Introduction
HtrA1, first described by Zumbrunn and Trueb
(1), is expressed in different
normal human tissues (2) and
appears to be involved in several physiological processes, through
inhibition of extracellular protein transforming growth factor β
(TGF-β) signaling in vivo and in vitro (3), as well as in the pathogenesis of
diseases such as amyloid degeneration, senile macular degeneration,
Alzheimer’s disease, osteoarthritis and pre-eclampsia (4–7). It
has also been hypothesized to play a role as a tumor suppressor.
The first clinical study was carried out on melanoma by Baldi and
colleagues (8), who found
significant HtrA1 upregulation in the primary tumor compared with
metastases, and suggested that HtrA1 expression could be an
indicator of disease progression. The hypothesis has subsequently
been tested in other neoplasms. A possible role for HtrA1 as a
prognostic factor for cancer has also been hypothesized.
Downregulation of HtrA1 protein is associated with poor survival in
mesothelioma (9), hepatocellular
carcinoma (10) and breast cancer
(11); in the latter study
node-positivity was associated with shorter survival. HtrA1
downregulation has also been observed to be associated with poor
chemotherapy response in patients with gastric cancer (12). These findings suggest a possible
prognostic role for HtrA1 expression. The present manuscript
reviews current cancer-related HtrA1 research from the
methodological and clinical standpoints and studies exploring the
potential role of HtrA1 as a tumor marker and/or prognostic factor
in a number of tumors.
Materials and methods
A MEDLINE search was conducted in August 2014 using
the terms: HtrA1 OR PRSS11 protein, human OR L56 protein, human OR
protease, serine, 11 (IGF binding) protein, human OR
high-temperature requirement factor A1, human OR HtrA serine
peptidase 1, human AND Neoplasm OR Tumors OR Tumor OR Neoplasia OR
Cancer OR Cancers. No restriction was applied in terms of date,
language or study design. The PRISMA method was used to select
studies (14). A search of Embase
and the Cochrane Systematic Review and Clinical Trial Register did
not yield additional studies. A manual search was also conducted.
The articles thus collected were examined and selected by two
independent reviewers (P.M.A. and A.L.); any disagreement was
resolved by a methodologist (E.A.). The methods and laboratory
techniques were reviewed by a laboratory researcher (D.M.).
Studies involving clinical studies and those where
HtrA1 was determined either as mRNA or as protein were included;
articles involving exclusively in vitro studies and
exclusively animal studies, and the methods applied to detect HtrA1
in each study: in situ hybridization (ISH) polymerase chain
reaction (PCR) immunohistochemistry (IHC) and western blotting (WB)
were determined.
Those published as congress proceedings were
excluded. Data are presented as tables. Table I documents the selected studies
including the date of publication, sample size, age, gender and
general characteristics of the patient population; HtrA1 values in
the tumor and control tissue, and any association found between
HtrA1 level, histological differentiation and TNM and/or clinical
stage.
 | Table ICharacteristics of the 15 studies
surveyed. |
Table I
Characteristics of the 15 studies
surveyed.
| Author, year
(ref.) | Cancer site | Patient
population
| HtrA1
expression
|
|---|
| Sample size | Gender/Age
(years) | Characteristics of
the selected study | Clinical
notice | Cancer tissue vs.
control tissue | Degree of
differentiation | Relationship with
TNM or clinical stage |
|---|
| D’Angelo et
al, 2014 (42) | Autonomic nervous
system (neuroblastoma ganglioneuroblastoma) | 60 | Male, 34 Female, 26
Age <12, 17 Age >12, 3 | Descriptive study
No control group Enrollment method: not random | Perioperative
treatment: none | Not evaluated | Not evaluated | Highest HtrA1
levels found in patients with stage I, II and IVS vs.
stage III-IV disease (P=0.003)a |
| Lorenzi et
al, 2013 (43) | Bladder | 152 | Healthy
individuals: Male, 6 Female, 32 Mean age, 65±5.5 Patients with
urothelial bladder cancer:: Male, 50 Female,18 Mean age, 68.2±7.0
Patients with bacterial cystitis: Male, 4 Female, 12 Mean age,
59.1±11.8 | Diagnostic study
with analysis of of sensibility and specificity for urinary HtrA1
Control: healthy control Enrollment method: consecutive | Perioperative
treatment: not declared | Lower expression of
HtrA1 protein (autocatalytic form) in cancer than in
normal-appearing tissue (P<0.001) | Not evaluated | Not evaluated |
| Lehner et
al, 2013 (11) | Breast | 131 | Female only Age
<50, 23 Age >50, 108 | Longitudinal study
Tissue selected with criteria (see text for detail) Survival
analysis (see Table II) Enrollment
method: not random | Unilateral cancer
Perioperative treatment: none | Not evaluated | Not statistically
significant | Low T (pT1–2)
associated with higher HtrA1 mRNA levels compared with high T
(pT3–4) (P=0.025)b No correlation
with lymph node status (P=0.439) (see text for details) |
| Yu et al,
2012 (44) | Esophagus | 63 | Male, 50 Female, 13
Mean age, 73.4 Range, 45–79 | Control:
normal-looking tissue. Enrollment method: probably consecutive | Perioperative
treatment: none | Significantly lower
HtrA1 levels in cancer than in normal-appearing tissue; mRNA
(P=0.004) expression and protein expression (P<0.05) | Stage of
differentiation also correlates with HtrA1 mRNA levels for mRNA
P=0.024) than protein expression (P<0.05) | Pathological stage
correlates with mRNA expression as well as protein expression
(P=0.013) |
| Xia et al,
2012 (45) | Esophagus | 115 | Male, 71 Female, 44
Age <60, 5 Age >60, 63 | Longitudinal study
Survival analysis (see Table II)
Enrollment method: not reported | Perioperative
treatment: not reported | Significantly lower
HtrA1 mRNA and protein in cancer specimens than in normal-appearing
tissue (P<0.05) | Not statistically
significant | Low HtrA1 levels,
as mRNA or protein, are associated with TNM stage and lymph node
metastases (P<0.05) |
| Catalano et
al, 2010 (12) | Stomach | 80 | Male, 51, Female,
29 Mean age, 64 Range, 32–82 | Longitudinal study,
Survival analysis (see Table II)
Enrollment method: consecutive | Patients with
recurrent or metastatic cancer subjected to first-line chemotherapy
(5-FU and cisplatin) | Not evaluated | Not evaluated | Not statistically
significant |
| Zhu et al,
2010 (10) | Liver | 50 | Male, 42 Mean age,
52.43±9.94 Female, 8 Mean age, 50.39±14.12 | Longitudinal study
Control: normal- appearing tissue Survival analysis (see Table II) Enrollment method: probably
consecutive | Perioperative
treatment: none | Expression of HtrA1
lower in HCC than in normal- appearing specimens (P=0.045) | HtrA1 expression
and staining score related to histological gradec (grade I-II higher than grade III-IV)
(P=0.036) | TNM or clinical
stage not evaluated No correlation with tumor and size, metastasis
(see text for detail) |
| Mullany et
al, 2010 (46) | Endometrium | 184 | Mean age,
66.1±11.3 | Longitudinal study
Survival analysis (see Table II)
Enrollment method: random | | Not evaluated | Significantly lower
HtrA1 levels in high-grade (3) than
in low-grade (1–2) tumors (P=0.016) | Not statistically
significant |
| Narkiewicx et
al, 2008 (47) | Endometrium | 124 | Age not
reported | Control: healthy
subjects.d Enrollment method: not
reported | Perioperative
treatment: Not reported | Significantly lower
HtrA1 mRNA and protein levels in cancer than in control specimens
(P<0.001) | Not evaluated | Not statistically
significant |
| Bowden et
al, 2006 (48) | Endometrium | 33 | Age not
reported | Control: healthy
subjects.d Enrollment method: not
reported | Perioperative
treatment: not reported | Significantly lower
HtrA1 mRNA in tumors of all grades than in normal endometrium
(P<0.001) | Lowest levels of
HtrA1 mRNA from G2 to G3 (P<0.01) | Not evaluated |
| Zurawa- Janicka
et al, 2012 (49) | Thyroid | 40 | Not reported | Control: healthy
subjects. Specimens from 40 patients: 20 with benign lesions and 20
with cancer Enrollment method: not reported | Perioperative
treatment: not reported | Not statistically
significant | Not evaluated
Evaluation of HtrA1 levels between two different histology (See
text for detail) | Not evaluated |
| Baldi et al,
2008 (9) | Pleura
(mesothelioma) | 70 | Female, 29 Male, 41
Median age, 65 Range, 45–81 | Longitudinal study
Survival analysis (see Table II)
Enrollment method: probably consecutive | All patients were
treated with radiotherapy or chemotherapy 44 patients were treated
with cyto- reductive surgery | Not evaluated | Not evaluated | Not evaluated |
| Narkiewicz et
al, 2008 (50) | Ovary | 98 | Not reported | Study with five
different groupse Enrollment
method: not reported | Perioperative
treatment: not reported | Significant
decrease in HtrA1 mRNA levels was observed in the malignant tumor
tissue group compared to the normal ovarian tissue (P<0.001).
Not significant for protein (see text for details) | Not statistically
significant | Not statistically
significant |
| Esposito et
al, 2006 (51) | Lung | 99 | Median age, 58
Male, 73, Female, 13 | No control group
Enrolment method: consecutive | Perioperative
treatment: none | Not evaluated | Non-significant
differences between histological grades (see text for detail) | Not statistically
significant |
| Baldi et al,
2002 (8) | Skin
(melanoma) | 11 | Not reported | No control group
Enrolment method: not reported | Perioperative
treatment: not reported | Not evaluated | Not evaluated | Correlation with T
(P=0.002) |
Table II shows the
longitudinal observational data relating HtrA1 to survival.
 | Table IIObservational longitudinal
studies. |
Table II
Observational longitudinal
studies.
| Author, year (ref.)
cancer type | Aim | Follow-up | Cohort Overall
survival (OS) | Disease-free
survival (DFS) | Hazard ratio
(HR) |
|---|
| Lehnert et
al, 2013 (11) Breast | Evaluation of HtrA1
mRNA expression on patient outcome
Two groups selected with a cut-off of 48 | 60 months
120 months | 60 months: 85±3.2%
SE
120 months: 66±4.9% SE | 60 months: 75±4.0%
SE
120 months: 54.5±5.2% SE | OS: HR 0.45 (95% CI
0.23–0.90) P=0.023
DFS: HR 0.55 (95% CI 0.32–0.94) P=0.028 Lymph node-positive
groups:
OS: HR 0.21 (95% CI 0.07–0.63) P=0.006
DFS: HR 0.29 (95% CI 0.13–0.65) P=0.003 |
| (see Table III for Laboratory techniques) | | | | Therapy
groups:
(none, endocrine, chemotherapy)
OS: HR 0.23 (95% CI 0.07–0.72) P=0.0012
DFS: HR 0.29 (95% CI 0.13–0.67) P=0.004 |
| Xia et al,
2013 (45) Esophagus | Correlation between
HtrA1 levels and survival | 60 months | The overall cohort
survival, disease-free survival and HR: were not reported. Reported
date included:
HtrA1 mRNA-positive: 13 of 39 patients died (33.3%)
HtrA1 mRNA-negative: 56 of 76 patients died (73.7%)
(P<0.05) |
| Catalano et
al, 2011 (12) Stomach | Determine HtrA1
expression as predictor of chemoresponse in patients with advanced
gastric cancer Two groups high/medium expression vs. low expression
(see Table III for laboratory
techniques) | 60 months | 17.0 months:
high/medium HtrA1 expression 9.5 months: low HtrA1 expression | Not reported, they
evaluated time to progression High and medium HtrA1 expression
compared with low: 0.52 (95% CI 0.29–0.93) P=0.027 | OS: HR 0.55 (95% CI
0.32–0.96) P=0.037 |
| Mullany et
al, 2011 (46) | Dichotomized into 2
groups high intensiy score vs. low | 200 months | Not reported | No significant
relationship between high-medium expression of HtrA1 and
survival |
| Endometrium Zhu
et al, 2010 (10) Liver | Dichotomized into 2
groups high intensity score vs. low | 72 months | HtrA1 (+++/++)
survival rate 46%, median survival 35.5 months
HtrA1 (+) survival rate 26%, median survival 15.6 months
Cohort OS, DFS and HR not reported | |
| Baldi et al,
2008 (9) Pleural mesothelioma | Analyzed the
potential prognostic value of expression of HtrA1 Three class of
HtrA1 expression: low medium high | 40 months | HtrA1 (+): median
survival time (months), 6.0 (95% CI 4.46–7.51) HtrA1 (++): median
survival time (months), 16.0 (95% CI 12.54–19.45) HtrA1 (+++):
median survival time (months), 24.0 (95% CI 20.50–27.49)
P<0.0001 | Not reported | HtrA1 (+): HR 1
(reference category) HtrA1 (++): HR 0.65 (95% CI 0.348–0.876) HtrA1
(+++): HR 0.26 (95% CI 0.122–0.454) P<0.001 |
Table III reports
the methods applied to detect HtrA1 in each study: in situ
hybridization (ISH), polymerase chain reaction (PCR),
immunohistochemistry (IHC) and western blotting (WB).
 | Table IIILaboratory methods used in the
studies of HtrA1 in cancer. |
Table III
Laboratory methods used in the
studies of HtrA1 in cancer.
| Authors/ref. | Involved organ | Methods
| mRNA
| Protein
|
|---|
| In
vitro | In vivo | ISH | PCR | IHC | WB |
|---|
| D’Angelo et
al (42) | Nerve cells | | √ | | | √ | √ |
| Lorenzi et
al (43) | Bladder | √ | √ | | √ | √ | √ |
| Lehner et al
(11) | Breast | √ | √ | | √ | | |
| Yu 2012 et
al (44) | Esophagus | √ | √ | | √ | √ | √ |
| Xia 2013 et
al (45) | Esophagus | √ | √ | √ | √ | √ | √ |
| Catalano et
al (12) | Stomach | | √ | | | √ | |
| Zhu et al
(10) | Liver | | √ | | | √ | |
| Mullany et
al (46) | Endometrium | √ | √ | | | √ | |
| Narkiewicz et
al (47) | Endometrium | | √ | | √ | | √ |
| Bowden et al
(48) | Endometrium | | √ | | √ | √ | √ |
| Zurawa et al
(49) | Thyroid | | √ | | | | √ |
| Baldi et al
(9) | Pleural
mesothelioma | √ | √ | | | √ | √ |
| Narkiewicz et
al (50) | Ovary | | √ | | √ | | √ |
| Esposito et
al (51) | Lung | | √ | | | √ | |
| Baldi et al
(8) | Skin cells
(Malignant melanoma) | √ | √ | | | √ | |
Table IV reviews
the current use of HtrA1 as a marker in oncology.
 | Table IVHtrA1 as a potential tumor
marker. |
Table IV
HtrA1 as a potential tumor
marker.
| Cancer type | Early marker | Prognostic
marker | Tumor marker | Author/ref. |
|---|
| Neuroblastoma | | | | |
|
Ganglioneuroblastoma | | | √ | D’Angelo et
al (42) |
| Bladder | √ | | √ | Lorenzi et
al (43) |
| Breast | | √ | √ | Lehner et al
(11) |
| Esophagus | | | √ | Yu et al
(44) |
| Esophagus | | √ | √ | Xia et al
(45) |
| Stomach | | √ | √ | Catalano et
al (12) |
| Liver | | √ | √ | Zhu et al
(10) |
| Endometrial | | √ | √ | Mullany (46) |
| Endometrial | | | √ | Narkiewicz et
al (47) |
| Endometrial | | | √ | Bowden (48) |
| Thyroid | | | √ | Zurawa et al
(49) |
| Pleural
mesothelioma | | √ | √ | Baldi et al
(9) |
| Ovary | | | √ | Narkiewicz et
al (50) |
| Lung | | | √ | Esposito et
al (51) |
| Malignant
melanoma | | | √ | Baldi et al
(8) |
Results
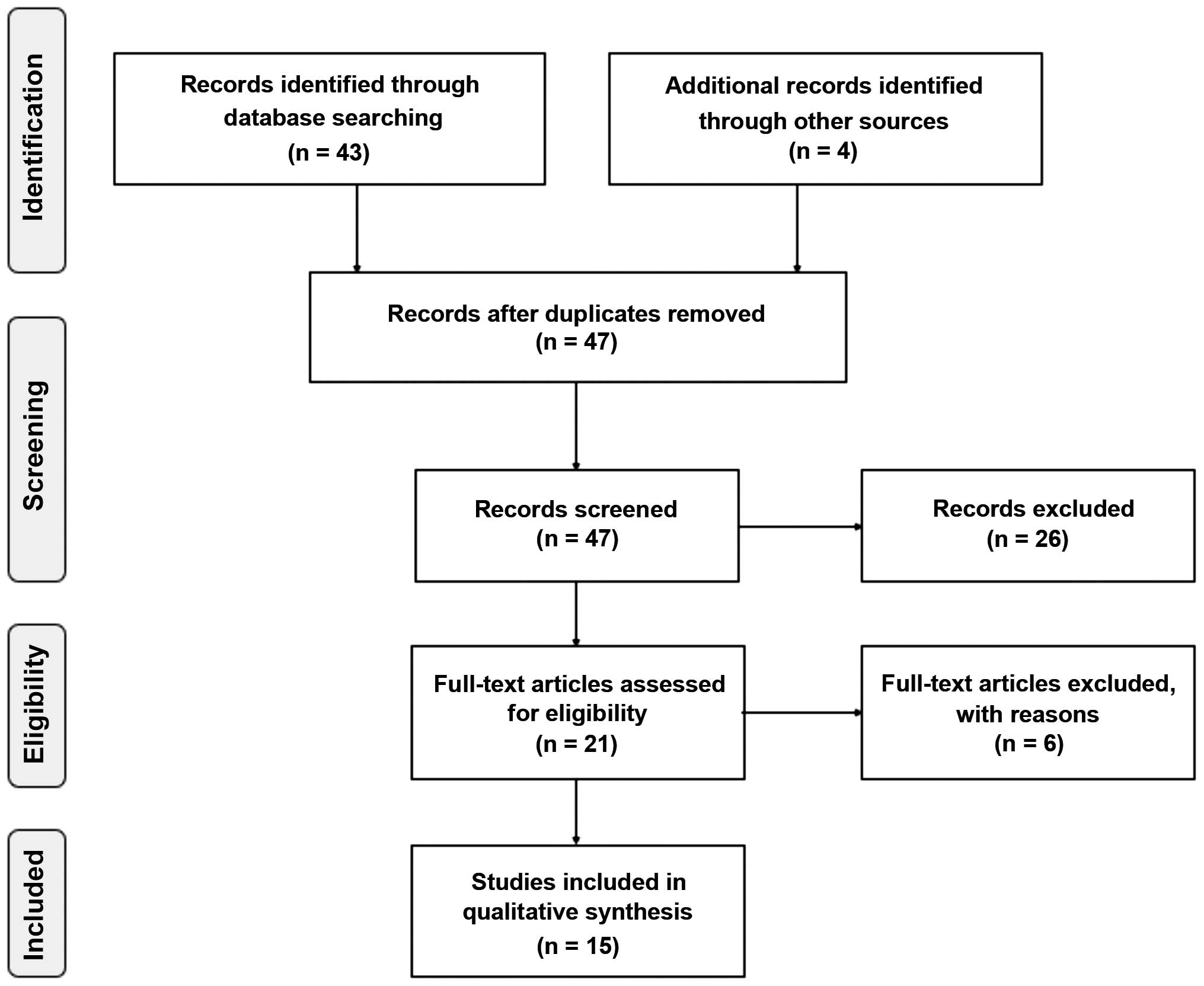
The database search conducted as described above
retrieved 43 studies, and the manual search retrieved 4 additional
studies. No duplicates were found (Fig.
1). Examination of the abstracts led to the exclusion of 26
studies; 1 regarded rheumatoid arthritis (15); 3 studied macular degeneration
(16–18); 2 investigated the placenta (7,19); 1
addressed tuberous sclerosis complex 2 (20); 3 were animal studies (21–23); 1
assessed amyloid precursor protein (4); 2 evaluated other biomarkers (24,25); 6
were exclusively in vitro studies (1;26–30); and 7 were
reviews (2,31–36),
thus leaving 21 studies. Examination of their full text led to the
exclusion of 6 further studies since they did not report clinical
studies (13,37–41).
Overall, 15 studies met the inclusion criteria and are reviewed
below.
Synthesis of the data of observational
studies and overview of laboratory techniques
The 15 studies included in the review describe
recent HtrA1 research in relation to cancer.
Neuroblastoma
There was a single investigation addressing neural
tissue tumors, i.e. neuroblastoma (NB) and ganglioneuroblastoma
(42). This is also the sole study
involving a pediatric population. Its aim was to test the value of
HtrA1 as a new biomarker of tumor differentiation and
aggressiveness by quantifying its expression and localization in
NB. D’Angelo et al (42)
assessed HtrA1 expression by semi-quantitative methods: by IHC
using the HSCORE value and by WB using Quantity One software,
neither of which provides an absolute value of protein
concentration. The authors examined 60 NB: 26 stage I-II; 14 stage
III; 16 stage IV; and 4 stage IV tumors. The statistical
distribution of the HtrA1 variable was not investigated. Higher
HtrA1 protein levels were found in stages associated with a more
favorable prognosis, but their possible correlation with gender or
age was not assessed.
Bladder cancer
The single study of bladder cancer by Lorenzi and
co-workers (43) examined HtrA1
expression in tissue and urine from 152 subjects, 38 healthy
individuals, 68 patients with urothelial cancer, and 16 subjects
with cystitis, to establish a possible association with urothelial
cancer. Two forms of HtrA1, a 50- and a 38-kDa autocatalytic form,
were detected in tissue specimens. The autocatalytic form was
downregulated in cancer tissue whereas significantly higher levels
of both forms were measured in urine from cancer patients compared
with healthy individuals. HtrA1 distribution was normal in urine
from all participants. The correlation of HtrA1 levels with gender
and age was not tested. Global test performance was assessed. The
authors applied molecular, morphological and biochemical
techniques. Since mRNA and protein levels were not measured in the
whole sample, but only in radical cystectomy tissue, the results
apply only to this group.
Breast cancer
Lehner and co-workers (11) assessed the impact of HtrA1 mRNA
expression on patient outcome in a cohort of 131 cancer patients
using molecular and in vitro techniques to measure HtrA1
mRNA and HtrA1 promoter hypermethylation, the latter as an HtrA1
silencing mechanism in breast tumors. HtrA1 protein expression was
not assessed. The statistical distribution of the HtrA1 variable
was not investigated. Significantly higher HtrA1 expression was
found in lower tumor stages, but no relationship was found with
grade (Table I). There were no
significant differences related to the expression of progesterone
and estrogen receptor; pre- or post-menopausal state; or type of
surgical management (lumpectomy vs. mastectomy). High levels of
HtrA1 mRNA were associated with longer overall survival (OS) and
disease-free survival (DFS) (Table
II).
Esophageal cancer
Yu and colleagues (44) explored the possible involvement of
HtrA1 levels in cell invasiveness, differentiation, stage and
metastasis formation in esophageal cancer tissue and adjacent
normal-appearing tissue from 63 patients. HtrA1 mRNA and protein
were measured using semi-quantitative biomolecular methods. The
statistical distribution of the HtrA1 variable was not examined.
Significantly higher HtrA1 mRNA (P=0.004) and protein (P<0.05)
levels were detected in normal-appearing tissue. The authors found
that HtrA1 mRNA and protein levels were higher in well and
moderately differentiated tumors than in poorly differentiated
tumors (respectively, mRNA P=0.024; protein P<0.05) as well as
in early compared with late disease stages (I-II vs. III-IV,
respectively, mRNA P=0.013; protein P<0.05). The data regarding
metastases are particularly important, since HtrA1 mRNA and protein
levels were lower in tumors with distant metastases that in those
without metastases (mRNA P<0.001; protein P<0.05). Similar
data were found for lymph node metastases (mRNA P=0.002; protein
P<0.05). In contrast HtrA1 mRNA was not found to correlate with
gender, age or size of the primary tumor.
Xia and co-workers (45) evaluated HtrA1 mRNA and protein in
cancer and normal-appearing tissue using respectively IHC and ISH,
two semi-quantitative techniques that were mainly applied to detect
the cell type expressing HtrA1. Staining (positive or negative) was
assessed in a blinded manner by two independent researchers. In
vitro analysis was used to detect the molecular targets of
HtrA1. The sample consisted of 115 patients with squamous cell
carcinoma (SCC) of the esophagus. The statistical distribution of
the HtrA1 variable was not examined. The authors found an
association between HtrA1 level and TNM stage (Table I), but not between tumor
differentiation, gender or age. Mean follow-up duration was 40
months. HtrA1 mRNA and protein levels correlated with survival
rate, since HtrA1 (mRNA and protein)-positive patients had a longer
survival than HtrA1-negative subjects (Table II).
Gastric cancer
Catalano and colleagues (12) mainly assessed whether HtrA1 levels
affect chemo-responsiveness in 80 patients with advanced gastric
cancer. They used only morphological analysis to evaluate HtrA1
expression in diseased tissue. Staining was evaluated by three
raters using an observational semi-quantitative method that does
not measure absolute protein value, but the change in staining
intensity. The statistical distribution of the HtrA1 variable was
not examined. HtrA1 expression was associated with
chemo-responsiveness, and higher HtrA1 levels correlated with
longer survival and time to progression. There were no associations
with gender, age or Lauren’s classification.
Hepatocellular carcinoma
Zhu et al (10) measured HtrA1 expression in liver
tissue from 50 patients with hepatocellular carcinoma to establish
whether it may have a role in cancer development and progression.
HtrA1 was assessed by IHC; staining intensity was evaluated by
light microscopy using a semi-quantitative score. HtrA1 levels
correlated with Edmondson and Steiner’s criteria and vascular
invasion (P=0.014) but not with gender, age, tumor size or presence
of metastases. The statistical distribution of the HtrA1 variable
was not examined. After a follow-up of 3 years, the survival was
longer in the HtrA1-positive (46%) than in the HtrA1-negative
patients (26%).
Endometrial cancer
Mullany and co-workers (46) investigated by an in vitro
model whether HtrA1 downregulation influences cancer cell invasion.
In addition, randomly selected patients from a historical cohort of
184 patients were used for IHC. Of these, 142 had tumor stages I-II
and 42 had stages III-IV; 65 had histological grade 1, 75
histological grade 2, and 44 histological grade 3; 171 had
endometroid or mucinous disease and 13 had non-endometroid cancer.
The authors did not examine the whole section, but used a
micro-array technique to analyze 3 endometrial samples 0.6-mm in
diameter from each patient. HtrA1 expression was evaluated as
staining intensity. The statistical distribution of the HtrA1
variable was not investigated. HtrA1 levels were lower in patients
with less differentiated tumors, whereas they did not correlate
with clinical stage, myometrial invasion, age or survival.
Narkiewicz and colleagues (47) studied HtrA1, HtrA2 and HtrA3 mRNA
and protein by two semi-quantitative techniques, RT-PCR (mRNA) and
WB (protein), in 124 women; 88 with endometrial cancer and 36 with
normal endometrium. The statistical distribution of the HtrA1
variable was not examined. They documented significant differences
in HtrA1 expression between normal and cancer tissue, whereas
differences in FIGO stage, histological type or grade or menopausal
state were not significant.
Bowden and colleagues (48) assessed HtrA1 and HtrA3 mRNA and
protein by semi-quantitative PCR, IHC and WB in 33 women. HtrA1
mRNA was significantly reduced in tumors compared with normal
tissue, whereas protein levels were not significantly different.
Neither HtrA1 distribution nor its relationship with age or
menopausal state were explored.
Thyroid cancer
Zurawa-Janicka et al (49) assessed HtrA1 in tissue extracts from
40 subjects; 20 with normal thyroid and 20 with follicular (n=12)
or papillary (n=8) tumors using WB. Protein band intensities
assessed by densitometry yielded relative values. There were no
differences between healthy and cancer tissue, whereas slightly
higher HtrA1 protein levels were found in follicular than in
papillary cancer (P=0.045). Neither HtrA1 distribution nor its
relationship with age or gender were examined.
Ovarian cancer
Narkiewicz and co-workers (50) assessed HtrA1 mRNA and protein in 98
women with various types of ovarian tumors (20 benign, 7
borderline, 44 malignant, and 8 Krukenberg tumors) or with healthy
ovaries (n=19) using densitometry, which provided relative values
of HtrA1 expression. Significant mRNA downregulation was found in
malignant tumors (P<0.001), whereas the protein level did not
exhibit significant differences. Histological type and grade did
not display significant differences. Neither HtrA1 distribution nor
its relationship with age were examined.
Pleural mesothelioma
Baldi et al (9) examined the prognostic role of HtrA1
and EGFR (epidermal growth factor receptor) in 70 mesotheliomas
whose histological type was epithelioid (n=45), mixed (n=14) and
sarcomatoid (n=11). Their T status was T1 (n=4); T2 (n=13); T3
(n=23); T4 (n=4), and TX (n=26); their N status was N0 (n=27); N1
(n=3); N2 (n=14); and NX (n=26). The authors investigated HtrA1
expression by IHC and measured its level by an observational
semi-quantitative method. The statistical distribution of the HtrA1
variable was not examined. HtrA1 was not assessed in relation to
gender or age. Findings showed that the HtrA1 level was closely
related to survival, highlighting a role for HtrA1 as a marker,
especially for prognosis.
Lung cancer
The study by Esposito and colleagues (51) evaluated HtrA1 expression in 99
patients with primary lung tumors and metastasis using IHC. The
intensity of HtrA1 staining was evaluated by an observational
method using a score. There were 43 patients with SCC; 45 with
adenocarcinoma and 11 with other histological types. Clinical stage
was I in 12 patients II in 34, IIIa in 27, and IIIB in 26 patients.
Management was surgical in 72 and non-surgical in 27 patients. The
statistical distribution of the HtrA1 variable was not
investigated. The authors found that HtrA1 was underexpressed in
metastases compared with the primary tumor. Differences regarding
histological type, TNM or stage were not significant. HtrA1 was not
assessed in relation to gender or age.
Malignant melanoma
Baldi and co-workers (8) investigated HtrA1 expression by IHC
using tissue array. Staining intensity was evaluated by an
observational semi-quantitative approach. This was the first study
reporting clinical data of patients with malignant melanoma and
demonstrated higher HtrA1 levels in primary tumors compared with
metastases. The statistical distribution of HtrA1 was not
tested.
The laboratory approaches applied to detect HtrA1 in
a variety of tumors are reported in Table III. Most articles described in
vivo studies using a morphological approach and IHC, whereas
protein expression was quantified as staining intensity scored by
two raters. Often the results were not comparable due to the
different rating scales. WB was used to detect HtrA1 protein both
in tissue and cell extracts. Protein band intensity was assessed by
densitometry using two different but comparable softwares, Quantity
One (Bio-Rad Laboratories, Hercules, CA, USA) and 1Dscan EX 3.0
(Scanalytics, Rockville, MD, USA). In vitro analysis was
performed mainly to explore the role of HtrA1 in cell
proliferation, migration and invasion. In addition, Baldi et
al (8), Xia et al
(45) and Lehner et al
(11) investigated the molecules
that may be involved in the regulation of HtrA1 activity. The data
regarding HtrA1 as a potential biomarker in oncology are reported
in Table IV.
Discussion
A role for HtrA1 in cell proliferation has been
described in a small number of studies (8,52,53)
for conditions such as macular degeneration (52) and skeletal osteoarthritis (31). Its involvement in proliferation
processes has suggested a possible role as a tumor suppressor. Even
though HtrA1 has been investigated in a variety of tumors for more
than a decade, the comparison of findings is hampered by
differences in design as well as in the clinical and laboratory
detection methods used. Such lack of uniformity is clearly apparent
in the 15 studies reviewed above, most of which use mainly a
semi-quantitative method of analysis that does not provide absolute
values; this entails that any differences that are found can be
compared only within each study. In addition, most studies use
tissue samples from pathological files, which have been shown to be
useful to diagnose early pathological changes, but are unsuitable
for large-scale screening of high-risk populations. A broader
clinical application of such approaches would have to rely on the
technical ability of individual clinical pathology
laboratories.
HtrA1: a possible tissue biomarker
The present study highlighted a consistent finding
that was reported by all of the studies that tested this aspect,
i.e. that HtrA1 levels are higher in healthy or normal-looking
control tissue than in diseased tissue from patients with a variety
of tumors. The difference was statistically significant in
urothelial bladder (43),
esophageal (44,45), liver (10), and endometrial (47,48)
cancer, but not in ovarian cancer (50). The remaining studies, which involved
NB (42), breast (11), gastric (12), endometrial (46) and lung cancer (51), pleural mesothelioma (9) and malignant melanoma (8), did not investigate the issue.
The correlation of HtrA1 with histological
differentiation was inconsistent, since it was demonstrated in
esophageal (44) and endometrial
(46,48) cancer and hepatocellular carcinoma
(10), but not in breast (11), esophageal (45), and ovarian (50) cancer, whereas it was not assessed in
NB (42), bladder (43), gastric (12), and endometrial cancer (47), pleural mesothelioma (9) and malignant melanoma (8).
Assessment of the relationship of HtrA1 with TNM and
clinical stage also yielded conflicting data, since a clear
relationship was found in NB (42)
and esophageal cancer (44,45), but not in gastric (12), endometrial (46,47)
ovarian (50), lung (51) or liver (10) tumors.
A correlation with tumor size was found only for
breast cancer (11) and malignant
melanoma (8). In the two clinical
studies of malignant melanoma and lung carcinoma (8,51)
analysis of HtrA1 levels in primary and metastatic tumors
demonstrated lower levels in metastasis, suggesting a role for
HtrA1 as a possible growth regulatory factor in the complex
controlling cell growth in normal and transformed cells. Further
investigation by a single study (45) failed to find significant
differences. Additional studies are therefore required to clarify
the role of HtrA1 in the growth of normal and tumor tissue
cells.
The current literature does not, therefore, provide
conclusive data on the role of HtrA1 as a tumor marker, but
documents the need for further research.
HtrA1:a possible diagnostic
biomarker
A single study (43)
assessed HtrA1 as a potential diagnostic marker. The study used
ELISA, which provided a continuous numerical value of protein, and
documented a good performance of HtrA1 in urothelial cancer
diagnosis. Since these data were obtained in a small sample and
have not been supported by other studies in biological fluids, they
require validation in a larger sample.
HtrA1: a possible prognostic
biomarker
The present review found six longitudinal
studies.
Catalano and colleagues (12) investigated the chemo-responsiveness
of gastric cancer in relation to HtrA1 levels in terms of OS and
DFS, hypothesizing that the HtrA1 level may be used as predictor of
responsiveness to chemotherapy.
HtrA1 levels were also evaluated by Lehner et
al (11) in breast cancer in
relation to OS and DFS. They found that patients with higher HtrA1
levels had a better prognosis.
A longer OS was also measured in patients with
pleural mesothelioma showing higher HtrA1 levels (9).
Finally, the investigation of liver (10) and esophageal (45) cancer also documented a longer OS in
patients with a greater HtrA1 expression, whereas the difference
found in endome-trial cancer was not significant (46). These data suggest that HtrA1 may be
used as a predictor of OS.
Research implications
The current HtrA1 research does not conclusively
support its role as a tumor suppressor. Clinical investigations
sharing similar approaches, especially in terms of study design,
are needed to produce comparable data.
In the light of the findings reviewed above, HtrA1
research should focus on its role as a marker for early diagnosis
in selected patients; notably, establishing a diagnostic gold
standard would enable early diagnosis especially of tumors for
which a screening test is not available. It would also be
interesting to explore whether HtrA1 has a role in those tumors for
which a screening test is available but has suboptimal sensitivity,
such as colorectal cancer. Colorectal cancer ranks first in
incidence and second in mortality in Europe for both genders
(53), yet, HtrA1 has never been
investigated as a possible diagnostic and/or prognostic marker in
this tumor.
Recent advances in RNA sequencing, circulating DNA
methylation profiling, and glycoproteins may allow the development
of non-invasive diagnostic biomarkers for routine monitoring.
Future studies should combine different classes of circulating
biomarkers in large-scale investigations to improve the predictive
power of the individual biomarkers. The development of assays for
circulating biomarkers providing absolute and reproducible values
would help conduct large-scale multi-center investigations and
promote the use of circulating biomarkers in routine clinical
practice.
References
|
1
|
Zumbrunn J and Trueb B: Primary structure
of a putative serine protease specific for IGF-binding proteins.
FEBS Lett. 398:187–192. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Luca A, De Falco M, Severino A,
Campioni M, Santini D, Baldi F, Paggi MG and Baldi A: Distribution
of the serine protease HtrA1 in normal human tissues. J Histochem
Cytochem. 51:1279–1284. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oka C, Tsujimoto R, Kajikawa M,
Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A,
Kanda H, et al: HtrA1 serine protease inhibits signaling mediated
by TGF beta family proteins. Development. 131:1041–1053. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grau S, Baldi A, Bussani R, Tian X,
Stefanescu R, Przybylski M, Richards P, Jones SA, Shridhar V,
Clausen T, et al: Implications of the serine protease HtrA1 in
amyloid precursor protein processing. Proc Natl Acad Sci USA.
102:6021–6026. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D,
Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, et al: A variant of
the HTRA1 gene increases susceptibility to age-related macular
degeneration. Science. 314:992–993. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grau S, Richards PJ, Kerr B, Hughes C,
Caterson B, Williams AS, Junker U, Jones SA, Clausen T and Ehrmann
M: The role of human HtrA1 in arthritic disease. J Biol Chem.
281:6124–6129. 2006. View Article : Google Scholar
|
|
7
|
Marzioni D, Lorenzi T, Altobelli E,
Giannubilo SR, Paolinelli F, Tersigni C, Crescimanno C, Monsurrò V,
Tranquilli AL, Di Simone N, et al: Alterations of maternal plasma
HTRA1 level in preeclampsia complicated by IUGR. Placenta.
33:1036–1038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baldi A, De Luca A, Morini M, Battista T,
Felsani A, Baldi F, Catricalà C, Amantea A, Noonan DM, Albini A, et
al: The HtrA1 serine protease is down-regulated during human
melanoma progression and represses growth of metastatic melanoma
cells. Oncogene. 21:6684–6688. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baldi A, Mottolese M, Vincenzi B, Campioni
M, Mellone P, Di Marino M, di Crescenzo VG, Visca P, Menegozzo S,
Spugnini EP, et al: The serine protease HtrA1 is a novel prognostic
factor for human mesothelioma. Pharmacogenomics. 9:1069–1077. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu F, Jin L, Luo TP, Luo GH, Tan Y and
Qin XH: Serine protease HtrA1 expression in human hepatocellular
carcinoma. Hepatobiliary Panceat Dis Int. 9:508–512. 2010.
|
|
11
|
Lehner A, Magdolen V, Schuster T, Kotzsch
M, Kiechle M, Meindl A, Sweep FC, Span PN and Gross E:
Downregulation of serine protease HTRA1 is associated with poor
survival in breast cancer. PLoS One. 8:e603592013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Catalano V, Mellone P, d’Avino A, Shridhar
V, Staccioli MP, Graziano F, Giordani P, Rossi D, Baldelli AM,
Alessandroni P, et al: HtrA1, a potential predictor of response to
cisplatin-based combination chemotherapy in gastric cancer.
Histopathology. 58:669–678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chien J, Aletti G, Baldi A, Catalano V,
Muretto P, Keeney GL, Kalli KR, Staub J, Ehrmann M, Cliby WA, et
al: Serine protease HtrA1 modulates chemotherapy-induced
cytotoxicity. J Clin Invest. 116:1994–2004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moher D, Liberati A, Tetzlaff J and Altman
DG; PRISMA Group: Preferred reporting items for systematic reviews
and meta-analyses: the PRISMA statement. J Clin Epidemiol.
62:1006–1012. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou Y, Lin H, Zhu L, Liu Z, Hu F, Shi J,
Yang T, Shi X, Guo H, Tan X, et al: The inhibitory effect of IFN-γ
on protease HTRA1 expression in rheumatoid arthritis. J Immunol.
193:130–138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cipriani V, Leung HT, Plagnol V, Bunce C,
Khan JC, Shahid H, Moore AT, Harding SP, Bishop PN, Hayward C, et
al: Genome-wide association study of age-related macular
degeneration identifies associated variants in the
TNXB-FKBPL-NOTCH4 region of chromosome 6p21.3. Hum Mol Genet.
21:4138–4150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morrison MA, Silveira AC, Huynh N, Jun G,
Smith SE, Zacharaki F, Sato H, Loomis S, Andreoli MT, Adams SM, et
al: Systems biology-based analysis implicates a novel role for
vitamin D metabolism in the pathogenesis of age-related macular
degeneration. Hum Genomics. 5:538–568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Y, Bhangale TR, Fagerness J, Ripke S,
Thorleifsson G, Tan PL, Souied EH, Richardson AJ, Merriam JE,
Buitendijk GH, et al: Common variants near FRK/COL10A1 and VEGFA
are associated with advanced age-related macular degeneration. Hum
Mol Genet. 20:3699–3709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marzioni D, Quaranta A, Lorenzi T, Morroni
M, Crescimanno C, De Nictolis M, Toti P, Muzzonigro G, Baldi A, De
Luca A, et al: Expression pattern alterations of the serine
protease HtrA1 in normal human placental tissues and in gestational
trophoblastic diseases. Histol Histopathol. 24:1213–1222.
2009.PubMed/NCBI
|
|
20
|
Campioni M, Severino A, Manente L, Tuduce
IL, Toldo S, Caraglia M, Crispi S, Ehrmann M, He X, Maguire J, et
al: The serine protease HtrA1 specifically interacts and degrades
the tuberous sclerosis complex 2 protein. Mol Cancer Res.
8:1248–1260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Spugnini EP, Cardillo I, Fanciulli M,
Crispi S, Vincenzi B, Boccellino M, Quagliuolo L and Baldi A:
Electroporation as a strategy to promote HtrA1 gene uptake and
chemotherapy efficacy in a mouse model of mesothelioma. Front
Biosci (Elite Ed). 5:974–981. 2013.
|
|
22
|
Zurawa-Janicka D, Kobiela J, Stefaniak T,
Wozniak A, Narkiewicz J, Wozniak M, Limon J and Lipinska B: Changes
in expression of serine proteases HtrA1 and HtrA2 during
estrogen-induced oxidative stress and nephrocarcinogenesis in male
Syrian hamster. Acta Biochim Pol. 55:9–19. 2008.PubMed/NCBI
|
|
23
|
Spugnini EP, Cardillo I, Verdina A, Crispi
S, Saviozzi S, Calogero R, Nebbioso A, Altucci L, Cortese G, Galati
R, et al: Piroxicam and cisplatin in a mouse model of peritoneal
mesothelioma. Clinic Cancer Res. 12:6133–6143. 2006. View Article : Google Scholar
|
|
24
|
Sahasrabuddhe NA, Barbhuiya MA, Bhunia S,
Subbannayya T, Gowda H, Advani J, Shrivastav BR, Navani S, Leal P,
Roa JC, et al: Identification of prosaposin and transgelin as
potential biomarkers for gallbladder cancer using quantitative
proteomics. Biochem Biophys Res Commun. 446:863–869. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tatenhorst L, Senner V, Püttmann S and
Paulus W: Regulators of G-protein signaling 3 and 4 (RGS3, RGS4)
are associated with glioma cell motility. J Neuropathol Exp Neurol.
63:210–222. 2004.PubMed/NCBI
|
|
26
|
Wang N, Eckert KA, Zomorrodi AR, Xin P,
Pan W, Shearer DA, Weisz J, Maranus CD and Clawson GA:
Down-regulation of HtrA1 activates the epithelial-mesenchymal
transition and ATM DNA damage response pathways. PLoS One.
7:e394462012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He X, Ota T, Liu P, Su C, Chien J and
Shridhar V: Downregulation of HtrA1 promotes resistance to anoikis
and peritoneal dissemination of ovarian cancer cells. Cancer Res.
70:3109–3118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clawson GA, Bui V, Xin P, Wang N and Pan
W: Intracellular localization of the tumor suppressor HtrA1/Prss11
and its association with HPV16 E6 and E7 proteins. J Cell Biochem.
105:81–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chien J, Staub J, Hu SI, Erickson-Johnson
MR, Couch FJ, Smith DI, Crowl RM, Kaufmann SH and Shridhar V: A
candidate tumor suppressor HtrA1 is downregulated in ovarian
cancer. Oncogene. 23:1636–1644. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baldi A, Battista T, De Luca A, Santini D,
Rossiello L, Baldi F, Natali PG, Lombardi D, Picardo M, Felsani A,
et al: Identification of genes down-regulated during melanoma
progression: a cDNA array study. Exp Dermatol. 12:213–218. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tiaden AN and Richards PJ: The emerging
roles of HTRA1 in musculoskeletal disease. Am J Pathol.
182:1482–1488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Skorko-Glonek J, Zurawa-Janicka D, Koper
T, Jarzab M, Figaj D, Glaza P and Lipinska B: HtrA protease family
as therapeutic targets. Curr Pharm Des. 19:977–1009. 2013.
View Article : Google Scholar
|
|
33
|
Singh N, Kuppili RR and Bose K: The
structural basis of mode of activation and functional diversity: a
case study with HtrA family of serine proteases. Arch Biochem
Biophys. 516:85–96. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zurawa-Janicka D, Skorko-Glonek J and
Lipinska B: HtrA proteins as targets in therapy of cancer and other
diseases. Expert Opin Ther Targets. 14:665–679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
MacDonald TJ, Pollack IF, Okada H,
Bhattacharya S and Lyons-Weiler J: Progression-associated genes in
astrocytoma identified by novel microarray gene expression data
reanalysis. Methods Mol Biol. 377:203–222. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chien J, Campioni M, Shridhar V and Baldi
A: HtrA serine proteases as potential therapeutic targets in
cancer. Curr Cancer Drug Targets. 9:451–468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu Y, Jiang Z, Zhang Z, Sun N, Zhang M,
Xie J, Li T, Hou Y and Wu D: HtrA1 downregulation induces cisplatin
resistance in lung adenocarcinoma by promoting cancer stem
cell-like properties. J Cell Biochem. 115:1112–1121. 2014.
View Article : Google Scholar
|
|
38
|
Neuhausen SL, Brummel S, Ding YC, et al:
Genetic variation in IGF2 and HTRA1 and breast cancer risk among
BRCA1 and BRCA2 carriers. Cancer Epidemiol Biomarkers Prev.
20:1690–1702. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He X, Khurana A, Maguire JL, Chien J and
Shridhar: HtrA1 sensitizes ovarian cancer cells to
cisplatin-induced cytotoxicity by targeting XIAP for degradation.
Int J Cancer. 130:1029–1035. 2012. View Article : Google Scholar
|
|
40
|
Folgueira MA, Carraro DM, Brentani H,
Patrão DF, Barbosa EM, Netto MM, Caldeira JR, Katayama ML, Soares
FA, Oliveira CT, et al: Gene expression profile associated with
response to doxorubicin-based therapy in breast cancer. Clin Cancer
Res. 11:7434–7443. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barros Filho MC, Katayama ML, Brentani H,
Abreu AP, Barbosa EM, Oliveira CT, Góes JC, Brentani MM and
Folgueira MA: Gene trio signatures as molecular markers to predict
response to doxorubicin cyclophosphamide neoadjuvant chemotherapy
in breast cancer patients. Braz J Med Biol Res. 43:1225–1231. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
D’Angelo V, Pecoraro G, Indolfi P,
Iannotta A, Donofrio V, Errico ME, Indolfi C, Ramaglia M, Lombardi
A, Di Martino M, et al: Expression and localization of serine
protease Htra1 in neuroblastoma: correlation with cellular
differentiation grade. J Neurooncol. 117:287–294. 2014. View Article : Google Scholar
|
|
43
|
Lorenzi T, Lorenzi M, Altobelli E,
Marzioni D, Mensà E, Quaranta A, Paolinelli F, Morroni M,
Mazzucchelli R, De Luca A, et al: HtrA1 in human urothelial bladder
cancer: a secreted protein and a potential novel biomarker. Int J
Cancer. 133:2650–2661. 2013.PubMed/NCBI
|
|
44
|
Yu Y, Shao W, Hu Y, Zhang J, Song H and
Zhu ZH: HtrA1 expression associated with the occurrence and
development of esophageal cancer. World J Surg Oncol. 10:1792012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xia J, Wang F, Wang L and Fan Q: Elevated
serine protease HtrA1 inhibits cell proliferation, reduces
invasion, and induces apoptosis in esophageal squamous cell
carcinoma by blocking the nuclear factor-κB signaling pathway.
Tumor Biol. 34:317–328. 2013. View Article : Google Scholar
|
|
46
|
Mullany SA, Moslemi-Kebria M, Rattan R,
Khurana A, Clayton A, Ota T, Mariani A, Podratz KC, Chien J and
Shridhar V: Expression and functional significance of HtrA1 loss in
endometrial cancer. Clin Cancer Res. 17:427–436. 2011. View Article : Google Scholar :
|
|
47
|
Narkiewicz J, Lapinska-Szumczyk S,
Zurawa-Janicka D, Skorko-Glonek J, Emerich J and Lipinska B:
Expression of human HtrA1, HtrA2, HtrA3 and TGF-beta1 genes in
primary endometrial cancer. Oncol Rep. 21:1529–1537.
2009.PubMed/NCBI
|
|
48
|
Bowden MA, Di Nezza-Cossens LA, Jobling T,
Salamonsen LA and Nie G: Serine proteases HTRA1 and HTRA3 are
down-regulated with increasing grades of human endometrial cancer.
Gynecol Oncol. 103:253–260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zurawa-Janicka D, Kobiela J, Galczynska N,
Stefaniak T, Lipinska B, Lachinski A, Skorko-Glonek J, Narkiewicz
J, Proczko-Markuszewska M and Sledzinski Z: Changes in expression
of human serine protease HtrA1, HtrA2 and HtrA3 genes in benign and
malignant thyroid tumors. Oncol Rep. 28:1838–1844. 2012.PubMed/NCBI
|
|
50
|
Narkiewicz J, Klasa-Mazurkiewicz D,
Zurawa-Janicka D, Skorko-Glonek J, Emerich J and Lipinska B:
Changes in mRNA and protein levels of human HtrA1, HtrA2 and HtrA3
in ovarian cancer. Clin Biochem. 41:561–569. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Esposito V, Campioni M, De Luca A,
Spugnini EP, Baldi F, Cassandro R, Mancini A, Vincenzi B, Groeger
A, Caputi M, et al: Analysis of HtrA1 serine protease expression in
human lung cancer. Anticancer Res. 26:3455–3459. 2006.PubMed/NCBI
|
|
52
|
Horie-Inoue K and Inoue S: Genomic aspects
of age-related macular degeneration. Biochem Biophysis Res Commun.
452:263–275. 2014. View Article : Google Scholar
|
|
53
|
Altobelli E, Lattanzi A, Paduano R,
Varassi G and di Orio F: Colorectal cancer prevention in Europe:
burden of disease and status of screening programs. Prev Med.
62:132–141. 2014. View Article : Google Scholar : PubMed/NCBI
|