Introduction
Epstein-Barr virus (EBV), a ubiquitous human γ
herpes virus, establishes persistent latent infections and has been
causally associated with a variety of B-cell and epithelial cell
malignancies, including Burkitt’s lymphoma (BL), Hodgkin’s
lymphoma, gastric carcinoma and nasopharyngeal carcinoma (NPC)
(1). EBV depends on Epstein-Barr
nuclear antigen 1 (EBNA1) to mediate critical processes in the
viral life cycle, and EBNA1 is the only viral protein that has been
identified in all EBV-associated malignant tissues. It is
well-known that EBNA1 expression is necessary for maintaining EBV
in latently proliferating cells, mediating synthesis of the EBV
genome, non-randomly partitioning the virus to daughter cells
(2–4) and regulating viral gene transcription
(5–9). The effects of EBNA1 on the host cell
remain to be elucidated (10,11),
although it has been suggested that EBNA1: is critical for the
continued survival of BL (12), may
affect the function of p53 by competing for the USP7
ubiquitin-specific protease (13,14),
promotes the development of B-cell lymphoma in transgenic mice
(15), counteracts Nm23-H1-mediated
suppression of cell migration (16,17),
serves as a potential regulator of signaling pathways (18–21),
and increases reactive oxygen species levels and genomic
instability in BL cell lines (22,23).
The aforementioned findings suggest that EBNA1 may play a direct
role in promoting the tumorigenesis of EBV-associated
malignancies.
The molecular weight of EBNA1 varies from 69 to 96
kDa, depending on the number of internal glycine-alanine repeats
(24–26), which contribute to downregulating
EBV-mediated immune responses by inhibiting antigen presentation
(27). Sequence variations in EBNA1
relative to that of the prototypic B95.8 strain were first
described in sequences N- and C-terminal to the internal repeat
regions of EBNA1 proteins isolated from EBV-positive NPC samples
obtained in Hong Kong (28).
Subsequently, numerous EBNA1 sequences have been identified from
healthy individuals and patients with various EBV-associated
diseases in different geographic regions (29). Gutiérrez et al (30) classified EBNA1 into the P-ala,
P-thr, V-val, V-leu and V-pro subtypes, which were termed according
to the amino acid (aa) at position 487 in EBNA1 (numbered according
to prototype B95.8 strain). Sub-variants of P-ala, P-thr and V-val,
have also been characterized (31–33).
Previously, it was shown that within a given geographic area,
tumor-associated EBNA1 subtypes were also prevalent in the general
population (34). For example,
V-val EBNA1 was found exclusively in southeastern Chinese NPC
samples, and was also the predominant subtype observed in this
general population (32,33,35–37).
However, most of the molecular studies on the biological functions
of full-length EBNA1 have used the sequence from prototype B95.8, a
strain originating from an infectious mononucleosis patient in
Africa, and whose EBNA1 belongs to the P-ala subtype (30). Thus it is important to investigate
whether any functional differences are associated with genetic
variations in EBNA1, and the manner in which they affect risk of
disease. Results of such an investigation may provide a better
understanding of the molecular mechanisms underlying the
associations between EBV-related disease and particular cell types
and/or genetic backgrounds.
Consequently, we cloned the full-length EBNA1
gene from CG3 cells (38). This
EBV-containing lymphoblastoid cell line was originally established
from the bone marrow cells of a chronic myeloid leukemic patient
from Taiwan, with V-val being the predominant EBNA1 subtype. As
expected, the EBNA1 detected in CG3 cells belonged to the V-val
subtype. Subsequently, we purified His-tagged, bacterially
expressed N- and C-terminal CG3 EBNA1 fusion proteins and used them
to raise EBNA1-reactive antibodies in rabbits. Additionally, we
successfully expressed the full-length CG3 EBNA1 in eukaryotic
cells, generating an experimental system that was subsequently used
to elucidate the oncogenic potential of this V-val EBNA1
subtype.
Materials and methods
Cell lines
CG3 (38) is a
spontaneous EBV-carrying lymphoblastoid cell line that was derived
from a Taiwanese patient with juvenile chronic myeloid leukemia
(kindly provided by Dr Y.S. Chang). B95.8 (39) is a marmoset lymphoid cell line that
was EBV-immortalized with a virus originating from an infectious
mononucleosis patient, sampled in Africa. Raji (40), P3HR1 (41) and Daudi (42) are EBV-containing human lymphoid cell
lines harboring viruses from Burkitt’s lymphoma patients. The cells
were maintained in RPMI-1640 supplemented with 10% fetal calf serum
(FCS), at 37°C in a humidified 5% CO2 atmosphere. The
293 cell line (a human embryonal kidney cell line) (43) was grown in DMEM supplemented with
10% FCS. The 293-EBNA cells (i.e., 293 cells stably expressing
B95.8 EBNA1; Invitrogen-Life Technologies, Carlsbad, CA, USA) were
maintained in DMEM-10% FCS containing 250 μg/ml of Geneticin (G418;
Gibco-Life Technologies, Carlsbad, CA, USA).
PCR amplification and sequence analysis
of the EBNA1 gene from cultured cells
DNA was extracted from CG3 and 293-EBNA cells using
a QIAamp DNA Mini kit (Qiagen, Valencia, CA, USA) according to the
manufacturer’s instructions. The full-length EBNA1 gene and
the sequence encoding its middle repeat region were PCR-amplified
using the GC-RICH PCR System (Roche Molecular Biochemicals)
according to the manufacturer’s instructions. The sequences of the
utilized primers are shown in Table
I, with the nucleotide numbering system used in this report
being in accordance with that of Baer et al (44). For all PCR reactions discussed
herein, the input DNAs were preheated at 95°C for 5 min prior to
cycling, and the PCR products were subjected to a final extension
for 10 min at 72°C. Briefly, 100 ng of DNA was combined with the
provided reaction mixture (containing 0.5 M GC-RICH resolution
solution) and subjected to PCR. To amplify the full-length
EBNA1 gene, the F1/F2 primers were used along with the
following reaction conditions: 30 cycles of 95°C for 1 min, 63°C
for 2 min 30 sec and 72°C for 1 min. Nested PCR was required to
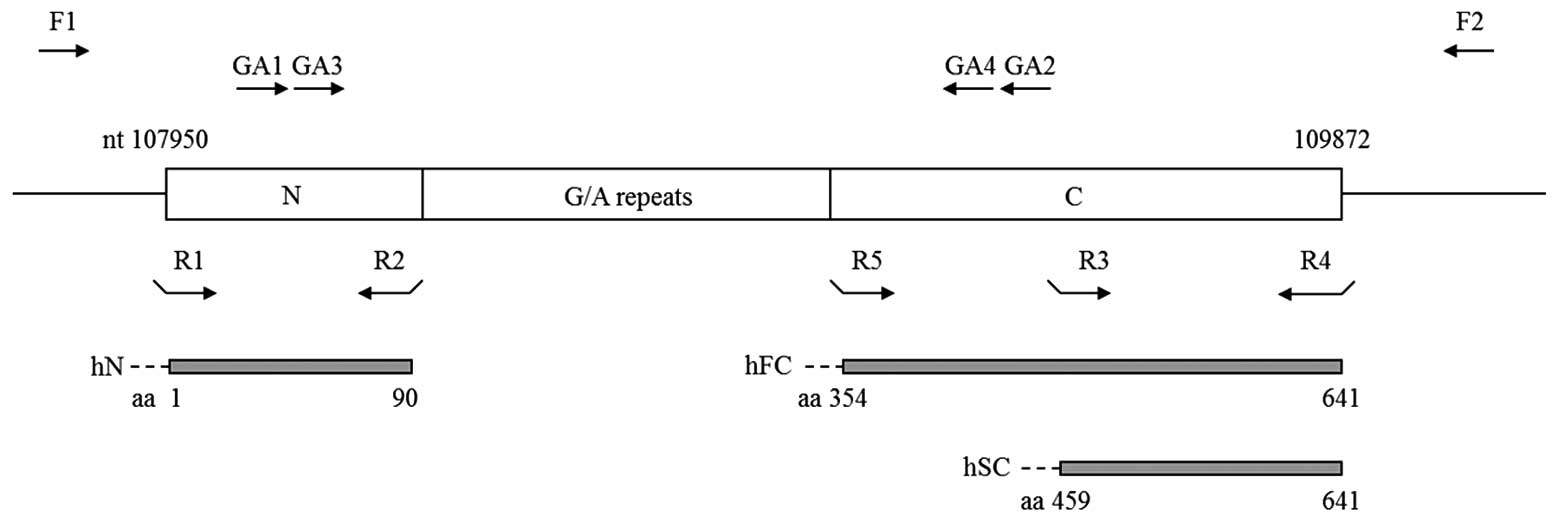
successfully amplify the internal repeat sequences of EBNA1
(Fig. 1). Primer pairs GA1/GA2 and
GA3/GA4 were used for the first and second round of PCR,
respectively, under the aforementioned reaction conditions. The PCR
products representing the full-length EBNA1 gene from CG3
and 293-EBNA cells were purified and cloned into a T-vector
(pCRII-TOPO; Invitrogen) and three independent clones were
sequenced (ABI377 DNA sequencing system; Perkin-Elmer/Applied
Biosystems). From the clone, plasmids containing consensus
sequences for pCRII-TOPO-CG3.1 and pCRII-TOPO-B95.8.1 were selected
and used to generate EBNA1 eukaryotic expression vectors that
expressed the CG3 and B95.8 EBNA1s, respectively (see below).
 | Table IList of primers used for PCR
amplification of EBNA 1 sequences. |
Table I
List of primers used for PCR
amplification of EBNA 1 sequences.
| Primers | Sequence
(5′-3′) | B95.8
coordinates |
|---|
| F1 |
CGTCCAGCAAAAAGGGGGACGAG | 107671-107693 |
| F2 |
GCGGAGCTGAGTGACGTGACAAC | 110062-110040 |
| GA1 |
CCATGGACGAGGACGGGGAAG | 108063-108083 |
| GA2 |
GCTTGGGCCTTCTCCTGGGTCAT | 109290-109266 |
| GA3 |
AGGACGAGGAGGCGGAAGACCAG | 108090-108112 |
| GA4 |
TGCGGGGCCCTGCTCTATCG | 109266-109247 |
| R1 |
CGCCTAGCATATGATGTCTGACGAGGGGCCA |
NdeI-107950-107967 |
| R2 |
CCGGGATCCTTATCCTGTTCCACCGTGGGTC |
BamHI-108219-108202 |
| R3 |
CGCCTAGCATATGAGGCGCAAAAAAGGAGGGTGG |
NdeI-109323-109344 |
| R4 |
CCGGGATCCTTAATATACGAACACACCGGCGAC |
BamHI-109875-109855 |
| R5 |
CGCCTAGCATATGCGGGGTAGAGGACGTGAAAGA |
NdeI-109009-109029 |
Expression and purification of
recombinant EBNA1 proteins in E. coli
pCRII-TOPO-CG3.1 served as the template for PCR
amplifying three fragments: the N fragment covering the N-terminus
of EBNA1 (aa 1-90), and fragments FC and SC corresponding to two
C-terminal regions downstream of the glycine-alanine repeat region
(aa 354-641 and 459-641, respectively) (Fig. 1). The N, SC and FC fragments were
amplified with primer pairs R1/R2, R3/R4 and R5/R4 (Table I and Fig. 1), respectively. For cloning
purposes, restriction enzyme sites were included in the primers.
The PCR products were then subjected to NdeI and
BamHI double digestion followed by gel purification. The
purified products were inserted in-frame at the C-terminus of the
His-tag in pET-15b plasmids (Novagen) which had been pretreated
with NdeI and BamHI. The resulting plasmids
containing the N, FC and SC inserts were designated as
pET15b-hN, pET15b-hFC and
pET15b-hSC, respectively, and their encoded fusion
proteins were referred to as hN, hFC and hSC, respectively. These
plasmids were used to transform BL21(DE3)pLysS (Novagen), and the
fusion proteins were induced and purified as per the manufacturer’s
instructions. Briefly, bacteria were grown to an OD600
of 0.6, 1 mM IPTG was added, and the incubation was continued for 3
h via agitation. The bacterial pellet was collected by
centrifugation at 2500 g for 15 min and the cells were lysed by
freeze-thawing. The soluble recombinant EBNA1 proteins were
purified with one-step metal chelation chromatography (Novagen).
The purified hFC was then concentrated using Ultracel-3K
(Millipore, Billerica, MA, USA). Equal amounts of the purified
fractions were collected, resolved by SDS-PAGE and subjected to
Coomassie blue staining. Western blot analysis, as previously
described (45), was performed
using an anti-His antibody (diluted 1:1,000; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA).
Immunization of animals
The backs of New Zealand White rabbits were
subcutaneously immunized with 300 μg of hN or hSC in 0.5 ml
complete Freund’s adjuvant (v/v 1:1) on day 0, and with the same
amount of the corresponding protein in incomplete Freund’s adjuvant
(v/v 1:1) (Sigma-Aldrich) on day 20 and were bled on day 27. The
obtained antisera were designated Ab-N and Ab-SC, respectively. The
animal experimental protocol was reviewed and approved by the Chang
Gung University Animal Ethics Committee. The antibody responses
were examined by western blot analysis using purified recombinant
proteins and extracts prepared from EBV-containing cell lines and
EBNA1-transfected 293 cells.
Expression of EBNA1 from eukaryotic
vectors
pCRII-TOPO-CG3.1 and pCRII-TOPO-B95.8.1 were
digested with BamHI, filled in with Klenow (New England
Biolabs, Beverly, MA, USA), and cleaved with XbaI to release
the inserts of interest. Plasmid pCMV-EBNA (Clontech), which
encoded EBNA1 of the B95.8 strain, was treated with PstI,
filled in with Klenow, and digested with XbaI to remove the
B95.8 EBNA1 gene, which served as the vector. The insert and
vector were gel purified and ligated to generate the eukaryotic
expression plasmids, pCMV-CG3EBNA1 and pCMV-B95.8EBNA1. These
plasmids were transfected into 293 cells. Briefly, 5×105
cells/wel) were seeded in 6-well (35-mm diameter) plates, grown for
18 h (~60–70% confluency), and then transfected with 4 μg DNA and
10 μl Lipofectamine (Invitrogen), according to the manufacturer’s
instructions. At 48 h post-transfection, the cell extracts were
collected, resolved by 10% SDS-PAGE, and subjected to western blot
analysis (45) with rabbit Ab-SC
antibodies. Proteins extracted from 293-EBNA (Invitrogen) and CG3
cells were used as positive controls. To generate 293 cells stably
expressing EBNA1 proteins of B95.8 or CG3 origin, or the
drug-resistant gene alone (as a control), EBNA1 expression vector-
and empty vector-transfected cells were selected in the presence of
500 μg/ml Geneticin (G418; Gibco) and then maintained in medium
containing 250 μg/ml Geneticin.
Serum withdrawal and cell survival
Stably transfected 293 cells (2×105
cells/well) were seeded in 6-well plates. At 16 h post-seeding, the
cells were washed twice with serum-free medium and then cultured in
the absence and presence of FCS. Cell viability was determined at
various time points using trypan blue exclusion. Proteins were also
extracted at different time points and subjected to western blot
analysis using antibodies against poly(ADP-ribose) polymerase
(PARP) and actin (diluted 1:600 and 1:5,000, respectively; both
from Millipore).
Results
The EBNA1 sequence in CG3 cells
Size polymorphisms and sequence variations have been
reported in EBNA1 proteins from different EBV isolates. CG3 is an
EBV-containing lymphoblastoid cell line established from a
Taiwanese patient with chronic myeloid leukemia (38). To the best of our knowledge, no
previous study has reported the size and sequence of its EBNA1
protein. Consequently, PCR was amplified with the full-length
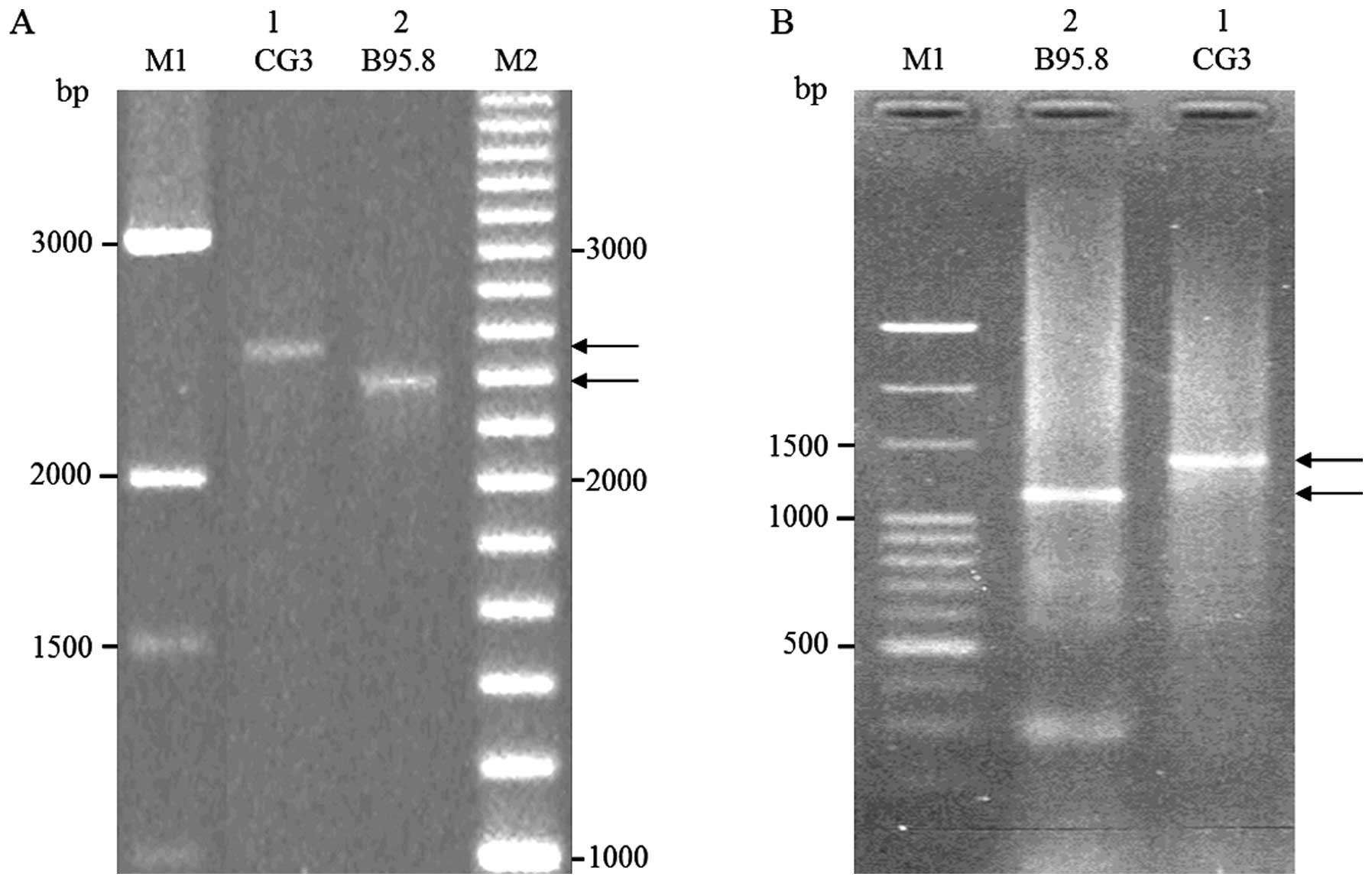
EBNA1 gene from CG3 cells. As shown in Fig. 1, PCR reactions using primers F1/F2
and nested primers GA1/GA2 and GA3/GA4 were performed to amplify
the full-length EBNA1 gene and its middle domain, respectively. DNA
extracted from B95.8 cells was amplified as a control. The
full-length gene and its middle region from B95.8 cells were
previously predicted to be 2392 and 1177 bp in length, respectively
(44). We obtained PCR products
consistent with those predicted sizes (Fig. 2A and 2B, lane 2). The PCR products
representing the full-length gene and the middle region of EBNA1
from CG3 cells (Fig. 2A and B, lane
1) appeared to be ~150 bp longer than those from B95.8 cells
(Fig. 2A and B, lane 2). These data
are consistent with previous studies that the molecular mass
polymorphisms of EBNA1 across different EBV isolates reflect
different numbers of internal glycine-alanine repeats (24–26).
Sequence analysis revealed that the N- and
C-terminal regions of the CG3 EBNA1 gene did not contain any
insertion or deletion, but harbored a series of nucleotide changes
relative to the B95.8 clone. The deduced amino acid sequences of
the CG3 clone relative to the B95.8 EBNA1 sequence are shown in
Fig. 3. Based on the amino acid at
position 487 and the characterized sequence variations at other
positions, we determined that the CG3 EBNA1 belongs to the V-val
subtype. We identified 4 (aa 16, 18, 20 and 85) and 12 (aa 364,
410, 411, 418, 439, 487, 499, 502, 524, 528, 533 and 594) amino
acid substitutions relative to the B95.8 sequence in the N- and
C-terminal regions, respectively, of the CG3 EBNA1. By contrast,
only three amino acid substitutions (aa 85, 364 and 410) were
identified between the CG3 EBNA1 and the EBNA1 isolated from NPC
patients of Hong Kong (28), which
was the first reported V-val EBNA1.
Isolation of EBNA1 fusion proteins and
generation of rabbit anti-EBNA1 antibodies
Recombinant CG3 EBNA1 proteins covering aa 1-90,
354-641 and 459-641 were expressed as His-tagged N-terminal fusion
proteins in E. coli, and were designated as hN-, hFC- and
hSC-EBNA1, respectively (Fig. 1).
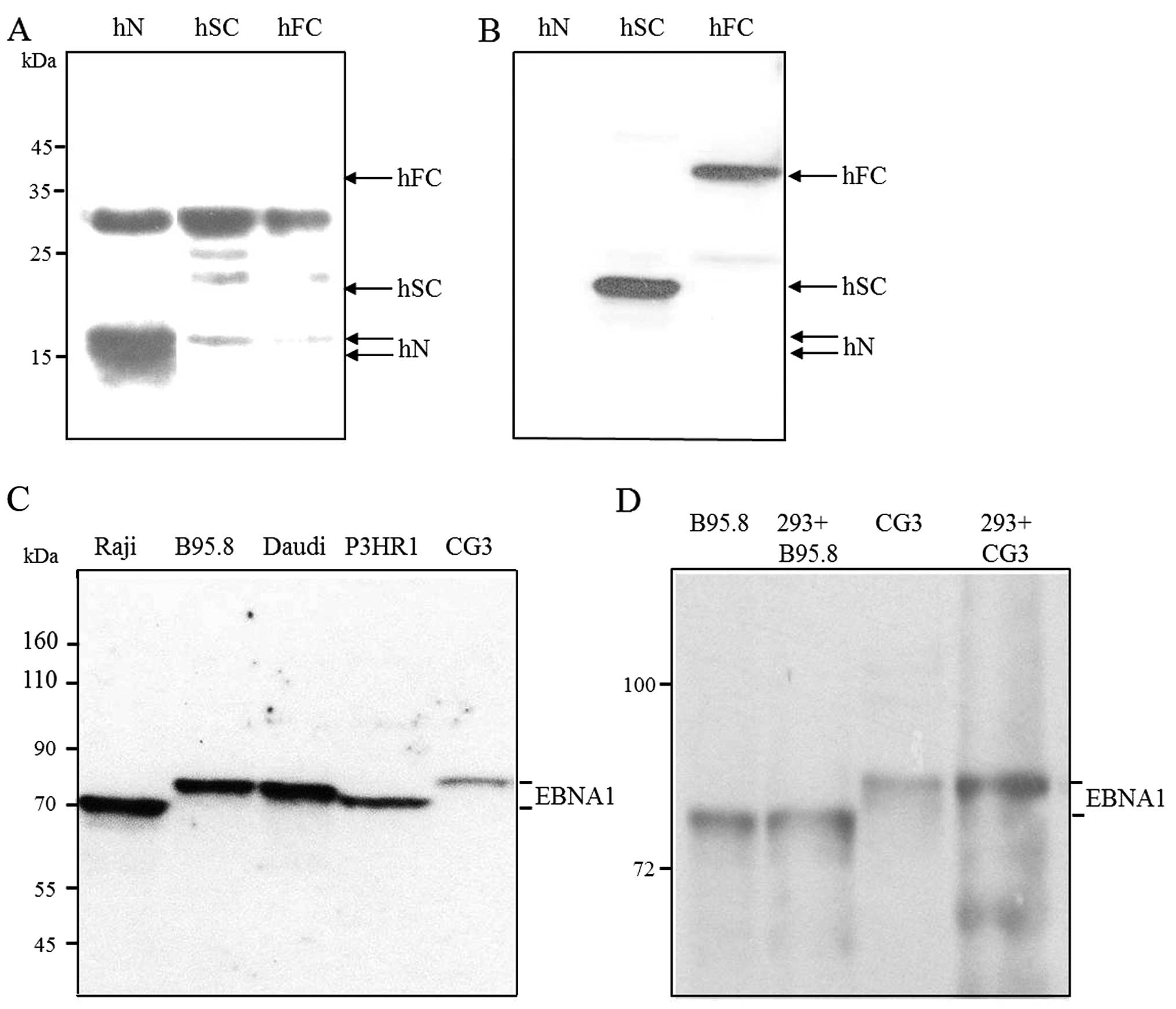
SDS-PAGE and Coomassie blue staining were used to examine the EBNA1
fusion proteins before (Fig. 4A–C,
lane C) and after (lanes 1–4) their purification by metal chelation
chromatography. The sizes of the EBNA1 fusions were as expected,
except that doublet bands were observed for hN (Fig. 4A, lanes 2 and 3). The molecular
nature of these doublets remains unknown. As shown in Fig. 4D, fraction 2 of hN (lane 1),
fraction 3 of hSC (lane 2), and concentrated pooled hFC (fractions
2 and 3, lane 3) were stained with Coomassie blue. These purified
EBNA1 fusions were then analyzed by western blotting using an
anti-His antibody (Fig. 4E).
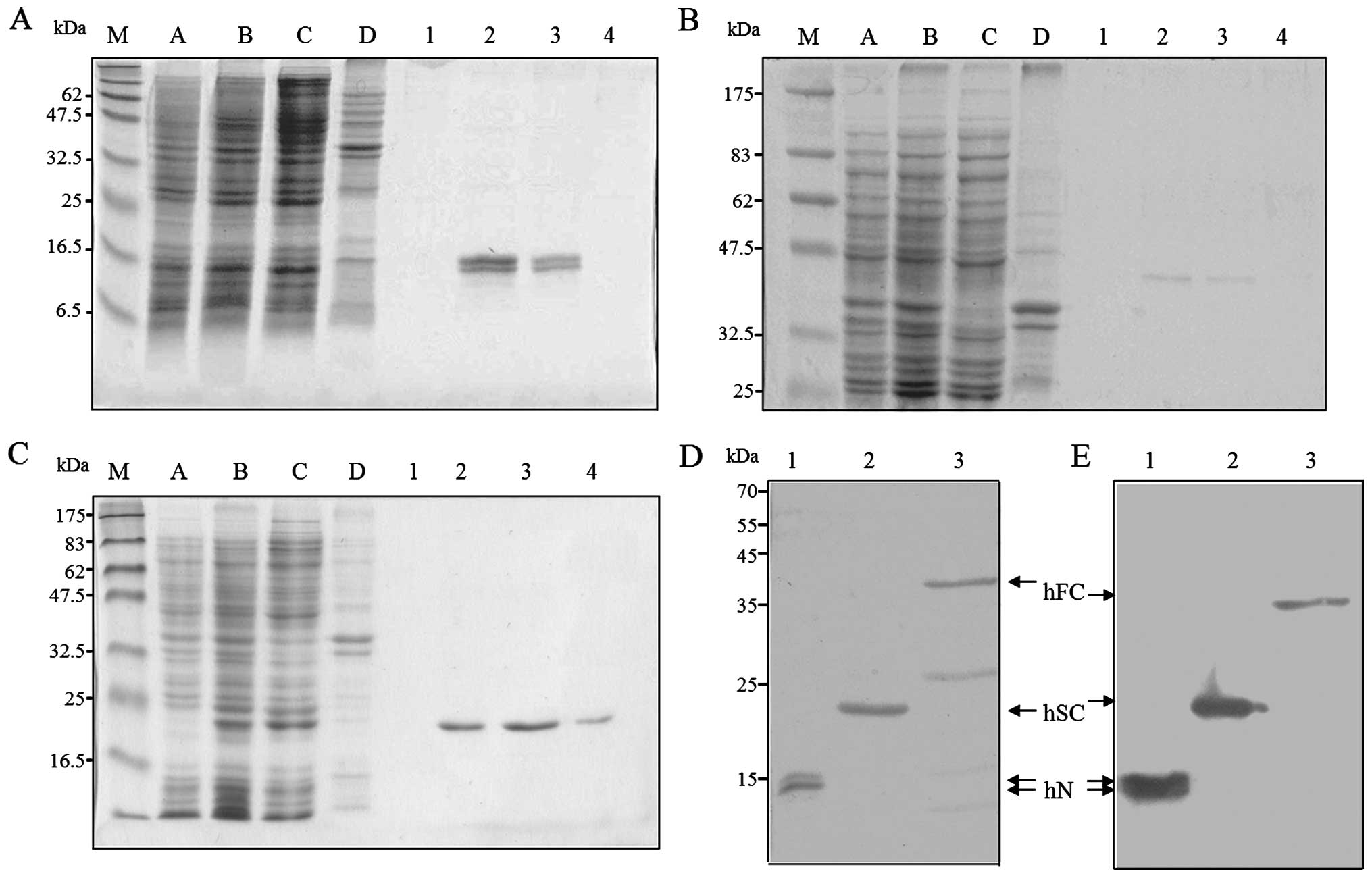 | Figure 4Expression and purification of
recombinant EBNA1s expressed in E. coli. The His-tagged
EBNA1 fusion proteins, (A) hN-, (B) hFC- and (C) hSC-EBNA1, were
expressed in E. coli and recovered by affinity purification.
The purified proteins were separated by 15, 10, and 12.5% SDS-PAGE,
respectively, and stained with Coomassie blue. Lanes: A and B,
protein extracts obtained before and after IPTG induction; C and D,
soluble and pellet fractions after induction; and 1-4, the purified
fractions. (D and E) The purified proteins were separated by 12%
SDS-PAGE followed by (D) Coomassie blue staining or (E) western
blot analysis using an anti-His antibody. Lanes: 1, hN-EBNA1; 2,
hSC-EBNA1; and 3, hFC-EBNA1. The mobilities of prestained protein
markers (GeneTeks BioScience Inc., Taipei, Taiwan) are indicated on
the left. |
The purified hN and hSC fusion proteins were used to
raise antibodies in rabbits, and the resulting antisera were
designated as Ab-N and Ab-SC, respectively. As expected, our
western blot analysis showed that Ab-N reacted with purified hN
(although a significant background was observed; Fig. 5A), while Ab-SC detected hSC and hFC
but not hN (Fig. 5B). We determiend
the ability of Ab-SC to detect EBNA1 from cell lines carrying
various EBNA1 subtypes. As reported in the present and other
studies (46,47), the EBNA1 proteins in the B95.8,
P3HR1, Daudi and CG3 cell lines correspond to the P-ala, V-leu,
P-thr and V-val subtypes, respectively. The EBNA1 protein in Raji
cells corresponds to the P-ala subtype, but has amino acid
substitutions with respect to B95.8 EBNA1 (47). As shown in Fig. 5C, the Ab-SC antibody reacted with
EBNA1 in all of the tested EBNA1-positive cells. As reported
previously (26), the molecular
weights of the EBNA1 proteins in Raji, B95.8, Daudi, and P3HR1
cells were 69, 79, 78, and 75 kDa, respectively. We obtained
results consistent with those prior findings (Fig. 5C), and found that the CG3 EBNA1 was
larger than the B95.8 EBNA1 in our western blot analysis (similar
to the gene-based findings described earlier and shown in Fig. 2). Collectively, these results showed
that our rabbit polyclonal anti-EBNA1 antibodies detected EBNA1
fusion proteins expressed in E. coli, and EBNA1 proteins of
various subtypes in different EBV-associated cell lines.
Cloning and expression of the full-length
CG3 V-val EBNA1 in cultured cells
Since most of the previous functional studies
involved EBNA1 cloned from the B95.8 prototype (which belongs to
the P-ala subtype), we subcloned the full-length V-val CG3
EBNA1 gene into a mammalian expression vector. The resulting
plasmid, pCMV-CG3EBNA1, was transfected into 293 cells, and EBNA1
expression was examined by western blot analysis using our rabbit
Ab-SC antibody (Fig. 5D). The
electrophoretic mobility of the expressed CG3 EBNA1 was retarded as
compared to that of B95.8 EBNA1, and the plasmid-expressed CG3 and
B95.8 EBNA1 proteins were indistinguishable from those of the
corresponding EBV-positive cells. These findings indicated that the
full-length EBNA1 gene was successfully amplified from
EBV-positive samples and expressed in the cultured cells, which in
turn should facilitate studies on the functional differences among
the different EBNA1 subtypes.
Expression of V-val CG3 EBNA1 supports
serum-independent cell proliferation and blocks apoptosis
Growth factor-independent proliferation is
considered to be a hallmark of cancer. To study the effect of CG3
EBNA1 on the cell proliferation and apoptosis of serum-starved
cells, we generated 293 cells stably expressing CG3 EBNA1 (V-val
subtype), B95.8 EBNA1 (P-ala subtype), or the drug-resistance gene
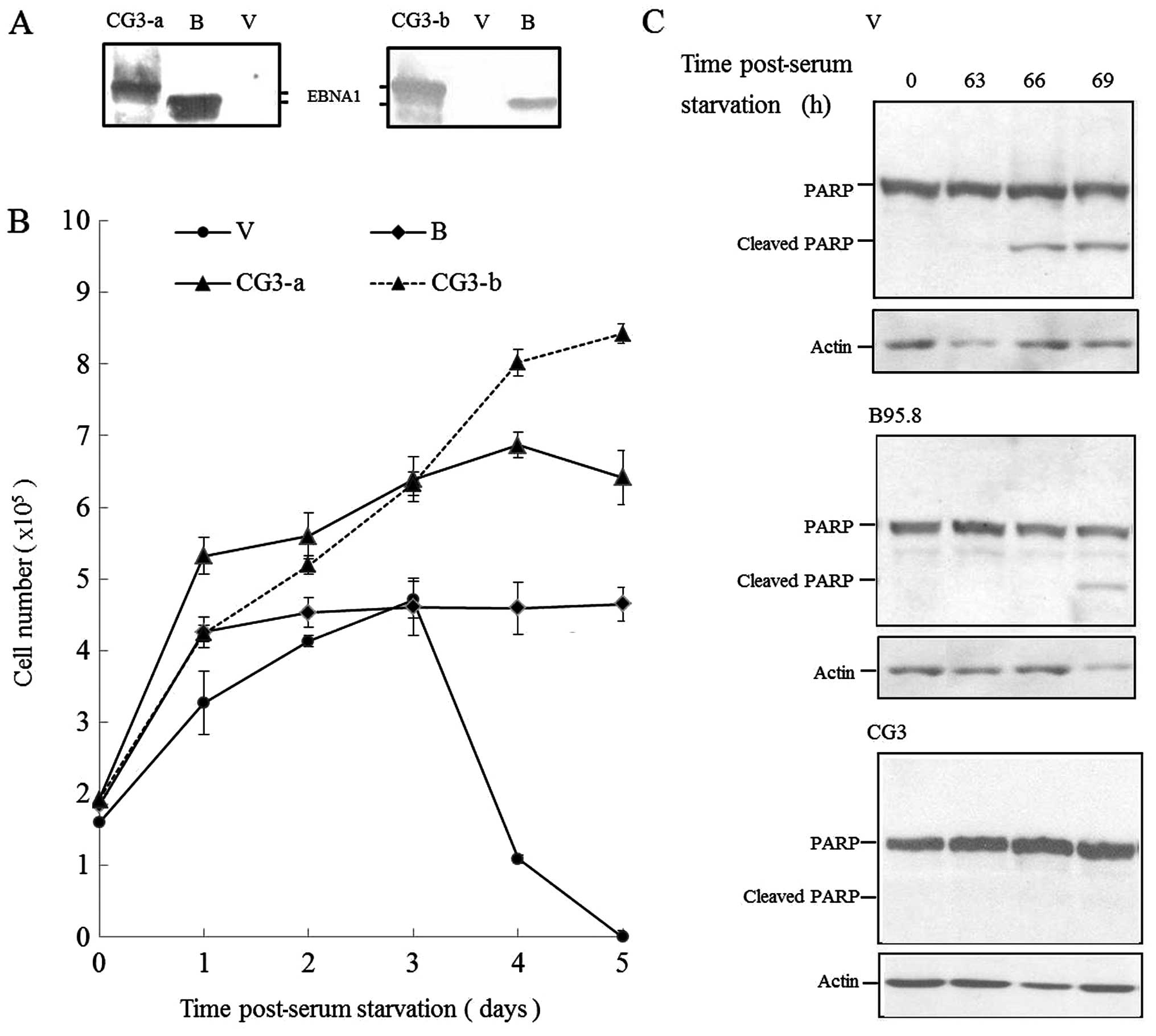
alone (empty vector, control). Expression of CG3 EBNA1 (from two
independent clones) and B95.8 EBNA1 was confirmed by western blot
analysis using our Ab-SC antibody (Fig.
6A). The cells were then cultured in the absence of FCS, and
cell viability was determined by trypan blue exclusion. As
expected, 293 cell-expressing empty vector demonstrated loss of
viability within three days of serum deprivation (Fig. 6B). Cells expressing B95.8 EBNA1 grew
exponentially only for one day after serum withdrawal, and then
plateaued through to the end of the experiment (day 5). As shown in
Fig. 6B, however, the 293 cells
expressing two independent clones of CG3 EBNA1 were able to
proliferate under serum starvation throughout the experimental
period. We also used western blotting to examine the status of
PARP, a substrate cleaved by caspases during apoptosis, in 293
cells expressing empty vector, B95.8 EBNA1, and CG3 EBNA1 following
serum withdrawal. As shown in Fig.
6C, cells expressing empty vector showed marginal induction of
PARP cleavage after 63 h and significant PARP cleavage after 66 h
of serum starvation. PARP cleavage was observed after 69 h of serum
starvation in cells expressing B95.8 EBNA1. Of note, 293 cells
expressing CG3 EBNA1 under serum starvation inhibited PARP cleavage
at all of the tested time points. Taken together, these results
suggested that CG3 EBNA1 blocked serum withdrawal-induced apoptosis
and promoted cell proliferation.
Discussion
EBV has been associated with different malignancies
in various geographic regions (1),
and EBNA1, which is the only viral protein observed in all
EBV-associated tumors, shows region-specific sequence variations
(29,34). The B85.8 EBNA1, which belongs to the
P-ala subtype, is the most commonly studied EBNA1. However, the
V-val subtype predominates in Taiwan. Thus, we cloned and expressed
a non-prototypic EBNA1 subtype from a CG3 cell line, an
EBV-carrying lymphoblastoid cell line that was derived from a
Taiwanese patient with juvenile chronic myeloid leukemia. The
GC-rich middle region of the EBNA1-encoding gene, which encodes the
glycine-alanine repeats, can complicate the PCR amplification of
the EBNA1 gene (28). To
clone the full-length V-val EBNA1 gene from CG3 cells, we
therefore used commercial resolution buffer to improve the
efficiency of the PCR reaction. Our data indicate that we
successfully PCR amplified the full-length EBNA1 gene from
CG3 cells. Using our protocol, therefore, the full-length EBNA1
genes of various subtypes may be readily amplified from
EBV-positive samples, potentially facilitating studies of their
functional differences (especially in the context of EBV-related
disease).
Variations in the EBNA1 sequences relative to the
prototype strain, B95.8, were first described in EBV isolates from
NPC samples obtained in Hong Kong (28). These variations were subsequently
classified into the V-val subtype, based on the system established
by Gutiérrez et al (30). In
the present study, we demonstrate that the EBNA1 protein expressed
in the CG3 cell line, which is associated with chronic myeloid
leukemia, also belongs to the V-val subtype. This finding is
consistent with previous studies that V-val EBNA1 is the
predominant subtype detected in healthy Taiwanese individuals and
Taiwanese patients with EBV-associated diseases (32,37,47).
Furthermore, our sequencing results revealed only minor changes in
the EBNA1 sequence (one and two amino acid substitutions in the N-
and C-terminal domains, respectively) relative to the EBNA1
sequences found in NPC samples from Hong Kong. In the future,
larger-scale sequence analyses and functional analyses of samples
collected from different tumors are required to elucidate the role
of these sequence variations in the tissue tropism of EBV.
To further characterize EBNA1 and establish a good
source of antigens and antibodies for use in functional studies and
as diagnostic reagents, we subcloned the N- and C-terminal regions
of the CG3 V-val EBNA1 into a bacterial expression vector to
produce His-tagged fusion proteins, and then raised the
corresponding antibodies in rabbits. The purified EBNA1 fusion
proteins and rabbit polyclonal antibodies constitute specific and
biologically safe reagents. They are good reagents for the
diagnosis of EBV infection, the immunodetection of EBNA1 in western
blot analysis, and potentially for a large-scale ELISA-based
screening program that may provide diagnostic and epidemiological
information on EBV-related diseases.
Most of the existing studies regarding the
functional roles of EBNA1 in the tumorigenesis of EBV have used the
cloned B95.8 EBNA1 (a P-ala EBNA1) or EBV-containing cell lines
carrying EBNA1 of unknown subtypes. In the present study, we
constructed a eukaryotic expression plasmid containing a V-val
EBNA1 gene originally derived from chronic myeloid leukemia.
We found that the expression of this V-val EBNA1 in 293 cells
supported cell growth and inhibited apoptosis (two important
characteristics for an oncogene) under serum starvation. This V-val
EBNA1 expression plasmid provides a good experimental tool to
determine, whether the V-val EBNA1 can promote oncogenesis in the
absence of other EBV sequences, and whether the V-val EBNA1 and the
P-ala prototype show functional differences in contributing to the
development of EBV-associated malignancies.
Although NPC has been identified worldwide, its
incidence varies among population groups, with the highest
incidence observed in southern Chinese males (48). V-val EBNA1 is the only viral protein
that has been observed in all NPC samples from southern China
(28,33,36,37,49).
Thus, future studies should be conducted to examine how this EBNA1
subtype may contribute to the pathogenesis of NPC in high-incidence
areas. Since we successfully established a protocol for amplifying
the full-length EBNA1 gene from EBV-positive samples, it
should now be possible to clone and express the V-val EBNA1 from
various NPC samples. Previous studies have indicated that some of
the variations observed between the P-ala and V-val EBNA1s lie in
the DNA-binding region (28,33,49).
EBNA1 and FR derived from Asian isolates induced higher
transcriptional activity than those from the B95.8 strain, as
assessed using a luciferase reporter system (33,35).
Findings of a previous study have indicated that the expression of
V-val EBNA1 isolated from an NPC biopsy had no effect on 293 cell
growth under normal growth conditions (49). However, the authors observed that
the fluorescence from GFP reporter was more intense in 293 cells
expressing a V-val-EBNA1-GFP plasmid compared with a
P-ala-EBNA1-GFP plasmid after long-term culture. V-val and P-thr
EBNA1s have been detected in NPC samples obtained from Beijing
Hospital (31) and North Africa
(28,30), whereas P-ala and P-thr (but not
V-val) were detected in NPC samples from Denmark, which is an area
of low NPC risk (31). The V-leu
subtype has not been found in NPC patients, even in areas in which
it is not a rare EBNA 1 subtype (30,46).
Since NPC differs clinically and molecularly in different regions
of the world (50), we conclude a
possible association of V-val with undifferentiated NPC in southern
China, and/or the possibility that yet-unidentified genetic
polymorphisms in EBNA1 (e.g., the number of internal
glycine-alanine repeats) may be important in the tumorigenesis of
NPC.
The results of the present study suggest that V-val
EBNA1 isolated from a chronic myeloid leukemia-derived cell line
potentially contribute to regulating proliferation and
anti-apoptosis in 293 cells. We also introduced a cloning strategy
that may be used to compare the biological functions of fulllength
EBNA1s isolated from specific populations, such as patients with
EBV-associated disease as compared to healthy individuals from the
same geographic region. Such investigations are likely to
considerably improve our understanding of the polymorphic patterns
of EBNA1 and their associations with malignancies in different
geographic regions.
Acknowledgements
This study was supported by the Chang Gung Memorial
Hospital (CMRPD32039, 160071-3, and 190111-2).
References
|
1
|
Young LS and Murray PG: Epstein-Barr virus
and oncogenesis: from latent genes to tumours. Oncogene.
22:5108–5121. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schepers A, Ritzi M, Bousset K, et al:
Human origin recognition complex binds to the region of the latent
origin of DNA replication of Epstein-Barr virus. EMBO J.
20:4588–4602. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sears J, Kolman J, Wahl GM and Aiyar A:
Metaphase chromosome tethering is necessary for the DNA synthesis
and maintenance of oriP plasmids but is insufficient for
transcription activation by Epstein-Barr nuclear antigen 1. J
Virol. 77:11767–11780. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nanbo A, Sugden A and Sugden B: The
coupling of synthesis and partitioning of EBV’s plasmid replicon is
revealed in live cells. EMBO J. 26:4252–4262. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sugden B and Warren N: A promoter of
Epstein-Barr virus that can function during latent infection can be
transactivated by EBNA-1, a viral protein required for viral DNA
replication during latent infection. J Virol. 63:2644–2649.
1989.PubMed/NCBI
|
|
6
|
Altmann M, Pich D, Ruiss R, Wang J, Sugden
B and Hammerschmidt W: Transcriptional activation by EBV nuclear
antigen 1 is essential for the expression of EBV’s transforming
genes. Proc Natl Acad Sci USA. 103:14188–14193. 2006. View Article : Google Scholar
|
|
7
|
Gahn TA and Sugden B: An EBNA-1-dependent
enhancer acts from a distance of 10 kilobase pairs to increase
expression of the Epstein-Barr virus LMP gene. J Virol.
69:2633–2636. 1995.PubMed/NCBI
|
|
8
|
Sample J, Henson EB and Sample C: The
Epstein-Barr virus nuclear protein 1 promoter active in type I
latency is autoregulated. J Virol. 66:4654–4661. 1992.PubMed/NCBI
|
|
9
|
Sung NS, Wilson J, Davenport M, Sista ND
and Pagano JS: Reciprocal regulation of the Epstein-Barr virus
BamHI-F promoter by EBNA-1 and an E2F transcription factor. Mol
Cell Biol. 14:7144–7152. 1994.PubMed/NCBI
|
|
10
|
Frappier L: Role of EBNA1 in NPC
tumourigenesis. Semin Cancer Biol. 22:154–161. 2012. View Article : Google Scholar
|
|
11
|
Westhoff Smith D and Sugden B: Potential
cellular functions of Epstein-Barr nuclear antigen 1 (EBNA1) of
Epstein-Barr virus. Viruses. 5:226–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kennedy G, Komano J and Sugden B:
Epstein-Barr virus provides a survival factor to Burkitt’s
lymphomas. Proc Natl Acad Sci USA. 100:14269–14274. 2003.
View Article : Google Scholar
|
|
13
|
Holowaty MN, Zeghouf M, Wu H, et al:
Protein profiling with Epstein-Barr nuclear antigen-1 reveals an
interaction with the herpesvirus-associated ubiquitin-specific
protease HAUSP/USP7. J Biol Chem. 278:29987–29994. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Holowaty MN, Sheng Y, Nguyen T, Arrowsmith
C and Frappier L: Protein interaction domains of the
ubiquitin-specific protease, USP7/HAUSP. J Biol Chem.
278:47753–47761. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wilson JB, Bell JL and Levine AJ:
Expression of Epstein-Barr virus nuclear antigen-1 induces B cell
neoplasia in transgenic mice. EMBO J. 15:3117–3126. 1996.PubMed/NCBI
|
|
16
|
Murakami M, Lan K, Subramanian C and
Robertson ES: Epstein-Barr virus nuclear antigen 1 interacts with
Nm23-H1 in lymphoblastoid cell lines and inhibits its ability to
suppress cell migration. J Virol. 79:1559–1568. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaul R, Murakami M, Choudhuri T and
Robertson ES: Epstein-Barr virus latent nuclear antigens can induce
metastasis in a nude mouse model. J Virol. 81:10352–10361. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
O’Neil JD, Owen TJ, Wood VH, et al:
Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription
factor pathway in nasopharyngeal carcinoma cells and enhances
angiogenesis in vitro. J Gen Virol. 89:2833–2842. 2008. View Article : Google Scholar
|
|
19
|
Wood VH, O’Neil JD, Wei W, Stewart SE,
Dawson CW and Young LS: Epstein-Barr virus-encoded EBNA1 regulates
cellular gene transcription and modulates the STAT1 and TGFbeta
signaling pathways. Oncogene. 26:4135–4147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Flavell JR, Baumforth KR, Wood VH, et al:
Down-regulation of the TGF-beta target gene, PTPRK, by the
Epstein-Barr virus encoded EBNA1 contributes to the growth and
survival of Hodgkin lymphoma cells. Blood. 111:292–301. 2008.
View Article : Google Scholar
|
|
21
|
Valentine R, Dawson CW, Hu C, et al:
Epstein-Barr virus-encoded EBNA1 inhibits the canonical NF-kappaB
pathway in carcinoma cells by inhibiting IKK phosphorylation. Mol
Cancer. 9:12010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gruhne B, Sompallae R, Marescotti D,
Kamranvar SA, Gastaldello S and Masucci MG: The Epstein-Barr virus
nuclear antigen-1 promotes genomic instability via induction of
reactive oxygen species. Proc Natl Acad Sci USA. 106:2313–2318.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gruhne B, Sompallae R and Masucci MG:
Three Epstein-Barr virus latency proteins independently promote
genomic instability by inducing DNA damage, inhibiting DNA repair
and inactivating cell cycle checkpoints. Oncogene. 28:3997–4008.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hennessy K and Kieff E: One of two
Epstein-Barr virus nuclear antigens contains a glycine-alanine
copolymer domain. Proc Natl Acad Sci USA. 80:5665–5669. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hennessy K, Heller M, van Santen V and
Kieff E: Simple repeat array in Epstein-Barr virus DNA encodes part
of the Epstein-Barr nuclear antigen. Science. 220:1396–1398. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Allday MJ and MacGillivray AJ:
Epstein-Barr virus nuclear antigen (EBNA): size polymorphism of
EBNA 1. J Gen Virol. 66:1595–1600. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Levitskaya J, Coram M, Levitsky V, et al:
Inhibition of antigen processing by the internal repeat region of
the Epstein-Barr virus nuclear antigen-1. Nature. 375:685–688.
1995. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Snudden DK, Smith PR, Lai D, Ng MH and
Griffin BE: Alterations in the structure of the EBV nuclear
antigen, EBNA1, in epithelial cell tumours. Oncogene. 10:1545–1552.
1995.PubMed/NCBI
|
|
29
|
Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim
A and Bhatia K: The extent of genetic diversity of Epstein-Barr
virus and its geographic and disease patterns: a need for
reappraisal. Virus Res. 143:209–221. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gutiérrez MI, Raj A, Spangler G, et al:
Sequence variations in EBNA-1 may dictate restriction of tissue
distribution of Epstein-Barr virus in normal and tumour cells. J
Gen Virol. 78:1663–1670. 1997.PubMed/NCBI
|
|
31
|
Sandvej K, Zhou XG and Hamilton-Dutoit S:
EBNA-1 sequence variation in Danish and Chinese EBV-associated
tumours: evidence for geographical polymorphism but not for
tumour-specific subtype restriction. J Pathol. 191:127–131. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang JT, Sheeng TS, Su IJ, Chen JY and
Chen MR: EBNA-1 sequence variations reflect active EBV replication
and disease status or quiescent latency in lymphocytes. J Med
Virol. 69:417–425. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Do NV, Ingemar E, Phi PT, et al: A major
EBNA1 variant from Asian EBV isolates shows enhanced
transcriptional activity compared to prototype B95.8. Virus Res.
132:15–24. 2008. View Article : Google Scholar
|
|
34
|
Habeshaw G, Yao QY, Bell AI, Morton D and
Rickinson AB: Epstein-Barr virus nuclear antigen 1 sequences in
endemic and sporadic Burkitt’s lymphoma reflect virus strains
prevalent in different geographic areas. J Virol. 73:965–975.
1999.PubMed/NCBI
|
|
35
|
Mai SJ, Xie D, Huang YF, et al: The
enhanced transcriptional activity of the V-val subtype of
Epstein-Barr virus nuclear antigen 1 in epithelial cell lines.
Oncol Rep. 23:1417–1424. 2010.PubMed/NCBI
|
|
36
|
Zhang XS, Wang HH, Hu LF, et al: V-val
subtype of Epstein-Barr virus nuclear antigen 1 preferentially
exists in biopsies of nasopharyngeal carcinoma. Cancer Lett.
211:11–18. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang WY, Chien YC, Jan JS, Chueh CM and
Lin JC: Consistent sequence variation of Epstein-Barr virus nuclear
antigen 1 in primary tumor and peripheral blood cells of patients
with nasopharyngeal carcinoma. Clin Cancer Res. 8:2586–2590.
2002.PubMed/NCBI
|
|
38
|
Chang YS, Shih LY, Peng CL, Chen SH and
Liu ST: Characterization of two newly established EBV-containing
lymphoblastoid cell lines from patients with myeloid leukemias.
Leuk Res. 14:309–320. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miller G, Shope T, Lisco H, Stitt D and
Lipman M: Epstein-Barr virus: transformation, cytopathic changes,
and viral antigens in squirrel monkey and marmoset leukocytes. Proc
Natl Acad Sci USA. 69:383–387. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pulvertaft JV: A study of malignant
tumours in Nigeria by shortterm tissue culture. J Clin Pathol.
18:261–273. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hinuma Y and Grace JT Jr: Cloning of
immunoglobulin-producing human leukemic and lymphoma cells in
long-term cultures. Proc Soc Exp Biol Med. 124:107–111. 1967.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arrand JR, Walsh-Arrand JE and Rymo L:
Cytoplasmic RNA from normal and malignant human cells shows
homology to the DNAs of Epstein-Barr virus and human adenoviruses.
EMBO J. 2:1673–1683. 1983.PubMed/NCBI
|
|
43
|
Graham FL, Smiley J, Russell WC and Nairn
R: Characteristics of a human cell line transformed by DNA from
human adenovirus type 5. J Gen Virol. 36:59–74. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Baer R, Bankier AT, Biggin MD, et al: DNA
sequence and expression of the B95-8 Epstein-Barr virus genome.
Nature. 310:207–211. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang TC and Chao M: Molecular cloning and
expression of the hepatitis delta virus genotype IIb genome.
Biochem Biophys Res Commun. 303:357–363. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bhatia K, Raj A, Guitierrez MI, et al:
Variation in the sequence of Epstein-Barr virus nuclear antigen 1
in normal peripheral blood lymphocytes and in Burkitt’s lymphomas.
Oncogene. 13:177–181. 1996.PubMed/NCBI
|
|
47
|
Chen MR, Tsai CH, Wu FF, Kan SH, Yang CS
and Chen JY: The major immunogenic epitopes of Epstein-Barr virus
(EBV) nuclear antigen 1 are encoded by sequence domains which vary
among nasopharyngeal carcinoma biopsies and EBV-associated cell
lines. J Gen Virol. 80:447–455. 1999.PubMed/NCBI
|
|
48
|
Niedobitek G: Epstein-Barr virus infection
in the pathogenesis of nasopharyngeal carcinoma. Mol Pathol.
53:248–254. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mai SJ, Ooka T, Li DJ, et al: Functional
advantage of NPC-related V-val subtype of Epstein-Barr virus
nuclear antigen 1 compared with prototype in epithelial cell line.
Oncol Rep. 17:141–146. 2007.
|
|
50
|
de Thé G: Nasopharyngeal carcinoma. Viral
Infections of Humans, Epidemiology and Control. Evans AS and
Kazslow RA: 4th edition. Plenum Medical Book Co; New York, NY: pp.
935–967. 1997, View Article : Google Scholar
|