: Promotion of tumor
cell metastasis and vasculogenic mimicry by way of transcription
coactivation by Bcl-2 and Twist1: a study of hepatocellular
carcinoma. Hepatology. 54:1690–1706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Che N, Zhao XL, Sun T, Zhao XM, Gu Q, Dong
XY, Zhao N, Liu YR, Yao Z and Sun BC: The role of Twist1 in
hepatocellular carcinoma angiogenesis: a clinical study. Hum
Pathol. 42:840–847. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu T, Sun B, Zhao X, Gu Q, Dong X, Yao Z,
Zhao N, Chi J, Liu N, Sun R and Ma Y: HER2/neu expression
correlates with vasculogenic mimicry in invasive breast carcinoma.
J Cell Mol Med. 17:116–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu TJ, Sun BC, Zhao XL, Zhao XM, Sun T,
Gu Q, Yao Z, Dong XY, Zhao N and Liu N: CD133+ cells
with cancer stem cell characteristics associates with vasculogenic
mimicry in triple-negative breast cancer. Oncogene. 32:544–553.
2013. View Article : Google Scholar
|
|
21
|
Sun D, Sun B, Liu T, Zhao X, Che N, Gu Q,
Dong X, Yao Z, Li R, Li J, Chi J and Sun R: Slug promoted
vasculogenic mimicry in hepatocellular carcinoma. J Cell Mol Med.
17:1038–1047. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and Slug
mediate radioresistance and chemoresistance by antagonizing
p53-mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nassour M, Idoux-Gillet Y, Selmi A, Côme
C, Faraldo ML, Deugnier MA and Savagner P: Slug controls
stem/progenitor cell growth dynamics during mammary gland
morphogenesis. PLoS One. 7:e534982012. View Article : Google Scholar
|
|
24
|
Guo W, Keckesova Z, Donaher JL, Shibue T,
Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell
G, Tam WL, Mani SA, van Oudenaarden A and Weinberg RA: Slug and
Sox9 cooperatively determine the mammary stem cell state. Cell.
148:1015–1028. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shih JY and Yang PC: The EMT regulator
Slug and lung carcinogenesis. Carcinogenesis. 32:1299–1304. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Basu-Roy U, Seo E, Ramanathapuram L, Rapp
TB, Perry JA, Orkin SH, Mansukhani A and Basilico C: Sox2 maintains
self renewal of tumor-initiating cells in osteosarcomas. Oncogene.
31:2270–2282. 2012. View Article : Google Scholar
|
|
27
|
Leis O, Eguiara A, Lopez-Arribillaga E,
Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R
and Martin AG: Sox2 expression in breast tumours and activation in
breast cancer stem cells. Oncogene. 31:1354–1365. 2012. View Article : Google Scholar
|
|
28
|
Ichida JK, Blanchard J, Lam K, Son EY,
Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K,
Huangfu D, Akutsu H, Liu DR, Rubin LL and Eggan K: A small-molecule
inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by
inducing nanog. Cell Stem Cell. 5:491–503. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grad I, Hibaoui Y, Jaconi M, Chicha L,
Bergström-Tengzelius R, Sailani MR, Pelte MF, Dahoun S, Mitsiadis
TA, Töhönen V, Bouillaguet S, Antonarakis SE, Kere J, Zucchelli M,
Hovatta O and Feki A: NANOG priming before full reprogramming may
generate germ cell tumours. Eur Cell Mater. 22:258–274.
2011.PubMed/NCBI
|