Introduction
Protein tyrosine kinases (PTKs) are a family of
enzymes regulating diverse cellular functions that are associated
with cancer hallmarks, such as cell survival (1). Many PTKs are plasma
membrane-associated signal transducers that transmit signals into
the nucleus in response to environmental cues. However,
accumulating evidence indicates that PTKs also translocate to the
nucleus and exert additional functions. Nuclear PTKs are known to
change the stability and activity of nuclear residing proteins as
well as to regulate nuclear gene expression (2). For example, epidermal growth factor
receptor (EGFR) can regulate nuclear activity either by functioning
as a transcription factor or by phosphorylating histone H4
(3,4). EGFR is present in the nuclei of
proliferating liver cells and breast cancer cells (5,6).
Nuclear EGFR binds to the cyclin D1 promoter region and
upregulates cyclin D1 expression to promote breast cancer cell
cycle progression (6). In breast
cancer cells, ErbB2 also interacts with and phosphorylates Cdc2 in
the nucleus to confer resistance to Taxol-induced apoptosis
(7). In addition to EGFR, other
receptor and non-receptor PTKs have been detected in the nuclei of
solid tumors (8,9). However, the role of nuclear PTKs in
blood cancer is largely unknown.
Lymphocyte-specific protein tyrosine kinase (Lck) is
a Src family kinase (SFK) predominantly expressed in T cells and
plays a pivotal role in normal T cell development and homeostasis
(10,11). The gene coding for Lck is
localized near the chromosomal region with a high frequency of
translocation (12). Overexpression
and hyperactivation of Lck have been reported in both acute and
chronic leukemias (13). Lck
overexpression is also linked to poor clinical outcome to
prednisone treatment in acute B lymphoblastic leukemia patients
(14). In addition to blood
malignancies, abnormally high expression and activity of Lck have
been reported in solid tumors, such as colorectal and prostate
cancers (15,16). Under physiological conditions, Lck
is associated with plasma membrane and propagates signals initiated
from the T cell receptors (17).
However, immunohistochemical analysis of specimens from breast
cancer patients revealed the presence of nuclear Lck (18). It suggests that nuclear localization
of Lck may also be associated with malignant progression of
hematopoietic cells.
Our previous study demonstrated that, in mouse LSTRA
leukemia, Lck upregulated the expression of the LIM domain only
2 gene through direct binding to its promoter region (19). We further provided evidence
supporting the mouse LSTRA leukemic cell line as a model for the
aggressive form of human large granular lymphocyte leukemia
(20). These findings led us to
hypothesize that Lck may also exhibit additional functions in the
nuclear compartment of human leukemic cells. In the present study,
we used the well-defined human T leukemic cell line Jurkat to
examine the biological outcome and underlying mechanism of Lck
nuclear translocation.
Materials and methods
Cell lines and reagents
Human Jurkat E6.1 and Jcam 1.6 T cell lines and the
mouse LSTRA T cell line were maintained as described previously
(21). The Jcam 1.6 cell line
transfected with an expression vector containing the wild-type Lck
(Jcam/Lck) was a generous gift from Dr Steven Burakoff (Icahn
School of Medicine at Mount Sinai, New York City, NY, USA).
CR6-interacting factor 1 (CRIF1)-knockdown stable cell lines were
generated from Jcam cells using lentiviral transduction. CRIF1
shRNA (sc-97804-V) and scrambled shRNA control (sc-108080)
lentiviral particles were purchased from Santa Cruz Biotechnology
(Dallas, TX, USA). After 24-h serum starvation, 104 Jcam
cells were harvested and resuspended in 50 μl of freshly
thawed virus mixture (2×105 infectious units). After a
6-h incubation, 500 μl of complete RPMI-1640 was added.
After 1 day of recovery, puromycin was added to a final
concentration of 14 μg/ml and cells were selected for 1
week.
Mouse splenocyte isolation
Splenocytes were isolated from BALB/c mouse spleens
as described previously (20).
Briefly, the spleen was dissociated by gently pressing it through a
70-μm cell strainer with a syringe plunger. Red blood cells
were lysed by resuspending the cells in ammonium-chloride-potassium
(ACK) lysis buffer at room temperature for 5 min. Splenocytes were
then washed with cold RPMI-1640 and kept on ice for further
analysis. The use of animals was approved by the Institutional
Animal Care and Use Committee (IACUC) of the university.
Pervanadate activation and dasatinib
treatment
Mouse splenocytes and human T cell lines were
stimulated with freshly prepared pervanadate at the final
concentration of 100 μM at 37°C for 5 min as described
previously (22). Dasatinib was
purchased from LC Laboratories (Woburn, MA, USA) and kept at −20°C
as a 10-mM stock solution in DMSO. For dasatinib treatment,
107 Jurkat cells were treated with DMSO as control or
500 nM of dasatinib at 37°C for 2 h. The reactions were stopped by
quick cool down on ice. Pervanadate, dasatinib and DMSO control
were then removed by washing cells in ice-cold phosphate-buffered
saline.
Subcellular fractionation
Cells were lysed by swelling in hypotonic buffer
followed by passing through a 27-gauge needle. Light microscopy was
used to ensure cell rupture before proceeding to the next step. The
nuclear fraction was collected by differential centrifugation as
described previously (19).
Fraction purity was verified by immunoblotting of specific
markers.
Immunoprecipitation and
immunoblotting
Whole cell and nuclear lysates were prepared by
solubilizing whole cell and nuclear pellets in RIPA buffer,
respectively. For co-immuno-precipitation experiments, proteins
were extracted from the nuclear pellets using high-salt buffer
(19). Target proteins were either
immunoprecipitated or directly detected from total lysates after
SDS-PAGE using specific antibodies according to manufacturers’
instructions. Antibodies specific for Lck, CRIF1 and epidermal
growth factor receptor substrate 15 (Eps15) were purchased from
Santa Cruz Biotechnology. Antibodies specific for phospho-Src
family (Tyr416) and glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) were purchased from Cell Signaling Technology (Danvers, MA,
USA). Anti-lamin B1 antibody was purchased from Abcam (Cambridge,
MA, USA). Appropriate secondary antibodies conjugated with
horseradish peroxidase were used to detect signals by enhanced
chemiluminescence system. For signal quantitation, the bands were
digitalized and analyzed by ImageJ software.
Confocal immunofluorescence
microscopy
The cells were adhered to 10-well slides, fixed, and
permeabilized as previously described (21). The cells were blocked with Image-iT
FX Signal Enhancer (Life Technologies, Grand Island, NY, USA) for
15 min at room temperature, and then either singly or doubly
stained with primary antibodies. Subsequent labeling with Alexa
Fluor-conjugated secondary antibodies and DAPI counterstain (Life
Technologies) were performed to visualize primary antibodies and
nuclei, respectively. Stained cells were viewed using the Olympus
FV10i fluorescence confocal microscope. Images were analyzed using
FluoView software (Olympus, Melville, NY, USA).
In situ proximity ligation assay (PLA)
microscopy
PLA was performed using the DuoLink PLA kit
(Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s
instructions. Briefly, 104 cells were seeded on each
well of 10-well slides. The adhered cells were fixed with 4%
paraformaldehyde and then permeabilized with 0.2% Triton X-100.
After treatment with DuoLink blocking buffer, the cells were
incubated with diluted primary antibodies for Lck and CRIF1. After
washing, the cells were incubated with species-specific PLA probes
and two additional oligonucleotides under hybridization conditions.
Hybridization occurs when PLA probes are in close proximity, which
can be subsequently ligated to form a closed circle. A
rolling-circle amplification step follows with polymerase to
generate a concatemeric product, which can be visualized with
fluorephore-labeled oligonucleotides after hybridization. The
slides were counterstained with DAPI and analyzed by fluorescence
microscope.
Transient transfection
Approximately 104 HEK293T cells were
plated in a 10-cm dish in 10 ml of DMEM supplemented with 10% fetal
bovine serum. Cells under 70% confluency were transiently
transfected with 18 μg of plasmids using calcium phosphate
precipitation protocol (23). The
cells were harvested between 42–46 h after transfection. Wild-type
Lck construct as well as constitutively active (Y505F), kinase dead
(Y505F/K273R), and basal kinase (Y505F/Y394F) Lck mutant constructs
have been described previously (23).
Cell number and viability
Human T cell lines were washed in plain RPMI-1640
and then incubated in serum-free RPMI-1640 at 37°C with 5%
CO2 for 48 h. Cell viability was determined by
incubation of a small fraction of cells in 0.1% trypan blue dye.
Both viable and dead cells were counted with a hemocytometer under
light microscope to calculate the percentage of dead cells.
Statistical analysis
Data are presented as mean ± SE from at least three
independent experiments. The significance of differences was
analyzed using a Student’s t-test (SigmaPlot 11; SPSS Inc.,
Chicago, IL, USA). Differences were considered significant at
P<0.05.
Results
Lck is present in the nuclear compartment
of leukemia cells
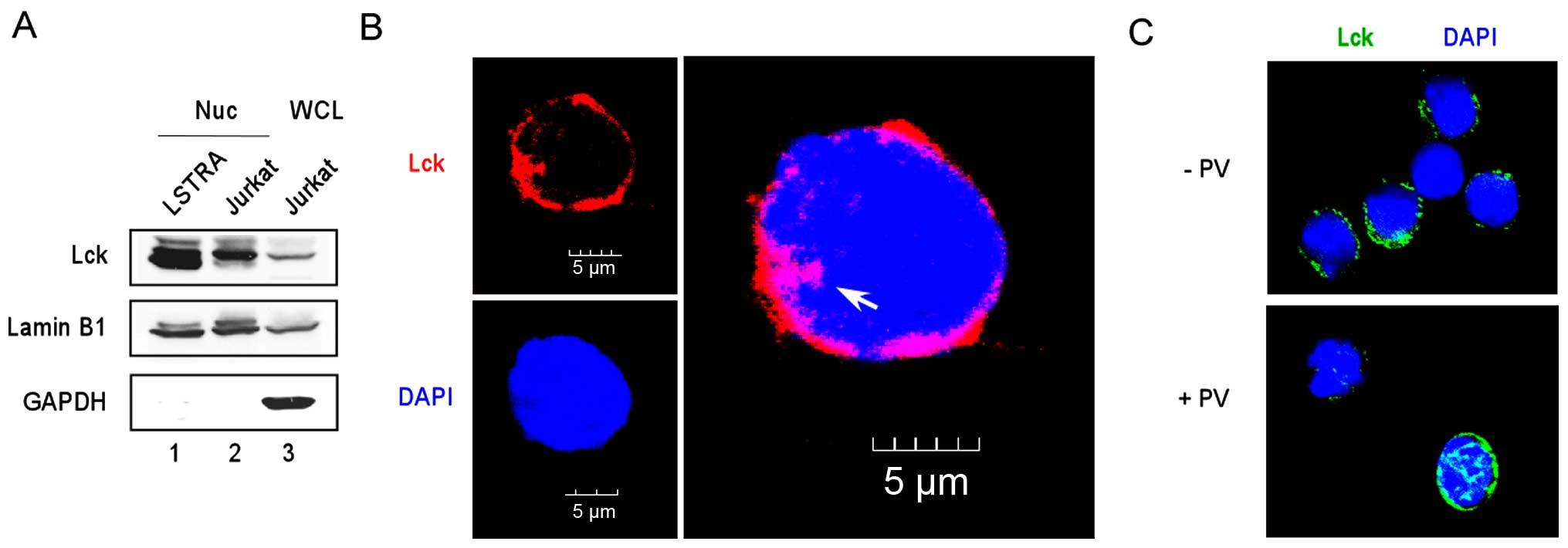
LSTRA is a mouse leukemic cell line that
overexpresses active Lck kinase (24). We previously showed that a large
percentage of endogenous Lck was accumulated in LSTRA nuclei
(Fig. 1A, lane 1) (19). To determine whether Lck
trans-locates to the nuclei in primary cells, we examined mouse
splenocytes by immunofluorescence microscopy using an anti-Lck
antibody. As shown in Fig. 1C, Lck
staining was mostly cytoplasmic in the resting cells (upper panel).
To maximally activate Lck tyrosine phosphorylation, we stimulated
mouse splenocytes with pervanadate, which is a potent tyrosine
phosphatase inhibitor. As shown in Fig.
1C, a significant amount of Lck translocated to the nucleus of
the pervanadate-stimulated cells (lower panel). This result
confirms the presence of nuclear Lck in primary cells and further
suggests the involvement of Lck phosphorylation in its nuclear
translocation.
The Jurkat cell line was isolated from an acute T
lympho-blastic leukemia patient and is one of the best
characterized human leukemic T cell lines (25). We examined the presence of nuclear
Lck by subcellular fractionation of Jurkat cells to isolate nuclear
proteins (Fig. 1A, lane 2).
Subsequent immunoblotting detected the presence of nuclear Lck (top
panel) in the absence of cytoplasmic contamination (bottom panel).
Confocal microscopy after staining with the anti-Lck antibody
further confirmed localization of endogenous Lck in the Jurkat
nucleus (Fig. 1B). These findings
support a potential role of nuclear Lck in both mouse and human T
cell leukemias.
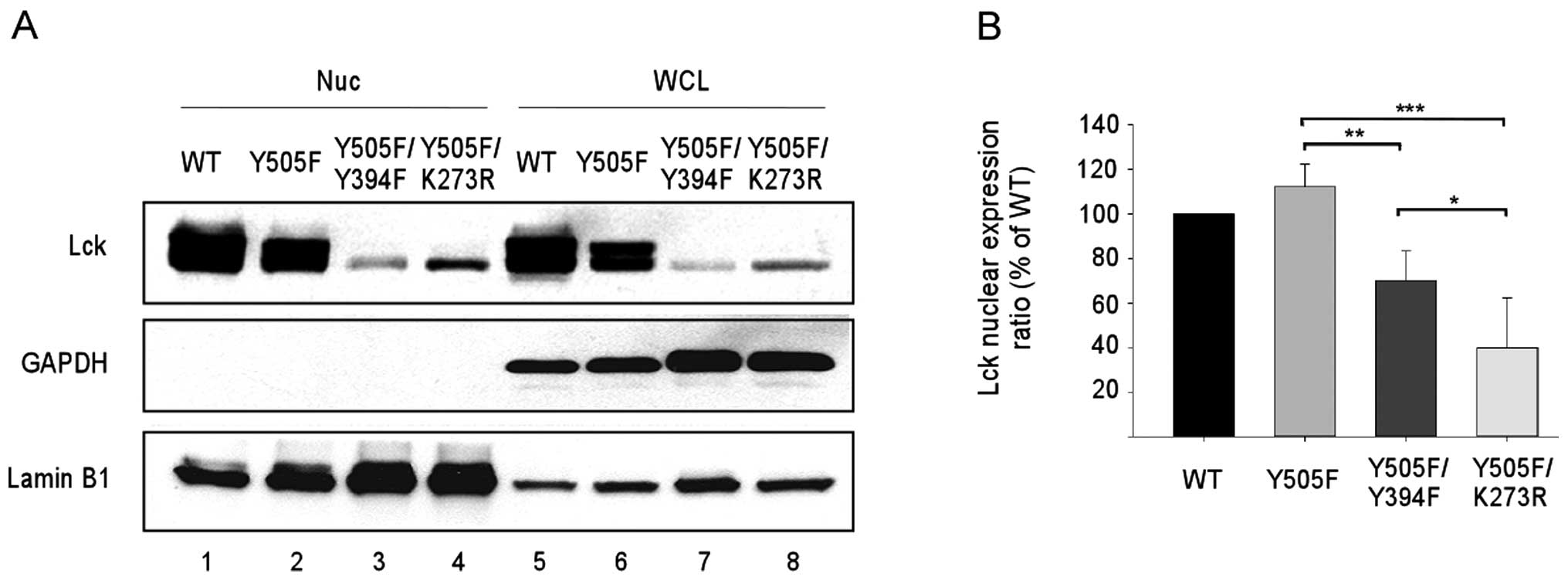
Kinase activity enhances Lck nuclear
translocation
Similar to other SFKs, Lck activity is tightly
regulated by phosphorylation of two key regulatory tyrosine
residues (26). Phosphorylation of
the negative regulatory Y505 results in a closed conformation and
reduced Lck kinase activity. On the other hand, phosphorylation of
the positive regulatory Y394 confers a fully active Lck kinase with
an open conformation. To establish a correlation between Lck kinase
activity and its nuclear translocation, we analyzed wild-type and
three mutant Lck proteins. The Y505F mutant is locked in an open
conformation and becomes constitutively active. Additional mutation
on Y394 (Y505F/Y394F) reduces the kinase activity; while mutation
on the key residue in kinase domain (K273R) abolishes the kinase
activity. All four constructs were transfected into HEK293T cells
that lack endogenous Lck. Lck immunoblotting of nuclear versus
total proteins (Fig. 2A) and
subsequent quantitation (Fig. 2B)
showed a step-wise reduction in nuclear Lck accumulation with
decreasing kinase activity.
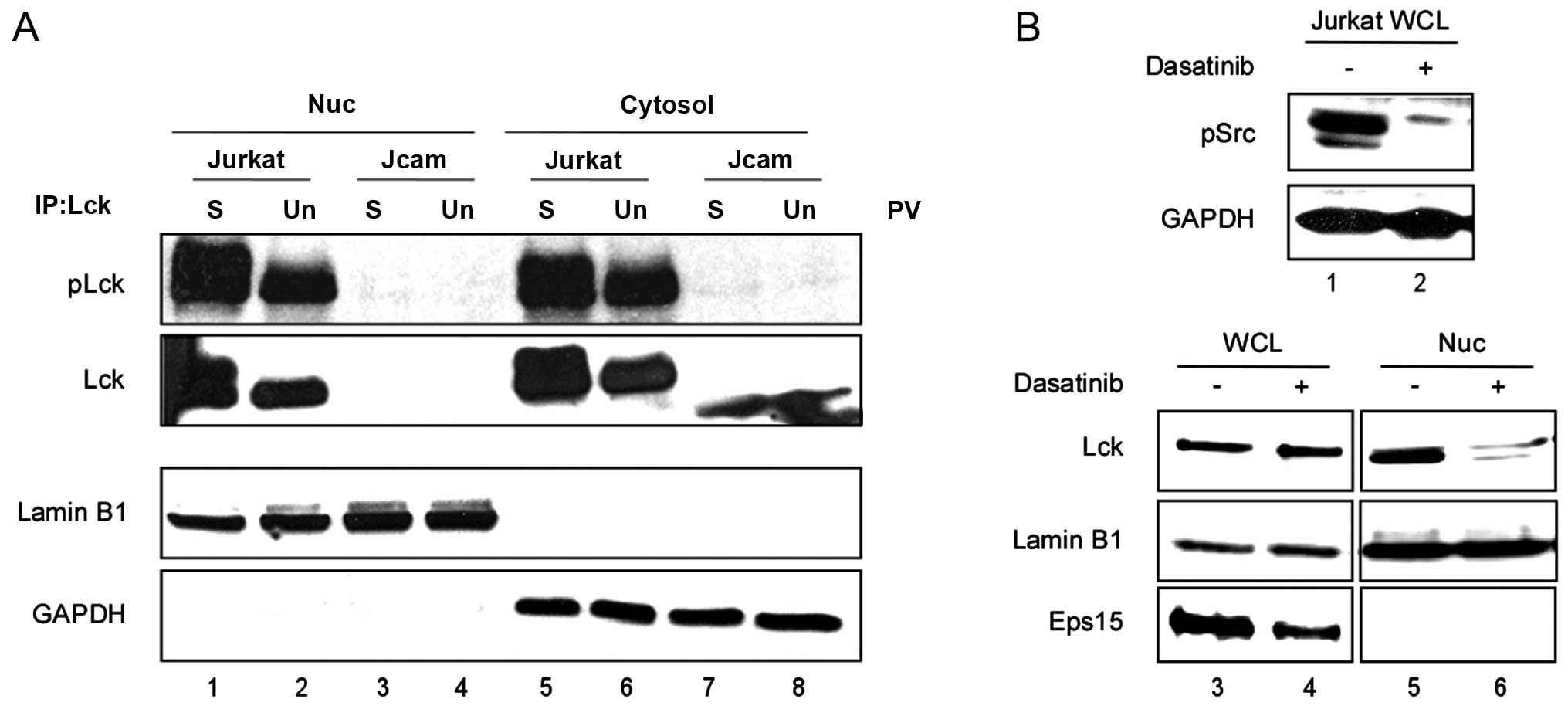
To further establish the role of Lck kinase activity
in its nuclear translocation in human T cells, we examined both
Jurkat and its Lck-deficient derivative Jcam cell lines. In the
Jcam cells, Lck is inactive due to truncation and is expressed at a
very low level (27).
Phosphorylation of the positive regulatory Y394 is indicative of
Lck kinase activity. We determined the level of Y394
phosphorylation by Lck immunoprecipitation and subsequent
immunoblotting using an antibody that specifically recognizes
phosphorylation of this conserved tyrosine residue in all SFKs. As
shown in Fig. 3A, in the Jcam
cells, the truncated kinase-dead Lck (lanes 7 and 8) could not
translocate to the nucleus either without (lane 4) or with (lane 3)
pervanadate stimulation. On the other hand, pervanadate stimulation
of Jurkat cells greatly enhanced Y394 phosphorylation and nuclear
translocation of Lck (Fig. 3A,
lanes 1 and 2). This was consistent with pervanadate-induced Lck
translocation into the nuclei of primary cells (Fig. 1C). As an independent confirmation,
we treated Jurkat cells with an SFK inhibitor, dasatinib. As shown
in Fig. 3B, in the absence of SFK
activity (compare lanes 1 and 2), the amount of nuclear Lck was
greatly diminished (compare lanes 5 and 6) in the Jurkat cells.
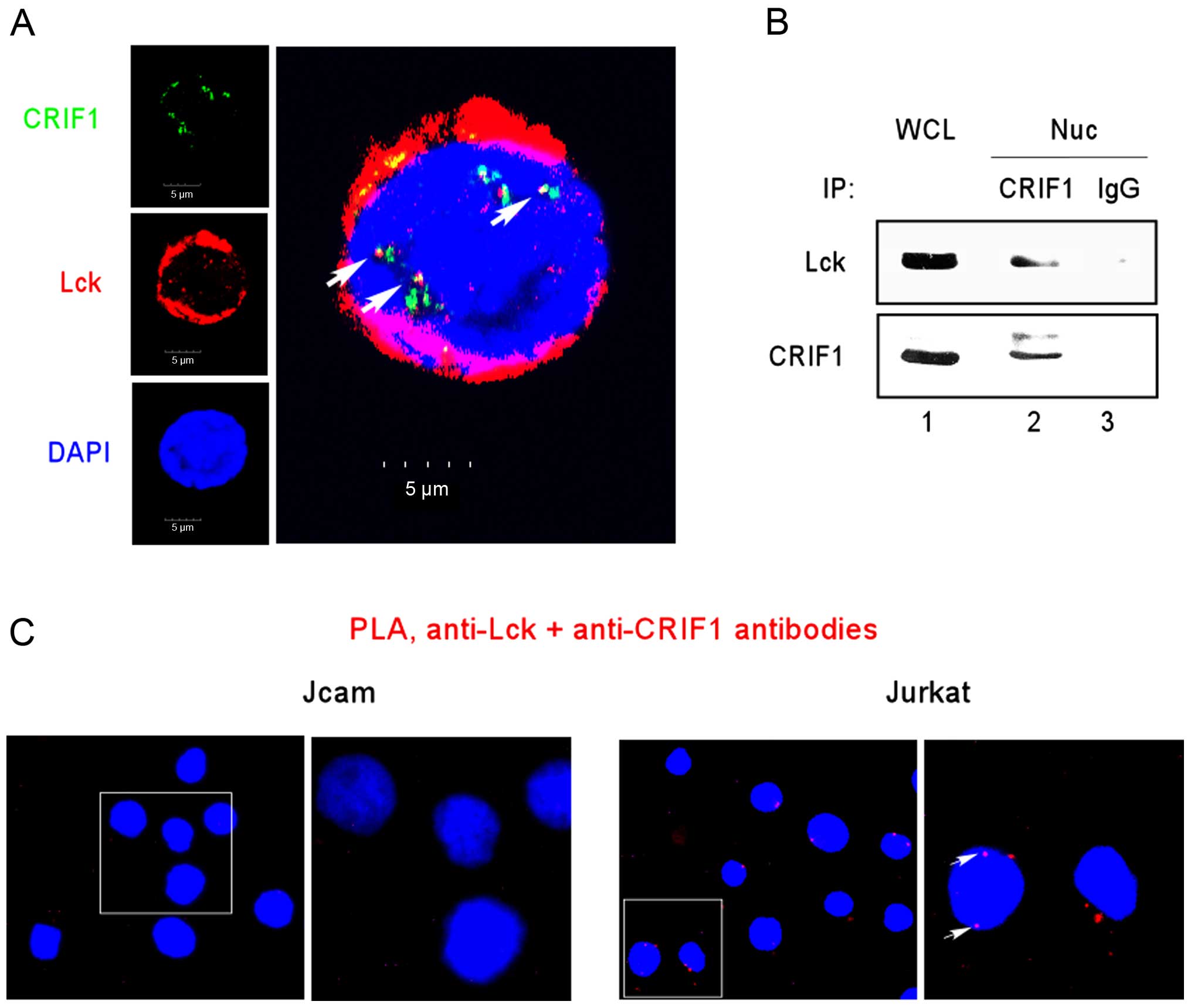
Lck interacts with CRIF1
We previously showed that Lck may function as a
transcription factor by binding to specific target genes (19). To identify additional
Lck-interacting partners, we performed mass spectrometry on Lck
immunoprecipitates prepared from LSTRA lysates. Our previous
proteomic analysis identified CRIF1 as one of the top candidates
(unpublished data). To confirm the interaction between Lck and
CRIF1 in human leukemic T cells, we performed immunofluorescence
microscopy and detected the co-localization of Lck and CRIF1 in the
Jurkat nucleus (Fig. 4A).
Co-immunoprecipitation between Lck and CRIF1 in nuclear extracts
prepared from Jurkat cells also confirmed the association of Lck
and CRIF1 (Fig. 4B, lane 2).
To further validate the close-range interaction
between Lck and CRIF1, we performed in situ PLA microscopy.
A positive PLA result relies on two molecules in the proximity of
16 nm or below, which reflects true protein-protein interaction. As
shown in Fig. 4C, a PLA signal was
detected in the Jurkat nucleus (right panels). Additional PLA
staining was observed outside the nuclei of Jurkat cells (Fig. 4C, right panels). This is consistent
with our earlier observation of Lck interaction with CRIF1 in
mitochondria (unpublished data). As a negative control, no PLA
signal was detected in the Lck-deficient Jcam cells (Fig. 4C, left panels). All together, these
results support a close interaction between Lck and CRIF1 in the
nuclear compartment of Jurkat cells.
Lck promotes leukemic T cell survival by
inhibiting CRIF1 function
Nuclear CRIF1 is known as a tumor suppressor by
blocking cell cycle progression and reducing cell survival. The
tumor-suppressing activities of CRIF1 are largely mediated by its
interaction with distinct nuclear proteins, such as
cyclin-dependent kinase 2 (CDK2) and Nur77 (28,29).
To determine the role of CRIF1 in human leukemic T cell survival in
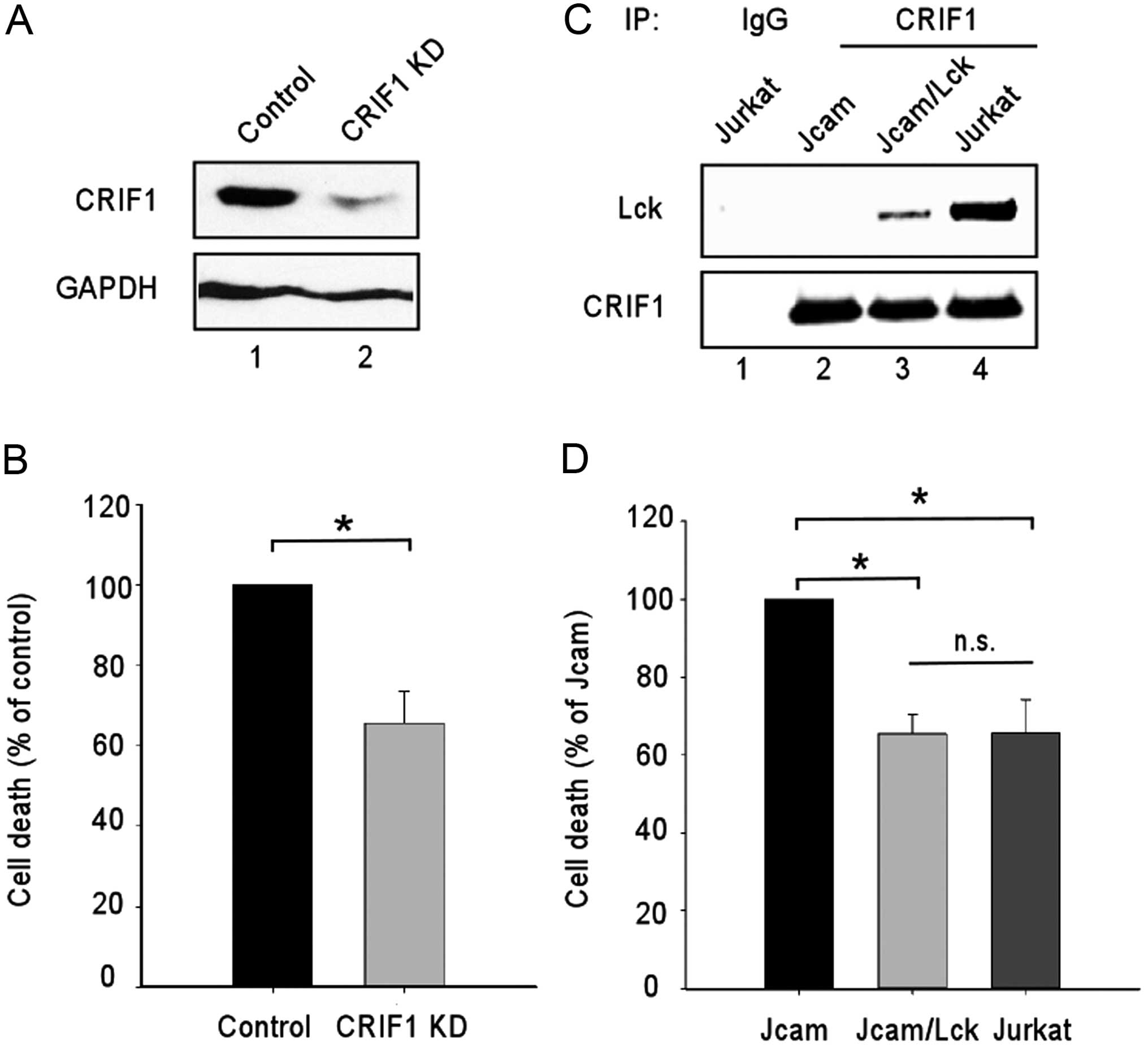
the absence of Lck, we knocked down CRIF1 expression in Jcam cells
by RNA interference (Fig. 5A, lane
2). Both the control and CRIF1-knockdown Jcam cells were subjected
to serum starvation to compare their sensitivity to growth factor
deprivation. Consistent with the role of CRIF1 as a tumor
suppressor, the CRIF1-knockdown Jcam cells were more resistant to
starvation-induced cell death (Fig.
5B).
Since Lck binds to CRIF1 in the nucleus (Fig. 4), we hypothesized that Lck
interaction with CRIF1 may interfere with CRIF1’s function as a
tumor suppressor. Indeed, similar to Jcam cells with CRIF1
knockdown (Fig. 5B), Jurkat cells
were more resistant to starvation-induced cell death as compared to
Jcam cells (Fig. 5D). To verify the
contribution of Lck, wild-type Lck was reconstituted in Jcam cells
and was capable of binding to CRIF1 (Fig. 5C, lane 3). The exogenous Lck confers
protection from starvation-induced cell death to a level similar to
endogenous Lck in Jurkat (Fig. 5D).
These results support the role of Lck in promoting cell survival by
its interaction with CRIF1 and, potentially, inhibition of CRIF1
activity.
Discussion
In summary, our present study revealed a novel
function of nuclear Lck in promoting cell survival through
interaction with a tumor suppressor. However, we do not exclude the
possibility that nuclear Lck may exert additional functions. For
example, our previous study demonstrated the role of Lck as a
nuclear transcription factor in regulating genes important in
oncogenesis (19). Other PTKs, such
as EGFR, also exhibit multiple functions in the nucleus. Nuclear
EGFR binds to the promoter regions of distinct target genes,
upregulates gene expression, and promotes cell cycle progression
(6,30). Interaction of EGFR with DNA protein
kinase in the nucleus, on the other hand, enhances DNA damage
repair and augments breast cancer cell’s resistance to cisplatin
and ionizing radiation treatment (31). Additionally, modulation of PCNA and
Cdc2 stability and activity through their association with nuclear
EGFR contributes to uncontrolled proliferation and DNA repair in
breast cancer cells (7,32). Our data further expand the
biological significance of nuclear PTKs from receptor PTK to
non-receptor PTK as well as from solid tumors to blood cancer.
Our results also support a positive effect of Lck
kinase activity on its nuclear translocation. Consistent with our
findings, kinase activity of EGFR is also important in its nuclear
translocation in breast cancer cells (33). It should be noted, however, that a
small amount of kinase-dead Lck (Fig.
2B) and inactive Lck (Fig. 3B)
can still be detected in the nuclear compartment. It is possible
that other kinase-independent mechanisms also contribute to Lck
nuclear translocation. While a nuclear localization signal (NLS)
has been identified in EGFR (34),
there is no discernible NLS in Lck. It remains to be determined
whether and how a non-classical NLS may mediate nuclear trafficking
of Lck.
Our mass spectrometry analysis identified CRIF1 as a
novel Lck-interacting partner. Consistent with its role as a tumor
suppressor, CRIF1 interacts with and inhibits CDK2 to induce cell
cycle arrest in leukemia cells (28). CRIF1-induced cell cycle arrest can
also be mediated by its interaction with other nuclear proteins,
including GADD45 and Nur77 (29,35).
Our CRIF1 knockdown data in Jcam cells further support the role of
CRIF1 in sensitizing leukemic cells to cell death, another
important cancer hallmark (36).
CRIF1 interaction with nuclear proteins may also contribute to its
role in cell death. More importantly, our data suggest that Lck can
bind to CRIF1 and inhibit its function as a tumor suppressor
(Fig. 5). It remains to be
determined whether Lck competes with other nuclear proteins in
binding to CRIF1.
Other than an important nuclear regulator, CRIF also
plays a critical role in the mitochondria (37). CRIF1 associates with the inner
membrane of mitochondria and participates in the synthesis of
oxidative phosphorylation polypeptides encoded by the mitochondrial
circular genome. CRIF1 also facilitates their insertion into the
inner membrane for the assembly of functional electron transport
chain complex. As expected, CRIF1 deficiency in the mitochondrial
compartment results in mitochondrial dysfunction. In brain-specific
CRIF1-knockout mice, fatal neurodegeneration is associated
with abnormal mitochondrial morphology (37). A recent study on an Alzheimer’s
disease animal model further suggests a protective role of
mitochondrial CRIF1 in neuronal cells against apoptosis (38). While the functions of nuclear CRIF1
are not fully addressed in neuronal cells, these studies do suggest
that CRIF1 can promote the survival of distinct cell types, such as
neuronal cells. In our T cell model, CRIF1 knockdown in Jcam cells
also resulted in slightly more dead cells before serum deprivation
(data not shown). However, under stress condition, CRIF1 promoted
Jcam cell death (Fig. 5B). The
overall effect of CRIF1 on cell survival, therefore, may depend on
the cell type and the experimental condition.
Consistent with the dual functions of CRIF1 in
different organelles, we also observed mitochondrial localization
of CRIF1 in Jurkat leukemia (unpublished data). Similarly, we
confirmed the presence of mitochondrial Lck in Jurkat T cells. The
concomitant localization of Lck in the cytoplasm, mitochondria and
nucleus further highlights the complexity of crosstalk between
different subcellular compartments. Other receptor and non-receptor
PTKs have been reported to exhibit diverse functions in the
cytoplasm, mitochondria and nucleus (39–41).
These findings of non-canonical signaling represent an important
paradigm shift of how PTKs coordinately regulate cellular
activities. They also provide critical insights in identifying
novel molecular targets in interfering with the oncogenic signal
transduction network.
Acknowledgments
This study was supported in part by funds from the
National Institute of Health R01 CA107210 (to C.-L.Y.) and the
RFUMS-H.M. Bligh Cancer Research Fund (to B.C.). The Midwest
Proteome Center (RFUMS) was supported in part by NIH S10
OD010662-01. We sincerely thank Xinli Yang, Patricia Loomis and
Virginie Bottero for the technical assistance concerning mass
spectrometry, confocal microscopy, and lentiviral transduction,
respectively. We also thank Srividya Venkitachalam for her input in
initiating this project.
Abbreviations:
|
PTK
|
protein tyrosine kinase
|
|
EGFR
|
epidermal growth factor receptor
|
|
SFK
|
Src family kinase
|
|
CRIF1
|
CR6-interacting factor 1
|
|
Eps15
|
epidermal growth factor receptor
substrate 15
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
PLA
|
proximity ligation assay
|
|
CDK2
|
cyclin-dependent kinase 2
|
|
NLS
|
nuclear localization signal
|
|
Lck
|
lymphocyte-specific protein tyrosine
kinase
|
References
|
1
|
Krause DS and Van Etten RA: Tyrosine
kinases as targets for cancer therapy. N Engl J Med. 353:172–187.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lo HW and Hung MC: Nuclear EGFR signalling
network in cancers: Linking EGFR pathway to cell cycle progression,
nitric oxide pathway and patient survival. Br J Cancer. 94:184–188.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin SY, Makino K, Xia W, Matin A, Wen Y,
Kwong KY, Bourguignon L and Hung MC: Nuclear localization of EGF
receptor and its potential new role as a transcription factor. Nat
Cell Biol. 3:802–808. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chou RH, Wang YN, Hsieh YH, Li LY, Xia W,
Chang WC, Chang LC, Cheng CC, Lai CC, Hsu JL, et al: EGFR modulates
DNA synthesis and repair through Tyr phosphorylation of histone H4.
Dev Cell. 30:224–237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marti U, Burwen SJ, Wells A, Barker ME,
Huling S, Feren AM and Jones AL: Localization of epidermal growth
factor receptor in hepatocyte nuclei. Hepatology. 13:15–20. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hadzisejdić I, Mustać E, Jonjić N,
Petković M and Grahovac B: Nuclear EGFR in ductal invasive breast
cancer: Correlation with cyclin-D1 and prognosis. Mod Pathol.
23:392–403. 2010. View Article : Google Scholar
|
|
7
|
Tan M, Jing T, Lan KH, Neal CL, Li P, Lee
S, Fang D, Nagata Y, Liu J, Arlinghaus R, et al: Phosphorylation on
tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and
is involved in resistance to taxol-induced apoptosis. Mol Cell.
9:993–1004. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Domingues I, Rino J, Demmers JA, de
Lanerolle P and Santos SC: VEGFR2 translocates to the nucleus to
regulate its own transcription. PLoS One. 6:e256682011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi A, Obata Y, Fukumoto Y, Nakayama
Y, Kasahara K, Kuga T, Higashiyama Y, Saito T, Yokoyama KK and
Yamaguchi N: Nuclear localization of Src-family tyrosine kinases is
required for growth factor-induced euchromatinization. Exp Cell
Res. 315:1117–1141. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Van Laethem F, Tikhonova AN, Pobezinsky
LA, Tai X, Kimura MY, Le Saout C, Guinter TI, Adams A, Sharrow SO,
Bernhardt G, et al: Lck availability during thymic selection
determines the recognition specificity of the T cell repertoire.
Cell. 154:1326–1341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Palacios EH and Weiss A: Function of the
Src-family kinases, Lck and Fyn, in T-cell development and
activation. Oncogene. 23:7990–8000. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Burnett RC, David JC, Harden AM, Le Beau
MM, Rowley JD and Diaz MO: The LCK gene is involved in the
t(1;7)(p34;q34) in the T-cell acute lymphoblastic leukemia derived
cell line, HSB-2. Genes Chromosomes Cancer. 3:461–467. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Majolini MB, Boncristiano M and Baldari
CT: Dysregulation of the protein tyrosine kinase LCK in
lymphoproliferative disorders and in other neoplasias. Leuk
Lymphoma. 35:245–254. 1999. View Article : Google Scholar
|
|
14
|
Accordi B, Espina V, Giordan M, VanMeter
A, Milani G, Galla L, Ruzzene M, Sciro M, Trentin L, De Maria R, et
al: Functional protein network activation mapping reveals new
potential molecular drug targets for poor prognosis pediatric
BCP-ALL. PLoS One. 5:e135522010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Veillette A, Foss FM, Sausville EA, Bolen
JB and Rosen N: Expression of the lck tyrosine kinase gene in human
colon carcinoma and other non-lymphoid human tumor cell lines.
Oncogene Res. 1:357–374. 1987.PubMed/NCBI
|
|
16
|
Robinson D, He F, Pretlow T and Kung HJ: A
tyrosine kinase profile of prostate carcinoma. Proc Natl Acad Sci
USA. 93:5958–5962. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chueh FY and Yu CL: Engagement of T-cell
antigen receptor and CD4/CD8 co-receptors induces prolonged STAT
activation through autocrine/paracrine stimulation in human primary
T cells. Biochem Biophys Res Commun. 426:242–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elsberger B, Fullerton R, Zino S, Jordan
F, Mitchell TJ, Brunton VG, Mallon EA, Shiels PG and Edwards J:
Breast cancer patients’ clinical outcome measures are associated
with Src kinase family member expression. Br J Cancer. 103:899–909.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Venkitachalam S, Chueh FY and Yu CL:
Nuclear localization of lymphocyte-specific protein tyrosine kinase
(Lck) and its role in regulating LIM domain only 2 (Lmo2) gene.
Biochem Biophys Res Commun. 417:1058–1062. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chueh FY, Cronk RJ, Alsuwaidan AN, Mallers
TM, Jaiswal MK, Beaman KD and Yu CL: Mouse LSTRA leukemia as a
model of human natural killer T cell and highly aggressive lymphoid
malignancies. Leuk Lymphoma. 55:706–708. 2014. View Article : Google Scholar
|
|
21
|
Chueh FY, Leong KF and Yu CL:
Mitochondrial translocation of signal transducer and activator of
transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated
cells. Biochem. Biophys Res Commun. 402:778–783. 2010. View Article : Google Scholar
|
|
22
|
Cooper JC, Shi M, Chueh FY, Venkitachalam
S and Yu CL: Enforced SOCS1 and SOCS3 expression attenuates
Lck-mediated cellular transformation. Int J Oncol. 36:1201–1208.
2010.PubMed/NCBI
|
|
23
|
Venkitachalam S, Chueh FY, Leong KF,
Pabich S and Yu CL: Suppressor of cytokine signaling 1 interacts
with oncogenic lymphocyte-specific protein tyrosine kinase. Oncol
Rep. 25:677–683. 2011.PubMed/NCBI
|
|
24
|
Yu CL, Jove R and Burakoff SJ:
Constitutive activation of the Janus kinase-STAT pathway in T
lymphoma overexpressing the Lck protein tyrosine kinase. J Immunol.
159:5206–5210. 1997.
|
|
25
|
Abraham RT and Weiss A: Jurkat T cells and
development of the T-cell receptor signalling paradigm. Nat Rev
Immunol. 4:301–308. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi M, Cooper JC and Yu CL: A
constitutively active Lck kinase promotes cell proliferation and
resistance to apoptosis through signal transducer and activator of
transcription 5b activation. Mol Cancer Res. 4:39–45. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Straus DB and Weiss A: Genetic evidence
for the involvement of the lck tyrosine kinase in signal
transduction through the T cell antigen receptor. Cell. 70:585–593.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ran Q, Hao P, Xiao Y, Xiang L, Ye X, Deng
X, Zhao J and Li Z: CRIF1 interacting with CDK2 regulates bone
marrow microenvironment-induced G0/G1 arrest of leukemia cells.
PLoS One. 9:e853282014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park KC, Song KH, Chung HK, Kim H, Kim DW,
Song JH, Hwang ES, Jung HS, Park SH, Bae I, et al: CR6-interacting
factor 1 interacts with orphan nuclear receptor Nur77 and inhibits
its transactivation. Mol Endocrinol. 19:12–24. 2005. View Article : Google Scholar
|
|
30
|
Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y
and Hung MC: Co-regulation of B-Myb expression by E2F1 and EGF
receptor. Mol Carcinog. 45:10–17. 2006. View Article : Google Scholar
|
|
31
|
Liccardi G, Hartley JA and Hochhauser D:
EGFR nuclear trans-location modulates DNA repair following
cisplatin and ionizing radiation treatment. Cancer Res.
71:1103–1114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu YL, Chou RH, Liang JH, Chang WJ, Su KJ,
Tseng YJ, Huang WC, Wang SC and Hung MC: Targeting the EGFR/PCNA
signaling suppresses tumor growth of triple-negative breast cancer
cells with cell-penetrating PCNA peptides. PLoS One. 8:e613622013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brand TM, Iida M, Li C and Wheeler DL: The
nuclear epidermal growth factor receptor signaling network and its
role in cancer. Discov Med. 12:419–432. 2011.PubMed/NCBI
|
|
34
|
Hsu SC and Hung MC: Characterization of a
novel tripartite nuclear localization sequence in the EGFR family.
J Biol Chem. 282:10432–10440. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chung HK, Yi YW, Jung NC, Kim D, Suh JM,
Kim H, Park KC, Song JH, Kim DW, Hwang ES, et al: CR6-interacting
factor 1 interacts with Gadd45 family proteins and modulates the
cell cycle. J Biol Chem. 278:28079–28088. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim SJ, Kwon MC, Ryu MJ, Chung HK, Tadi S,
Kim YK, Kim JM, Lee SH, Park JH, Kweon GR, et al: CRIF1 is
essential for the synthesis and insertion of oxidative
phosphorylation polypeptides in the mammalian mitochondrial
membrane. Cell Metab. 16:274–283. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Byun J, Son SM, Cha MY, Shong M, Hwang YJ,
Kim Y, Ryu H, Moon M, Kim KS and Mook-Jung I: CR6-interacting
factor 1 is a key regulator in Aβ-induced mitochondrial disruption
and pathogenesis of Alzheimer’s disease. Cell Death Differ. Nov
7–2014.Epub ahead of print. View Article : Google Scholar
|
|
39
|
Demory ML, Boerner JL, Davidson R, Faust
W, Miyake T, Lee I, Hüttemann M, Douglas R, Haddad G and Parsons
SJ: Epidermal growth factor receptor translocation to the
mitochondria: Regulation and effect. J Biol Chem. 284:36592–36604.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tibaldi E, Brunati AM, Massimino ML,
Stringaro A, Colone M, Agostinelli E, Arancia G and Toninello A:
Src-tyrosine kinases are major agents in mitochondrial tyrosine
phosphorylation. J Cell Biochem. 104:840–849. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding Y, Liu Z, Desai S, Zhao Y, Liu H,
Pannell LK, Yi H, Wright ER, Owen LB, Dean-Colomb W, et al:
Receptor tyrosine kinase ErbB2 translocates into mitochondria and
regulates cellular metabolism. Nat Commun. 3:12712012. View Article : Google Scholar : PubMed/NCBI
|