Introduction
Hepatocellular carcinoma (HCC) is the third most
frequent cause of cancer-related death worldwide (1,2). There
are several treatment modalities for HCC, including surgical
resection, radiofrequency ablation, transcatheter arterial
chemoembolization and percutaneous ethanol injection therapy.
Recent advances in these treatment modalities have improved the
prognosis of HCC, yet prognosis for advanced HCC remains poor
(1). Several studies investigated
the factors associated with refractory HCC have shown that CD133
and epithelial cell adhesion molecule (EpCAM) are related to the
poor prognosis (3–5).
Stemness is an essential characteristic of stem
cells to renew themselves and differentiate with multipotency.
EpCAM is a cell surface marker expressed on human hepatic stem
cells and is a known marker to identify cancer stem cells (CSCs) in
HCC (6). EpCAM is related to
tumorigenesis and metastasis, and its expression is a prognostic
factor for HCC (3–5). EpCAM is a downstream effector of the
Notch signaling cascade. Activation of Notch signaling enhances
cellular stemness and epithelial-mesenchymal transition (EMT) by
inducing EpCAM expression in both pancreatic cancer (7) and HCC (8). Moreover, other investigators have
reported that EpCAM plays an important role in EMT (9,10).
Cancer progression is related to genetic changes
within cancer cells as well as in the microenvironment. Stromal
cells in the cancer microenvironment facilitate the development of
cancer EMT (11). Cancer cells that
undergo EMT also acquire a stem cell-like phenotype (12), suggesting that cancer
cell-microenvironment interactions are important in cancer
progression. In various types of cancers, such as intrahepatic
cholangiocarcinoma (13),
oligodendroglioma (14), pancreatic
cancer (15), in vitro
co-culture assays of cancer cell lines and cells in the cancer
microenvironment increases EMT. In the present study, we
hypothesized that the microenvironment associated with HCC enhances
EMT. Hepatic stellate cells (HSCs) are liver-specific mesenchymal
cells located in perisinusoidal and portal areas. HSCs play an
important role in the stem cell niche for hepatic progenitor cells
and hepatocytes. In addition, HSCs are known to present
histopathologically among HCC tissue (16), and are thought to make a niche for
hepatic cancer cells. Therefore, in the present study, we
investigated the interaction between HSCs and HCC cells.
Materials and methods
Cell lines and culture
The human HCC cell lines HepG2, Hep3B, HuH-7 and
PLC/PRF/5 were obtained from American Type Culture Collection
(ATCC; Manassas, VA, USA). Immortalized human HSC cells (TWNT-1)
were a generous gift from Dr Naoya Kobayashi from the Department of
Gastroenterological Surgery, Okayama University School of Medicine.
Cells were maintained in high glucose Dulbecco's modified Eagle's
medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10%
heat-inactivated fetal bovine serum (FBS), 1% non-essential amino
acids, penicillin/streptomycin solution (both from Sigma-Aldrich,
St. Louis, MO, USA). Cells were cultured at 37°C in an atmosphere
of 5% CO2 and 95% air. The cells were treated under
restricted serum conditions with 0.5% dialyzed FBS for 24 h before
the experiment when necessary.
Direct co-culture of hepatic cancer cells
and HSCs
HCC cell lines [400,000 cells/well (HepG2), 200,000
cells/well (Hep3B, HuH-7 and PLC/PRF/5)] and TWNT-1 (50,000
cells/well) were seeded in 6-well culture plates (353046; Corning,
Corning, NY, USA) in DMEM supplemented with 0.5% dialyzed FBS and
1% supplements as previously described, and incubated for 3 days.
If required, HSCs were pre-treated with mitomycin C before they
were used for assays in order to inhibit self-proliferation. After
this, cells were seeded and cultured in this manner in case of
direct co-culture unless otherwise specified.
Indirect co-culture of hepatic cancer
cells and HSCs
HCC cell lines [400,000 cells/well (HepG2), 200,000
cells/well (Hep3B, HuH-7 and PLC/PRF/5)] were seeded in 6-well
culture plates in DMEM supplemented with 0.5% dialyzed FBS and 1%
supplements as previously described. TWNT-1 (50,000 cells/well)
were seeded into the Cell Culture Insert™ of 1.0-µm pore
size (353102; Corning) in the same medium as used for HCC cells,
inserted into the 6-well plates where HCC cells were seeded, and
incubated for 3 days. After this, cells were seeded and cultured in
this manner in case of indirect co-culture unless otherwise
specified.
Immunoblot analysis
HCC cells were seeded in 6-well culture plates and
uni-cultured and indirectly co-cultured with TWNT-1, and grown to
confluence. The HCC cells were washed twice with cold Dulbecco's
phosphate-buffered saline (DPBS) and lysed in 100 µl of
sample buffer [100 mM Tris-HCl (pH 6.8), 10% glycerol, 4% sodium
dodecyl sulfate (SDS), 1% bromophenol blue and 10%
β-mercaptoethanol]. The samples were resolved by SDS-polyacrylamide
gel electrophoresis and transferred to a polyvinylidene difluoride
(PVDF) membrane (Bio-Rad, Hercules, CA, USA). The membranes were
blocked using PVDF blocking reagent Can Get Signal™ (Toyobo, Osaka,
Japan) for 1 h. The membranes were then incubated with antibodies
against cleaved Notch [#2421; Cell Signaling Technology (CST),
Danvers, MA, USA], Hes1 (ab49170; Abcam, Cambridge, UK), Twist1
[sc-6070; Santa Cruz Biotechnology (SCBT), Dallas, TX, USA],
E-cadherin (#3195), N-cadherin (#13116), EpCAM (#2929) and β-actin
(#4967S) (all from CST) for 1 h at room temperature. The membranes
were washed 3 times with TBS-T and probed with horseradish
peroxidase-conjugated secondary antibody (#7074S; CST) before being
developed with an ECL blotting detection system (Amersham
Biosciences, Piscataway, NJ, USA) using enhanced
chemiluminescence.
Gene silencing of TGF-β and HB-EGF with
small interfering RNA (siRNA)
TWNT-1 was transfected with scrambled negative
control siRNA and TGF-β1/2/3, and HB-EGF siRNA, respectively
(sc-37007, sc-44146 and sc-39420; SCBT). siRNAs were transfected
into cells using a siRNA transfection reagent and siRNA
transfection medium (sc-45064; SCBT). Cells were seeded at 200,000
cells/well in 6-well culture plates and incubated for 24 h before
the 5 h transfection with scrambled negative control, TGF-β1/2/3
and HB-EGF siRNA, and then used for assays after additional
incubation for 48 h.
MTT assay
Cell proliferation ability was evaluated with the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. HCC cell lines were seeded at 40,000 cells/well in 500
µl of DMEM supplemented with 1% dialyzed FBS in 24-well
culture plates (353047; Becton-Dickinson, Franklin Lakes, NJ, USA).
TWNT-1 (30,000 cells/well) were seeded into the Cell Culture
Insert™ of 1.0-µm pore size (353104; Corning) in the same
medium as used for HCC cells, and plated into the 24-well plates
where the HCC cells were seeded. After 48 h of quiescence, 50
µl of MTT (5 mg/ml in PBS) was added to each well, and the
wells were incubated for an additional 3.5 h at 37°C. The
purple-blue MTT formazan precipitate was dissolved in 500 µl
of dimethylsulfoxide (Sigma-Aldrich) and applied in 100 µl
volumes in a 96-well culture plate in the fourth replicate. The
activity of mitochondria, used as a measure of cellular growth and
viability, was evaluated by measuring optical density at 570 nm
with a microplate reader (Model 680 microplate reader;
Bio-Rad).
Migration assay
Cell migration ability was evaluated by an in
vitro wound healing assay. HCC cell lines (HepG2, Hep3B, HuH-7
and PLC/PRF/5) were seeded at 500,000, 600,000, 200,000 and 600,000
cells/well, respectively, in 6-well culture plates then
uni-cultured, directly and indirectly co-cultured with TWNT-1
(50,000/well) in DMEM supplemented with 10% FBS. TWNT-1 was
pre-treated with mitomycin C before use in the direct co-culture
assays to inhibit self-proliferation. After cells grew to
confluence, the cell monolayer was mechanically scratched with a
sterile 200 µl pipette tip and incubation was continued for
an additional 48 h in DMEM medium supplemented with 0.5% dialyzed
FBS. Images were captured at 0, 24 and 48 h, and the distance
traveled by the cells at the acellular front was measured in 3
places.
Flow cytometric analysis
Cells were seeded in 6-well culture plates and
uni-cultured, directly or indirectly co-cultured with HSCs in DMEM
medium supplemented with 0.5% dialyzed FBS for 3 days. They were
subsequently washed twice with DPBS, collected and re-suspended in
100 µl of S-PBS (DPBS supplemented with 2% FBS and 10 mM
HEPES buffer saline solution). Vio-blue-anti-CD44,
PE-vio770-anti-CD133, APC-anti-EpCAM, APC-vio770-anti-E-cadherin
(130-104-270, 130-104-155, 130-091-254 and 130-101-095; Miltenyi
Biotechnology, Bergisch Gladbach, Germany), FITC-anti-CD90
(IM1839U; Beckman Coulter, Brea, CA, USA), and PE-anti-N-cadherin
antibodies (350806; BioLegend, San Diego, CA, USA) were added, and
cells were incubated on ice for 15 min in the dark. After washing
once, cells were re-suspended in 500 µl of S-PBS
supplemented with 5 µl of propidium iodide (PI) solution
(P378; Dojindo, Kumamoto, Japan) and analyzed using a flow
cytometer (MACSQuant, Miltenyi Biotechnology).
Statistical analysis
The results are expressed as mean ± SD. Experiments
were performed at least 3 times. Differences between the groups
were evaluated by the Student's t-test. A p-value <0.05 was
considered to indicate a statistically significant result.
Results
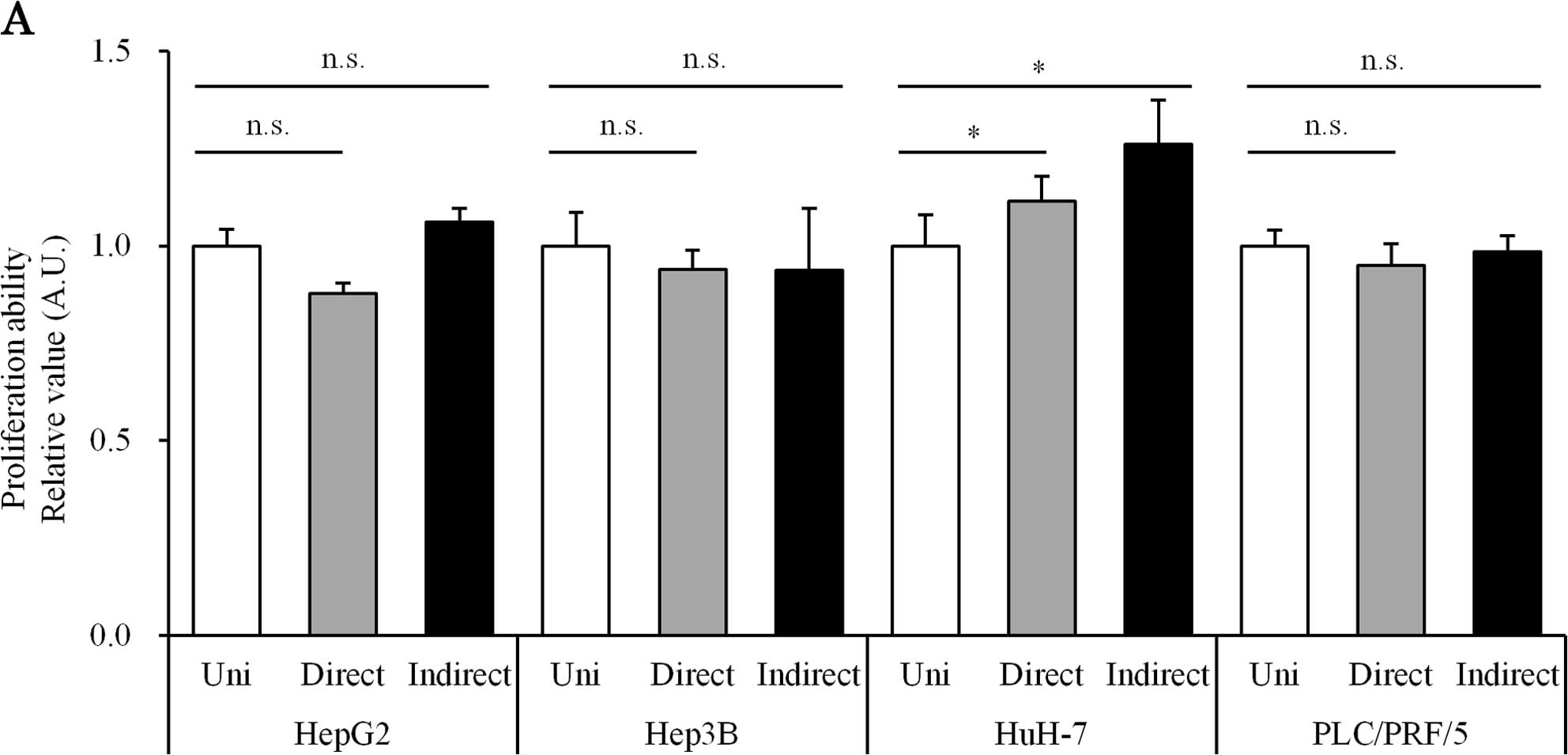
Direct or indirect co-culture with HSCs
does not increase proliferation in HCC cell lines
To assess the biological effect of co-culture with
HSCs, the proliferative activities of HCC cell lines were assessed
by flow cytometry. This technique allowed us to evaluate the number
of viable HCC cells in the mixture. The proliferation ability of
the co-culture group was not significantly different from that of
the uni-culture group (Fig.
1A).
Direct and indirect co-culture with HSCs
promotes migration in HCC cell lines
We evaluated the migration ability of the uni- and
co-culture groups using an in vitro wound healing assay. The
migration activity under co-culture conditions was higher than that
under uni-culture condition in all four HCC cell lines (Fig. 1B). This effect was observed in both
direct and indirect co-culture conditions (Fig. 1B).
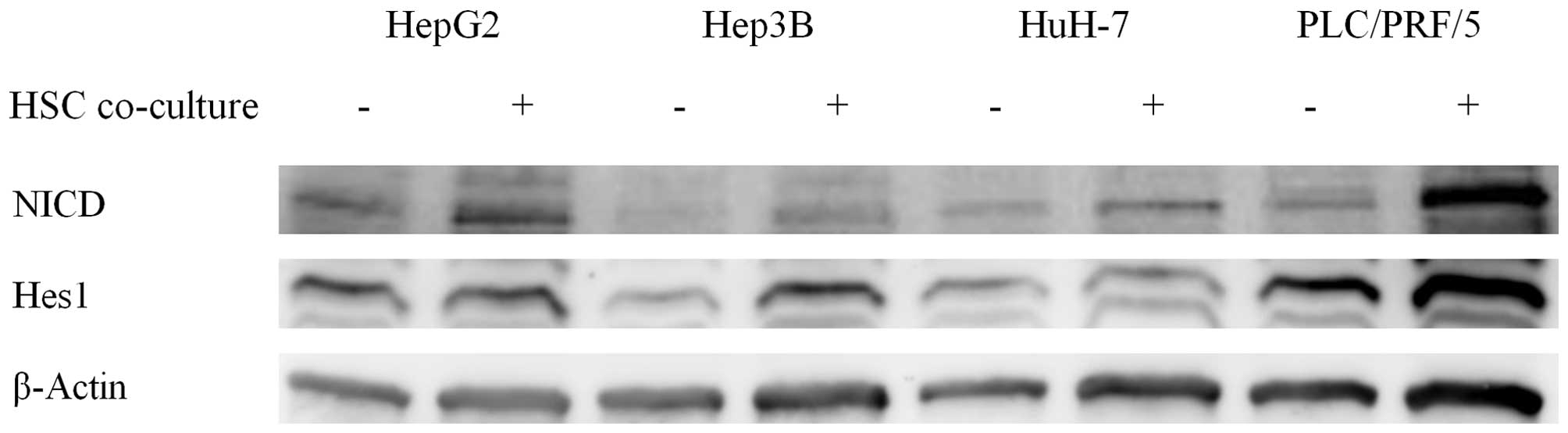
Indirect co-culture with HSCs upregulates
Notch signaling in HCC cell lines
As cell migration ability was increased by
co-culture with HSCs, we hypothesized that the co-culture condition
enhanced EMT in HCC cell lines. The Notch signaling pathway exists
upstream of EMT and regulates it. Therefore, we evaluated the
expression of proteins in the Notch signaling pathway by
immunoblotting. Notch intracellular domain (NICD) and Hes1
expression were increased in the co-culture group, indicating
activation of the Notch signaling pathway (Fig. 2).
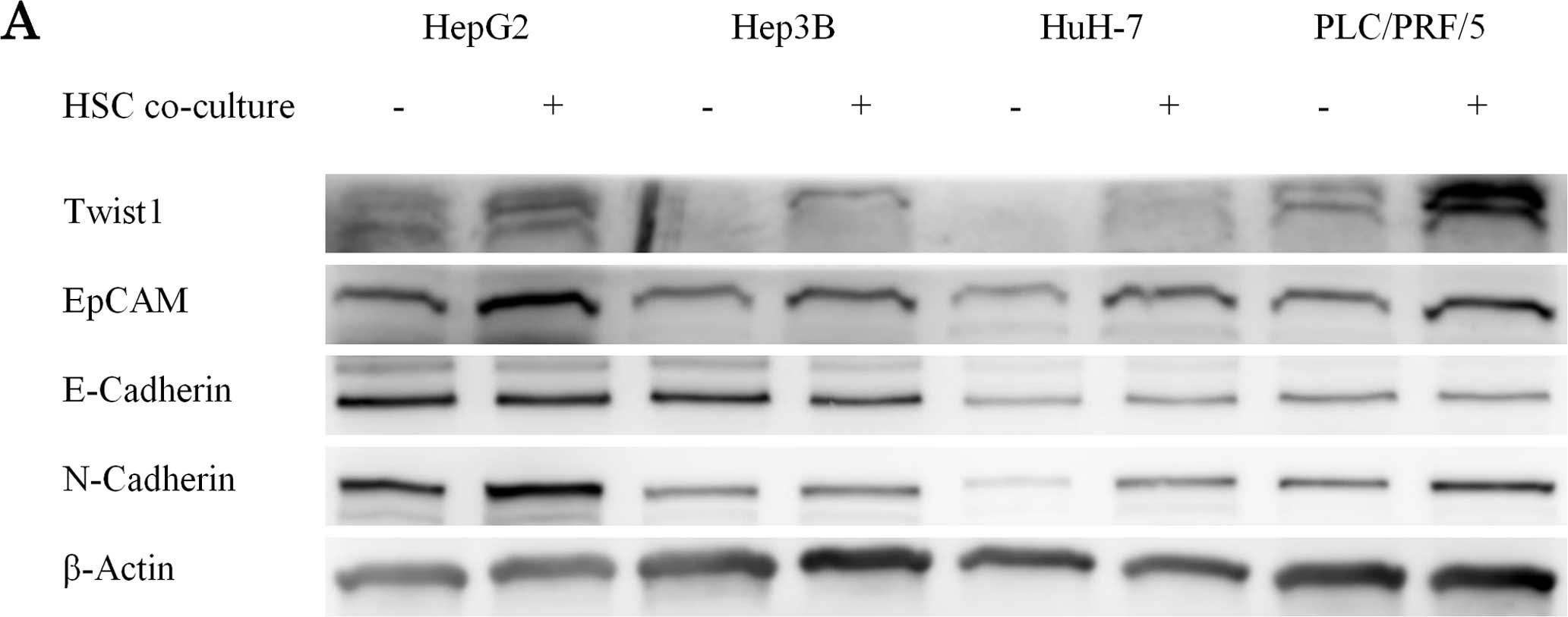
Indirect co-culture with HSCs promotes
EMT in HCC cell lines
It is known that the expression of EpCAM and Twist
are increased in EMT (17,18). We evaluated the expression of the
EMT markers Twist1, E-cadherin, N-cadherin and EpCAM by
immunoblotting in the uni- and co-culture groups. Both EpCAM and
Twist1 expression were increased in the co-culture group (Fig. 3A).
Direct and indirect co-culture with HSCs
increases the proportion of EpCAM-positive HCC cells
We evaluated the expression of E-cadherin and
N-cadherin, both of which are associated with EMT, by
immunoblotting in the uni- and co-culture groups. The co-culture
reduced E-cadherin expression in HepG2 and Hep3B, and induced
N-cadherin expression in HepG2, Hep3B, HuH-7 and PLC/PRF/5. As we
suspected that only a portion of the cells changed their EMT-marker
expression, we evaluated the expressions of EpCAM, E-cadherin and
N-cadherin by flow cytometry in both groups. In the co-culture
group, we found that the ratio of EpCAM-positive cells increased
(Fig. 3B). Furthermore, the
percentage of low E-cadherin and high N-cadherin cells increased,
suggesting the progression of EMT in the co-culture group (Fig. 3C).
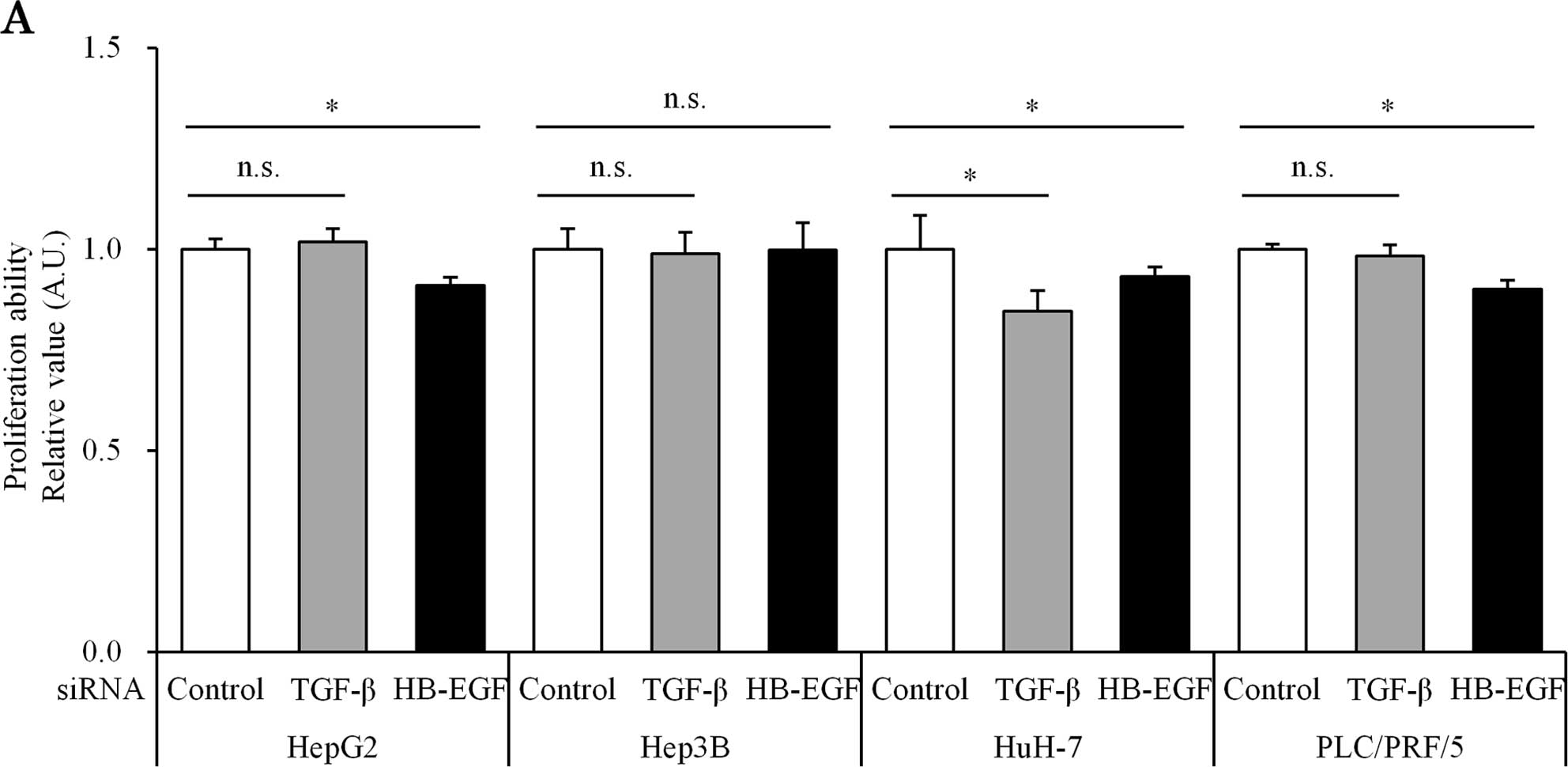
Suppression of TGF-β and HB-EGF inhibits
migration, Notch signaling
Enhanced migration activity and EMT progression were
observed in both direct and indirect co-culture groups, changes
that we suspected were caused by liquid factors. With the aim of
reducing the liquid factors secreted from HSCs, such as TGF-β and
HB-EGF, we treated HSCs with siRNA against these two molecules.
The siRNA against TGF-β and HB-EGF successfully
suppressed TGF-β (51.7±20.2% inhibition) and HB-EGF (69.2±12.3%
inhibition) expression in HSC cell lines, respectively. Both TGF-β
and HB-EGF siRNA had a minimal effect on cell proliferative
activity in HCC cell lines co-cultured with HSCs (Fig. 4A). However, both TGF-β and HB-EGF
siRNA reduced the cell migration potential of HCC cell lines under
co-culture with HSCs (Fig. 4B).
The expression of proteins in the Notch signaling
pathway was analyzed by immunoblotting. Migration activity, and
expression of proteins associated with the Notch signaling pathway
were decreased in HCC cells treated with siRNA against TGF-β and
HB-EGF compared to the control (Fig.
4C).
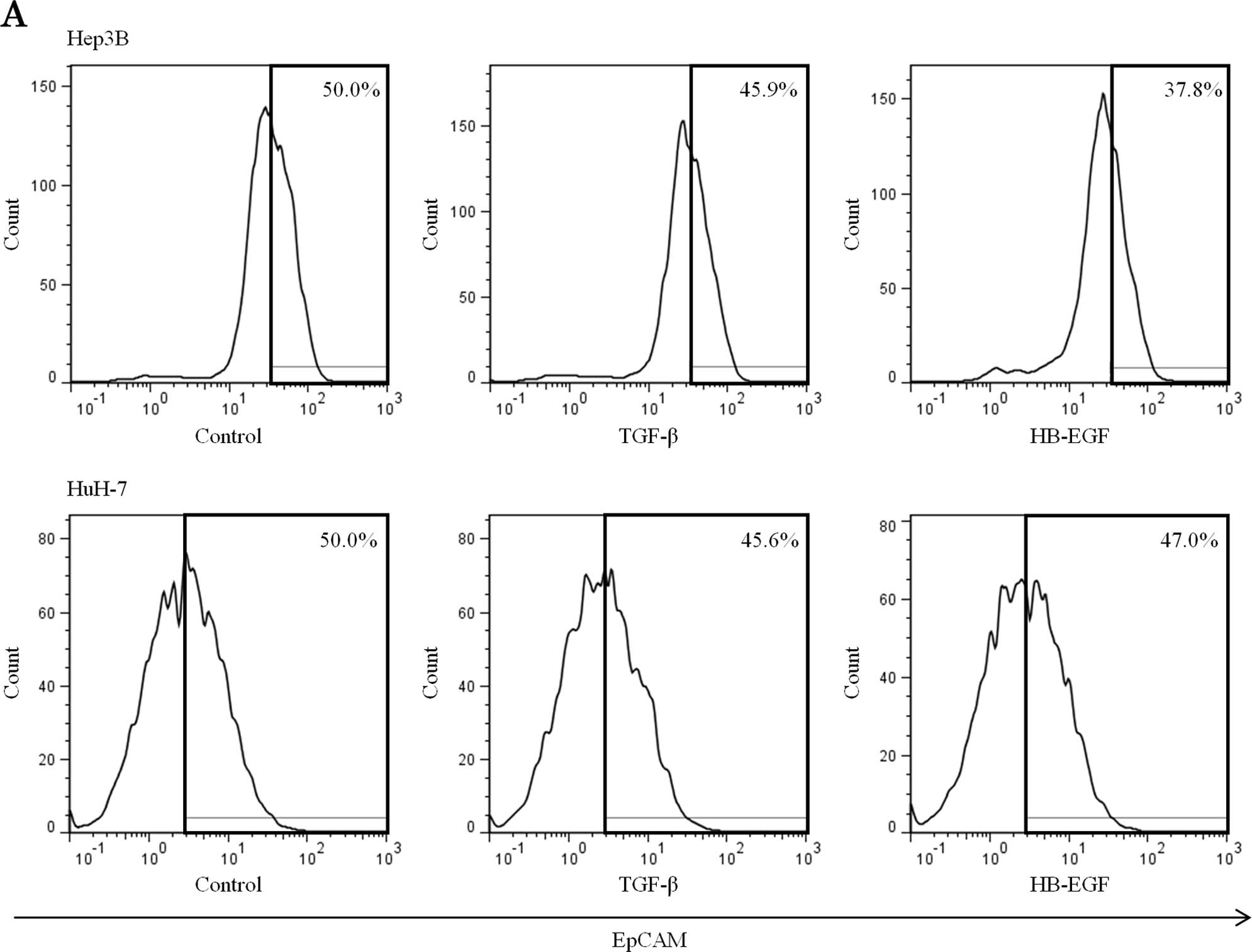
Suppression of TGF-β and HB-EGF reduces
EpCAM-positive HCC cells and inhibits EMT
We performed flow cytometry to evaluate EMT
capability and stemness of HCC cell lines co-cultured with HSCs
treated with siRNA against TGF-β and HB-EGF. Both TGF-β and HB-EGF
siRNA decreased the population of EpCAM-positive HCC cells
(Fig. 5A). The population of HCC
cells expressing N-cadherin but lacking E-cadherin occurring in EMT
was decreased (Fig. 5B).
Discussion
Cancerous tumors consist of heterogeneous cancer and
stromal cells. The stromal cell-associated cancer microenvironment
is known to play critical roles in cancer growth and progression
(11,19). In the present study, we demonstrated
that HSCs induce EMT in HCC cells. HSCs were chosen among various
hepatic stromal cells as they secrete various growth factors and
directly affect hepatocytes (20).
The immunohistochemical analyses revealed that HSCs were present in
the stroma of HCC (16).
Furthermore, the activation of HSCs in the medium promoted
tumorigenicity of HCC (21). It is
also reported that activated peritumoral HSCs are connected to high
and early tumor recurrence, and increased death rate (22). Collectively, the evidence suggests
that HSCs contribute to HCC progression.
We first assessed the biological impact on HCC cells
under co-culture conditions with HSC (Fig. 1). We observed that the co-culture
did not affect the proliferation of HCC cells (Fig. 1A), while the migration ability of
HCC cells was significantly enhanced (Fig. 1B). This effect was observed in both
direct and indirect co-culture experiments, suggesting that this
effect was induced by soluble factors.
It is reported that glioblastoma and
cholangiocarcinoma cells with their respective astrocytes enhance
EMT and stemness in cancer cells (13,14).
Pancreatic cancer cell lines are also reported to enhance EMT when
cultured with pancreatic stellate cells (15). The growing body of evidence
indicates that the stromal microenvironment contributes to cancer
progression. In accordance with these studies, the HSC-related
microenvironment induced EMT in HCC cells in the present study.
EMT plays an important role in cancer progression as
well as normal embryogenesis (23).
The cancer microenvironment contributes to cancer progression and
induces the occurrence of EMT in cancer cells (11,24).
As the migration ability is closely associated with EMT, we
conducted an immunoblot analysis for EMT-related molecules
(Figs. 2 and 3A). The co-culture condition decreased the
expression of E-cadherin in HCC cells and increased the expression
of N-cadherin. Notably, the co-culture condition also induced the
expression of Twist, a strong EMT inducer and EpCAM in HCC cells
(Fig. 3A).
Our previous study demonstrated that the expression
of EpCAM is suppressed by the expression of runt-related
transcription factor 3 (RUNX3) and that the Notch signaling pathway
is reduced by RUNX3 expression (8).
This evidence suggests that EpCAM expression and Notch signaling is
closely related. We subsequently assessed the effect of co-culture
on Notch signaling (Fig. 2).
Co-culture conditions activated a downstream signaling molecule of
Notch in HCC cells, as expected (Fig.
2).
In order to analyze the population of EMT-induced
cells, flow cytometric analysis was conducted, which revealed that
the co-culture increased the ratio of EpCAM-positive cells in Hep3B
and HuH-7 (Fig. 3B). The population
of cells with low expression of E-cadherin and high expression of
N-cadherin was increased in co-culture, compared to the uni-culture
condition (Fig. 3C). These results
indicated that the co-culture with HSC activated the Notch
signaling pathway and induced EMT in HCC cells. In these
experiments, both direct and indirect co-culture condition
demonstrated similar results, indicating that the Notch signal
activation and EMT were induced by soluble factors.
A number of soluble mitogens are secreted by HSCs
(20). As TGF-β and HB-EGF are
strong EMT inducers, we performed gene silencing experiments using
siRNAs against them. These siRNA-treated HSCs were cultured with
HCC, and their proliferation and migration abilities were assessed
(Fig. 4). The siRNAs successfully
silenced the expression of TGF-β and HB-EGF, resulting in decreased
cell migration ability in the co-culture (Fig. 4B).
Flow cytometric analysis confirmed that the siRNA
also reduced EMT in HCC cells under the co-culture with HSCs
(Fig. 5). The results indicate that
the interactions between HSCs and HCC cells were mainly induced by
the soluble factors secreted by HSCs.
The effect of HSC co-culture was relatively weak in
the immunoblot analysis, presumably as a result of the low
concentration of the soluble factors. The Cell Culture Insert™
allowed us to assess the effect of soluble factors in the
co-culture system while excluding the effect of cell-cell contact.
The limitation of this co-culture system, however, is the
relatively low concentration of soluble factors. The co-culture
system required at least 3 ml of medium, which generates a low
concentration of the soluble factors produced by 5×104
HSC cells.
Another limitation of the present study is the use
of immortalized HSCs. HSC cell line TWNT-1 may behave differently
from natural HSCs, although TWNT-1 is known to have features
similar to those exhibited by activated HSCs (25). This may be an advantage for
elucidating the mechanism of HCC progression, since the active form
of HSC contributes to it (21,26).
Further studies are necessary to elucidate the role of HSC in
vivo.
In advanced and metastatic HCC, the conventional
treatment modality cannot achieve a curative effect. EMT and cancer
stemness contributed to the poor prognosis in HCC (3–5,12,27).
The activation of Notch signaling contributes to both EMT and
stemness. Our results showed that HSCs activate the Notch signaling
pathway and induce EMT, which indicates that HSCs may also activate
CSCs. However, EpCAM-positive HCC cells were shown to have stem
cell properties such as self-renewal, differentiation and
tumor-initiating ability (6). It
was also reported that higher EpCAM expression was related to the
development of chemoresistance in HCC (28). Taken together, therefore, EpCAM has
become recognized as a CSC marker in HCC (6,28). The
interaction between HSCs and HCC cells may induce cancer
progression by activating EMT and stemness. CSCs are resistant to
the conventional therapeutic modality for HCC (29). A number of studies have demonstrated
that high expression levels of CSC markers are related to the poor
prognosis of HCC (3–5). Others have uncovered that EpCAM
expression is also associated with poor prognosis (3–5). Thus,
the interactions between HSC and HCC cells could be a therapeutic
target for HCC. Further studies are necessary, however, to
elucidate the effects induced by HSCs, particularly on CSCs.
In conclusion, we evaluated the effect of the cancer
microenvironment on HCC by in vitro co-culture assay with
HSCs. TGF-β and HB-EGF secreted by HSCs were shown to promote the
acquirement of EMT and a stem cell phenotype by HCCs. TGF-β and
HB-EGF could therefore be promising new therapeutic targets for
CSC-based therapy.
Acknowledgments
The authors wish to thank Dr Naoya Kobayashi for his
generous gift of immortalized human HSC cells (TWNT-1).
References
|
1
|
European Association For The Study Of The
Liver; European Organisation For Research And Treatment Of Cancer:
EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 56:908–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Venook AP, Papandreou C, Furuse J and de
Guevara LL: The incidence and epidemiology of hepatocellular
carcinoma: A global and regional perspective. Oncologist. 15(Suppl
4): S5–S13. 2010. View Article : Google Scholar
|
|
3
|
Bae JS, Noh SJ, Jang KY, Park HS, Chung
MJ, Park CK and Moon WS: Expression and role of epithelial cell
adhesion molecule in dysplastic nodule and hepatocellular
carcinoma. Int J Oncol. 41:2150–2158. 2012.PubMed/NCBI
|
|
4
|
Schulze K, Gasch C, Staufer K, Nashan B,
Lohse AW, Pantel K, Riethdorf S and Wege H: Presence of
EpCAM-positive circulating tumor cells as biomarker for systemic
disease strongly correlates to survival in patients with
hepatocellular carcinoma. Int J Cancer. 133:2165–2171. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan AW, Tong JH, Chan SL, Lai PB and To
KF: Expression of stemness markers (CD133 and EpCAM) in
prognostication of hepatocellular carcinoma. Histopathology.
64:935–950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamashita T, Ji J, Budhu A, Forgues M,
Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad
A, Banerjee S, Azmi AS, Miele L and Sarkar FH: Notch-1 induces
epithelial-mesenchymal transition consistent with cancer stem cell
phenotype in pancreatic cancer cells. Cancer Lett. 307:26–36. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishina S, Shiraha H, Nakanishi Y, Tanaka
S, Matsubara M, Takaoka N, Uemura M, Horiguchi S, Kataoka J,
Iwamuro M, et al: Restored expression of the tumor suppressor gene
RUNX3 reduces cancer stem cells in hepatocellular carcinoma by
suppressing Jagged1-Notch signaling. Oncol Rep. 26:523–531.
2011.PubMed/NCBI
|
|
9
|
van der Gun BT, Melchers LJ, Ruiters MH,
de Leij LF, McLaughlin PM and Rots MG: EpCAM in carcinogenesis: The
good, the bad or the ugly. Carcinogenesis. 31:1913–1921. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patriarca C, Macchi RM, Marschner AK and
Mellstedt H: Epithelial cell adhesion molecule expression (CD326)
in cancer: A short review. Cancer Treat Rev. 38:68–75. 2012.
View Article : Google Scholar
|
|
11
|
Wu SD, Ma YS, Fang Y, Liu LL, Fu D and
Shen XZ: Role of the microenvironment in hepatocellular carcinoma
development and progression. Cancer Treat Rev. 38:218–225. 2012.
View Article : Google Scholar
|
|
12
|
Brabletz T: To differentiate or not -
routes towards metastasis. Nat Rev Cancer. 12:425–436. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okamoto K, Tajima H, Nakanuma S, Sakai S,
Makino I, Kinoshita J, Hayashi H, Nakamura K, Oyama K, Nakagawara
H, et al: Angiotensin II enhances epithelial-to-mesenchymal
transition through the interaction between activated hepatic
stellate cells and the stromal cell-derived factor-1/CXCR4 axis in
intra-hepatic cholangiocarcinoma. Int J Oncol. 41:573–582.
2012.PubMed/NCBI
|
|
14
|
Kim SW, Choi HJ, Lee HJ, He J, Wu Q,
Langley RR, Fidler IJ and Kim SJ: Role of the endothelin axis in
astrocyte- and endothelial cell-mediated chemoprotection of cancer
cells. Neuro Oncol. 16:1585–1598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kikuta K, Masamune A, Watanabe T, Ariga H,
Itoh H, Hamada S, Satoh K, Egawa S, Unno M and Shimosegawa T:
Pancreatic stellate cells promote epithelial-mesenchymal transition
in pancreatic cancer cells. Biochem Biophys Res Commun.
403:380–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hellerbrand C: Hepatic stellate cells -
the pericytes in the liver. Pflugers Arch. 465:775–778. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Osta WA, Chen Y, Mikhitarian K, Mitas M,
Salem M, Hannun YA, Cole DJ and Gillanders WE: EpCAM is
overexpressed in breast cancer and is a potential target for breast
cancer gene therapy. Cancer Res. 64:5818–5824. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Coulouarn C, Corlu A, Glaise D, Guénon I,
Thorgeirsson SS and Clément B: Hepatocyte-stellate cell cross-talk
in the liver engenders a permissive inflammatory microenvironment
that drives progression in hepatocellular carcinoma. Cancer Res.
72:2533–2542. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amann T, Bataille F, Spruss T, Mühlbauer
M, Gäbele E, Schölmerich J, Kiefer P, Bosserhoff AK and Hellerbrand
C: Activated hepatic stellate cells promote tumorigenicity of
hepatocellular carcinoma. Cancer Sci. 100:646–653. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ju MJ, Qiu SJ, Fan J, Xiao YS, Gao Q, Zhou
J, Li YW and Tang ZY: Peritumoral activated hepatic stellate cells
predict poor clinical outcome in hepatocellular carcinoma after
curative resection. Am J Clin Pathol. 131:498–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Samatov TR, Tonevitsky AG and Schumacher
U: Epithelial-mesenchymal transition: Focus on metastatic cascade,
alternative splicing, non-coding RNAs and modulating compounds. Mol
Cancer. 12:1072013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tse JC and Kalluri R: Mechanisms of
metastasis: Epithelial-to-mesenchymal transition and contribution
of tumor microenvironment. J Cell Biochem. 101:816–829. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Watanabe T, Shibata N, Westerman KA,
Okitsu T, Allain JE, Sakaguchi M, Totsugawa T, Maruyama M,
Matsumura T, Noguchi H, et al: Establishment of immortalized human
hepatic stellate scavenger cells to develop bioartificial livers.
Transplantation. 75:1873–1880. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang N, Gores GJ and Shah VH: Hepatic
stellate cells: Partners in crime for liver metastases? Hepatology.
54:707–713. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hashiguchi M, Ueno S, Sakoda M, Iino S,
Hiwatashi K, Minami K, Ando K, Mataki Y, Maemura K, Shinchi H, et
al: Clinical implication of ZEB-1 and E-cadherin expression in
hepatocellular carcinoma (HCC). BMC Cancer. 13:5722013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kimura O, Kondo Y, Kogure T, Kakazu E,
Ninomiya M, Iwata T, Morosawa T and Shimosegawa T: Expression of
EpCAM increases in the hepatitis B related and the
treatment-resistant hepatocellular carcinoma. BioMed Res Int.
2014:1729132014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oishi N, Yamashita T and Kaneko S:
Molecular biology of liver cancer stem cells. Liver Cancer.
3:71–84. 2014. View Article : Google Scholar : PubMed/NCBI
|