Introduction
Breast cancer is one of the most common types of
cancer originating from breast tissues, and is the second most
notorious cause of cancer-related deaths after lung cancer.
Currently, the prognosis of breast cancer is encouraging in virtue
of the progress in diagnosis and effective systemic therapy,
including surgical operation, chemotherapy and radiotherapy.
However, such as for other solid tumors, the death rate of breast
cancer is more than 90% due to distant metastases (1). Numerous factors affect the occurrence
and progression of breast cancer, such as inactivation of
tumor-suppressor genes and activation of oncogenes by DNA
hypermethylation or histone deacetylase (2–4).
Previous studies indicate that the membrane-associated protein AMOT
is one of the most important biochemical characteristics of breast
cancer (5,6). An angiostatin binding protein AMOT was
first discovered in 2001 (7) and
plays an important role in the regulation of endothelial cell
migration and tube formation (7,8).
Research has demonstrated that the expression of AMOT is
upregulated in breast cancer tissues (5). However, the mechanism involved in the
upregulation of AMOT in breast cancer remains unclear.
MicroRNAs (miRNAs) are a class of endogenous
non-coding small RNAs of ~18–25 nucleotides long that play an
important role in post-transcriptional regulation and are found in
all eukaryotic cells. miRNAs usually have imperfect binding to the
3′-untranslated regions (3′-UTRs) of target mRNAs, resulting in
translational silencing or mRNA degradation (9,10), and
only a handful of miRNAs are able to enhance the expression of
mRNAs (11). miRNAs play important
roles in multifarious cellular processes, such as apoptosis and
proliferation (12), and are
frequently upregulated or downregulated in human types of cancers
(13), acting as either tumor
suppressors or oncogenes (14). The
expression of microRNA-205 (miR-205) was found to be downregulated
(15) or upregulated (16) in tumor tissues when compared with
matched adjacent normal tissues. Various studies have found that
miR-205 is downregulated in breast cancer tissues and in human
breast cancer cell lines (17–19).
However, the exact role of miR-205 in breast cancer at present
remains unclear.
In order to lay the foundation for later study, we
performed quantitative real-time PCR and western blot analysis of
AMOT in human breast cancer tissues, and found that the expression
of AMOT was significantly upregulated in tumor tissues when
compared with that in matched adjacent normal tissues both at the
protein and mRNA levels, consistent with Holmgren et al
(5). The aim of the present study
was to investigate the potential involvement of miR-205 in the
regulation of membrane-associated protein AMOT expression on the
mechanism of occurrence and development of breast cancer. In the
present study, we showed that overexpression of miR-205
significantly suppressed the proliferation and invasion of breast
cancer cells. Furthermore, we identified that miR-205 directly
targets AMOT by binding to its 3′-UTR, resulting in inhibition of
AMOT translation. Moreover, overexpression of AMOT reversed the
inhibitory effect of miR-205 on the growth and the invasion of
MCF-7 cells, in part.
Materials and methods
Tissues and cell lines
Tumor tissues and matched adjacent normal tissues
(20 breast cancer tissue samples and 20 normal adjacent tissue
samples) were obtained from breast cancer patients at the
Department of General Surgery, Sun Yat-Sen University Cancer
Center, from 2010 to 2013. All of the patients had an accurate
histological diagnosis according to the clinicopathological
criteria of the International Union for Cancer Control (UICC). The
median age of the patients was 49.6 years (range, 34–72 years). All
of the malignant tissues were from stage II-III tumors, according
to the International Federation of Gynecology and Obstetrics (FIGO)
classification. All patients provided consent for the use of their
specimens in research, and this use was approved by the
Institutional Research Ethics Committee of Sun Yat-sen University
Cancer Center.
Homo sapiens breast cancer SKBR-3,
MDA-MB-435S and MCF-7 cell lines were purchased from the American
Type Culture Collection (ATCC; Manassas, VA, USA). The breast
cancer cells were maintained according to the vendor's
instructions. In brief, SKBR-3 cells were cultured in McCoy's 5A
medium (modified) (ATCC) with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml
streptomycin). MDA-MB-435S cells were cultured in Leibovitz's L-15
medium (ATCC) with 10% FBS, 0.01 mg/ml bovine insulin, 0.01 mg/ml
glutathione and 1% penicillin-streptomycin (100 U/ml penicillin and
100 µg/ml streptomycin). MCF-7 cells were cultured in
Eagle's minimal essential medium (EMEM) (ATCC) with 10% FBS and 1%
penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml
streptomycin). All cells were cultured and maintained in a
humidified incubator under standard conditions.
Plasmid construction
AMOT 3′-UTR containing putative binding sites for
miR-205 were amplified from the normal human genome DNA and cloned
into downstream of the psi-CHECK2 vector (Promega, Madison, WI,
USA), and named AMOT-3′-UTR-WT. AMOT mutant 3′-UTR recombi-nant
plasmid was generated by the QuikChange Site-Directed Mutagenesis
kit (Stratagene, USA), which generated a mutation of 7 bp from
ATGAAGG to TACGGTC in the predicted miR-205 target binding site,
named as AMOT-3′-UTR-MUT. The full length cDNA encoding human AMOT
was amplified and the recombinant plasmid, pcDNA3.1/AMOT was
constructed. All plasmids were confirmed by DNA sequencing.
miRNA mimics and siRNA transfection
The miR-205 mimics, and the scrambled sequence
pre-miR negative control (NC) were purchased from a commercial
manufacturer (RiboBio, China) and small interfering RNA (siRNA)
targeting AMOT was obtained from Santa Cruz Biotechnology (Santa
Cruz, CA, USA). Cells (1×105/well) were seeded in
24-well plates and incubated for 24 h, and then the cells were
transfected with miRNA mimics (50 nM) or miR-NC (50 nM) or siRNA
(50 nM) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in
serum-free medium in accordance with the manufacturer's
instructions.
Quantitative reverse
transcriptase-polymerase chain reaction (qRT-PCR) analysis of miRNA
and Mrna
Total RNA and miRNA were extracted from the tissues
or the cultured cells using TRIzol reagent (Invitrogen). The
relative expression of miRNA-205 was quantitated using a SYBR
PrimeScript miRNA RT-PCR kit (Takara, China) in accordance with the
manufacturer's instructions, and the relative amount of miRNA was
normalized to U6 using the comparative threshold cycle method. For
quantitative analysis of mRNA expression, 2 µg of total RNA
was used to synthesize complementary DNA (cDNA) using M-MLV reverse
transcriptase (Promega, USA), and the corresponding cDNA was used
for quantitative PCR using SYBR-Green Real-Time Master Mix (Toyobo,
Japan), and a constitutive expression gene, GAPDH, was used as an
internal control to verify the fluorescent real-time RT-PCR
reaction. The sequence-specific primers were synthesized by Sangon
(Shanghai, China) (Table I). qPCR
was performed using the Applied Biosystems 7500 Sequence Detection
system (ABI, USA).
 | Table IPrimer sequences used for miRNA and
mRNA expression analysis. |
Table I
Primer sequences used for miRNA and
mRNA expression analysis.
| Name | Primer sequence
(5′-3′) |
|---|
| miR-205-RT |
CTCAACTGGTGTCGTGGAGTCG
GCAATTCAGTTGAGCAGACTCC |
| U6-RT |
CGCTTCACGAATTTGCGTGTCAT |
| U6-F |
CTCGCTTCGGCAGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
| miR-205-F |
ACACTCCAGCTGGGTCCTTCATTCCA |
| Universal-R |
GTGCAGGGTCCGAGGT |
| AMOT-F |
ATACGGTGATGGAGAAACAG |
| AMOT-R |
TGAAGAACTGCGACTGTG |
| GAPDH-F |
ACACCCACTCCTCCACCTTT |
| GAPDH-R |
TTACTCCTTGGAGGCCATGT |
Protein extraction and western
blotting
Total protein was extracted with SDS lysis buffer
(Beyotime, China) and the concentration of total proteins was
determined with the BCA protein assay kit (Pierce, USA). Equal
amounts of total proteins were separated on 10% SDS polyacrylamide
gels and transferred to polyvinylidene difluoride (PVDF) membranes
(Millipore, USA). The membranes were subjected to overnight
blocking in Tris-buffered saline (TBS) containing 5% non-fat dried
milk and incubated with a primary antibody [AMOT, 1:1,000; and
GAPDH, 1:3,000 (both from Abcam)] for 1 h at 37°C, and then the
membranes were washed and subsequently treated with the secondary
antibody (goat anti-rabbit IgG; Boster) at a 15,000 dilution for 1
h at room temperature and visualized by chemiluminescence.
Luciferase assays
For the reporter assay, the MCF-7 cells were
cultured in 24-well plates one day before transfection.
AMOT-3′-UTR-WT or -MUT vectors (100 ng) were co-transfected with
100 nM miR-205 mimics or negative control into MCF-7 cells using
Lipofectamine 2000 reagent as previously described. After
forty-eight hours of transfection, luciferase activities were
measured using a Dual-Luciferase Reporter assay system (Promega)
according to the manufacturer's instructions. Firefly luciferase
activity was normalized to Renilla luciferase activity.
Three independent experiments were performed, and the data are
presented as the mean ± SD.
MTT assay
The cell proliferation activities of the MCF-7 cells
with miRNA mimics or siRNA duplexes or miR-205 and AMOT were
determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT; Sigma, USA) assay at 0, 24, 48, 72 and 96 h. After a
24-h transfection, the MCF-7 cells were seeded into 96-well culture
plates at 1×104/well in a final volume of 100 µl.
At the indicated time points, 20 µl of MTT (5 mg/ml) was
added into each well for a 4-h incubation at 37°C. Following
removal of the culture medium, the remaining crystals were
dissolved in 200 µl dimethylsulfoxide (DMSO) (Sigma) and the
absorbance at 450 nm was measured. Three independent experiments
were performed.
Flow cytometry
Cell apoptosis was evaluated by flow cytometry using
an Annexin V-FITC/PI apoptosis detection kit (BestBio, China) in
accordance with the manufacturer's instructions. Briefly, the cells
were harvested with 0.5% trypsin, and washed with cold PBS, and
then centrifuged at 200 × g for 5 min. The cells (1×106)
were resuspended and stained using the Annexin V-FITC/PI apoptosis
detection kit according to the manufacturer's instructions. Then,
samples were analyzed using a Becton-Dickinson flow cytometer. Each
experiment was performed three times, and data are shown as mean ±
SD.
Cell invasion assays
For the cell invasion assay, 24-well Transwell
chambers (Millipore) were used according to the manufacturer's
protocol. In brief, 1×105 cells in 200 µl
serum-free EMEM were added into the upper chamber of the
Matrigel-coated inserts, and 500 µl of EMEM containing 10%
FBS was added to the lower chamber for culture. The cells were
cultured at 37°C in 5% CO2 in a humidified atmosphere
for 36 h, and the non-invading cells were removed. Then, the cells
were fixed, and stained using 0.5% crystal violet (Sigma), and the
invasive cells were counted under an inverted microscope (Olympus
IX71; Japan).
Statistical analysis
For quantitative data, all experiments were
performed at least three times, and all samples were tested in
triplicate. All data are expressed as the mean ± SD. Statistical
significance between groups was determined using one-way analysis
of variance (ANOVA) or an unpaired Student's t-test using SPSS 18.0
(SPSS, Inc., USA). A P-value of <0.05 was considered to indicate
a statistically significant result.
Results
AMOT is upregulated in breast cancer
tissues
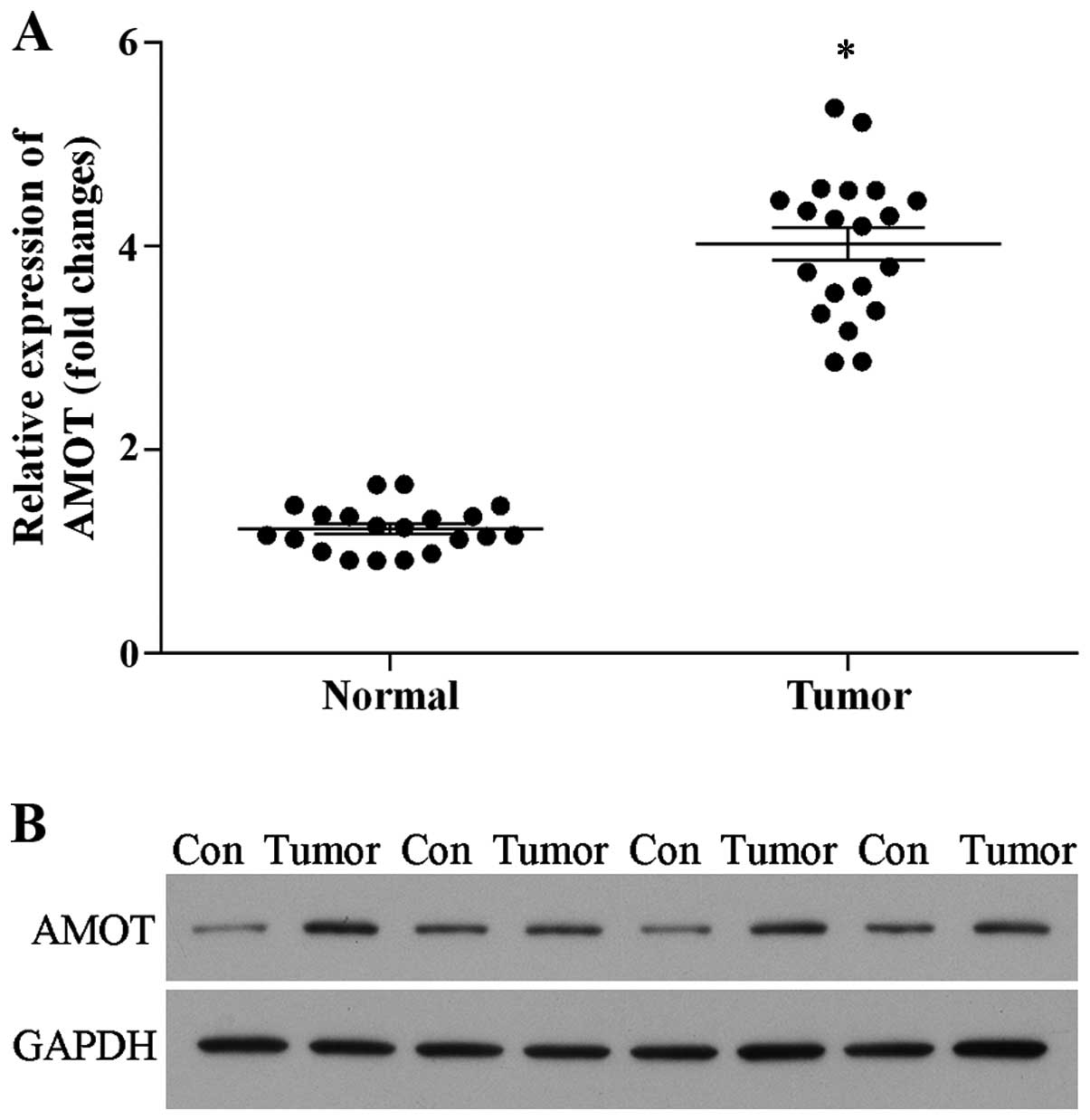
We first detected the expression level of AMOT by
real-time PCR and western blotting. As shown in Fig. 1, the expression of AMOT was
significantly upregulated in the tumor tissues when compared with
that of the matched normal tissues both at the mRNA (Fig. 1A) and protein levels (Fig. 1B). The data revealed that AMOT is
overexpressed in breast cancer tissues, suggesting that increased
AMOT protein expression contributes to breast cancer
development.
AMOT 3′-UTR is a predicted target of
miRNA-205
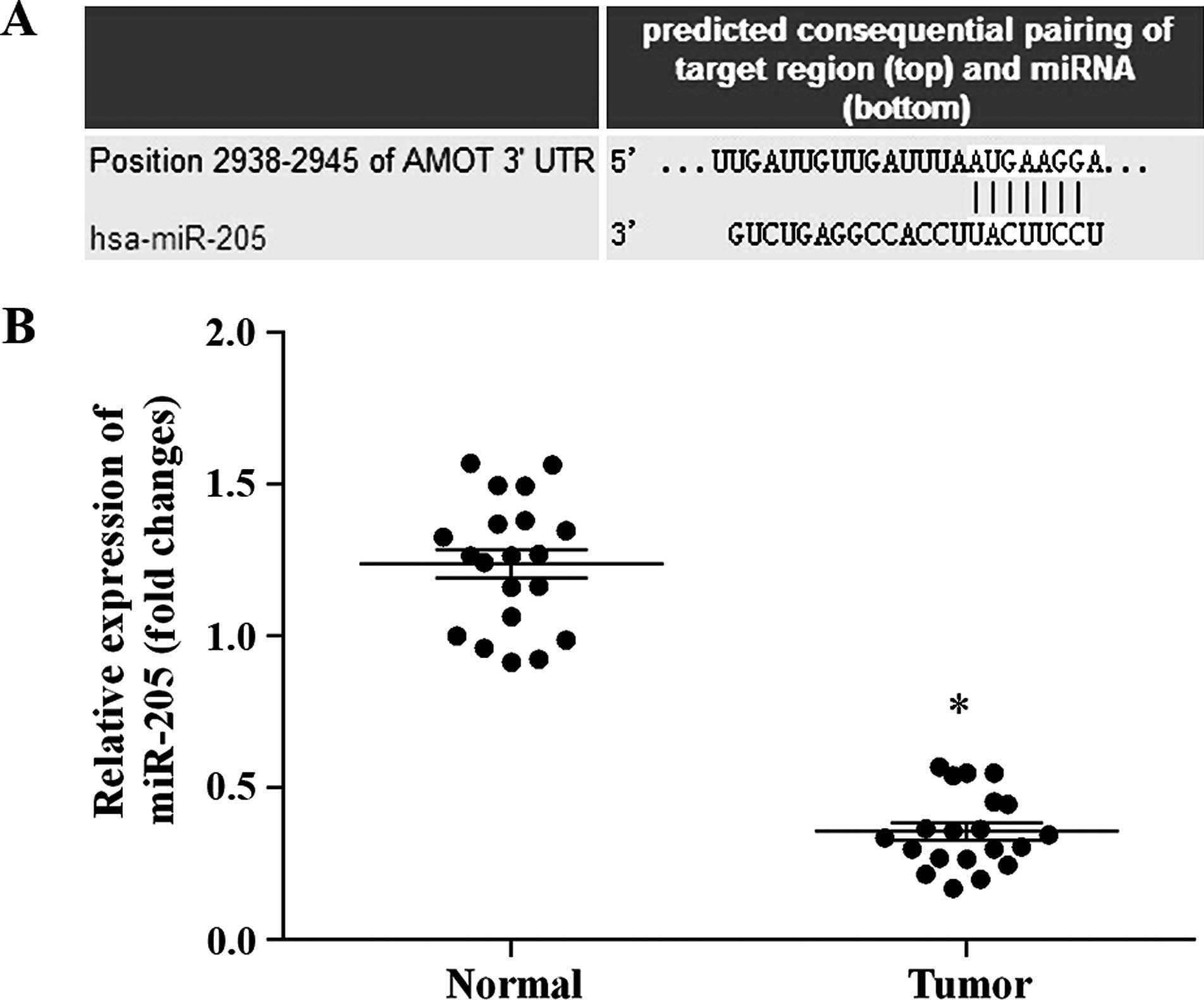
In order to determine whether miR-205 regulates AMOT
expression in breast cancer, miRNA target predication databases
were used for computational analyses, including TargetScan
(www.targetscan.org), miRDB (http://mirdb.org/miRDB) and microRNA (www.microrna.org). miR-205 was found to have one
predictive target site in the human AMOT 3′-UTR (Fig. 2A). The expression of mature miR-205
in the tumor and matched normal tissues was examined by real-time
PCR. As shown in Fig. 2B, miR-205
was significantly down-regulated in the cancer tissues when
compared with that in the matched normal tissues. The results
showed that the expression of miR-205 and AMOT in the tumor tissues
is inversely correlated. Therefore, miR-205 was selected to analyze
the role of AMOT in breast cancer.
AMOT is a direct target of miR-205 and is
downregulated by miR-205
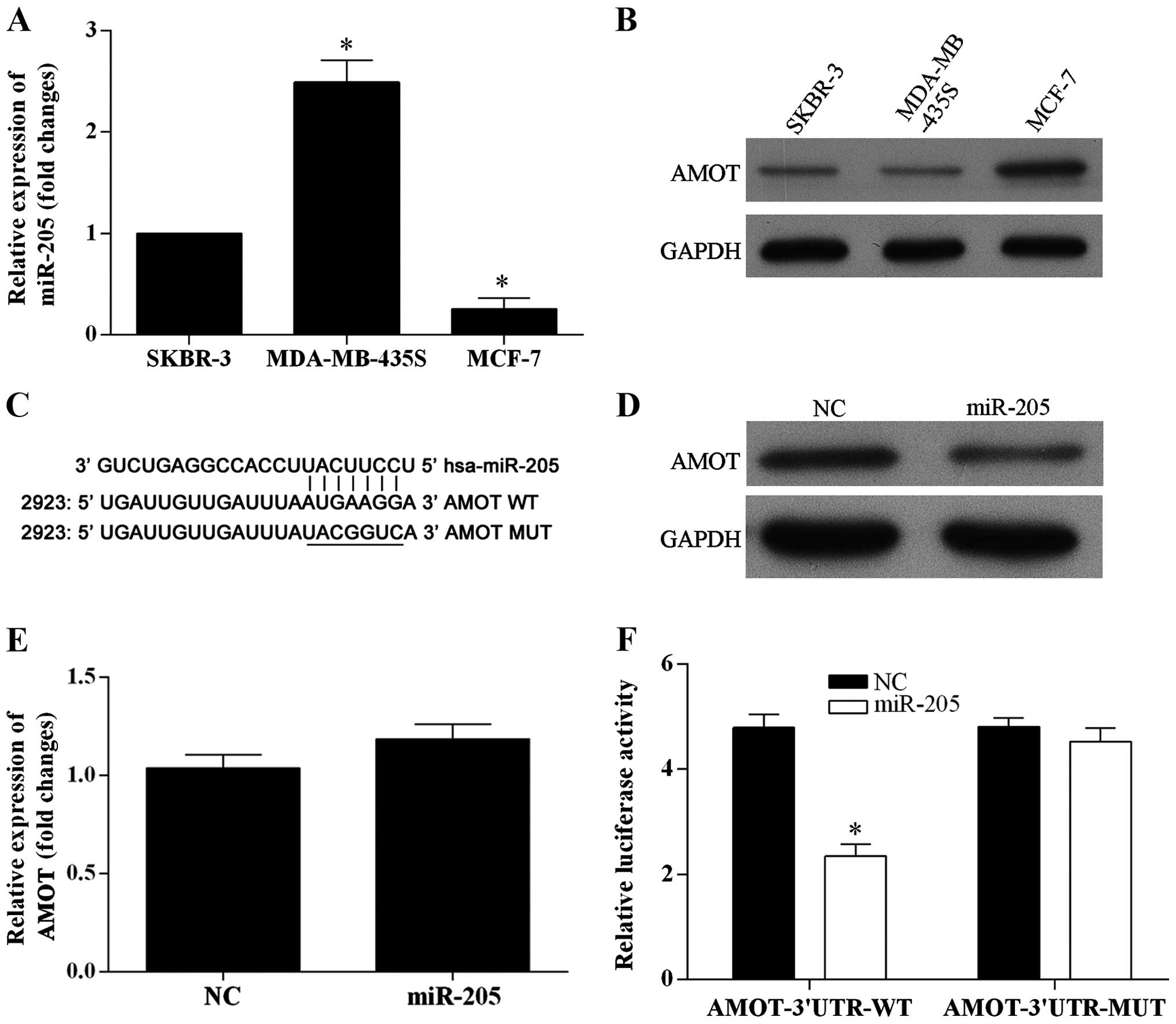
To investigate whether or not miR-205 inhibits the
expression of the endogenous AMOT protein, the expression level of
miR-205 and AMOT in three breast cancer cell lines were examined by
real-time PCR and western blotting, respectively. miR-205 was
significantly expressed at a low level in the MCF-7 cells (Fig. 3A), and the expression level of AMOT
was significantly higher in the MCF-7 cells (Fig. 3B). Thus, the MCF-7 cell line was
chosen to be used in the present study. The dual-luciferase
reporter vectors were constructed containing wild-type or
mutant-type seed sequences in the 3′-UTR of AMOT (Fig. 3C). Overexpression of miR-205
markedly reduced the expression of AMOT at the protein level in the
MCF-7 cells (Fig. 3D), yet did not
affect the AMOT mRNA level (Fig.
3E). The MCF-7 cells were co-transfected with AMOT-3′UTR-WT and
miR-205 mimics which resulted in a significantly reduced activity
of the luciferase reporter gene, yet the luciferase activity was
not significantly attenuated in the target region of the mutated
AMOT-3′UTR-MUT construct (Fig. 3F).
Based on the results, miR-205 directly targets the 3′UTR of AMOT
and downregulates AMOT expression.
Overexpression of miR-205 suppresses the
proliferation and invasion of MCF-7 cells in vitro
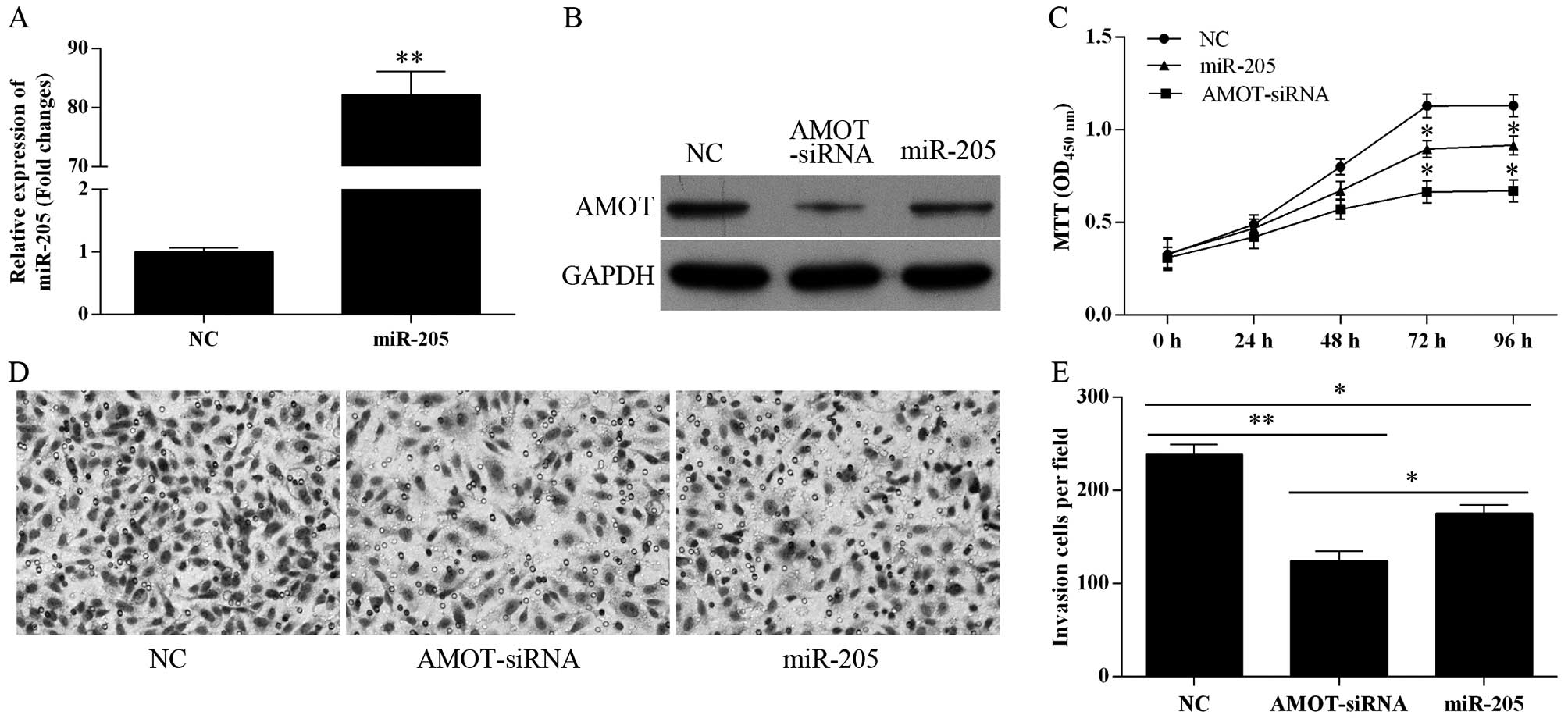
To evaluate the role of miR-205 in tumor cell
proliferation and invasion, miR-205 was overexpressed in the MCF-7
cells by transfection with miR-205 mimics (Fig. 4A). Overexpression of miR-205 as well
as transfection with AMOT-siRNA markedly reduced the expression
level of the AMOT protein (Fig.
4B). As shown in Fig. 4C, the
growth of miR-205- and AMOT-siRNA-transfected cells was
significantly suppressed compared with the NC-transfected MCF-7
cells (P<0.05 at 72 and 96 h). The role of miR-205 in tumor cell
invasion was detected using Transwell assay. miR-205- and
AMOT-siRNA-transfected cells exhibited much less invasive ability
when compared with the negative control cells (Fig. 4D and E). When MCF-7 cells were
transfected with miR-205 inhibitors, cell proliferation and
invasion were not affected (data not shown). These results indicate
that the miR-205-mediated growth and inhibition of invasion occur
in an AMOT-dependent manner in MCF-7 cells.
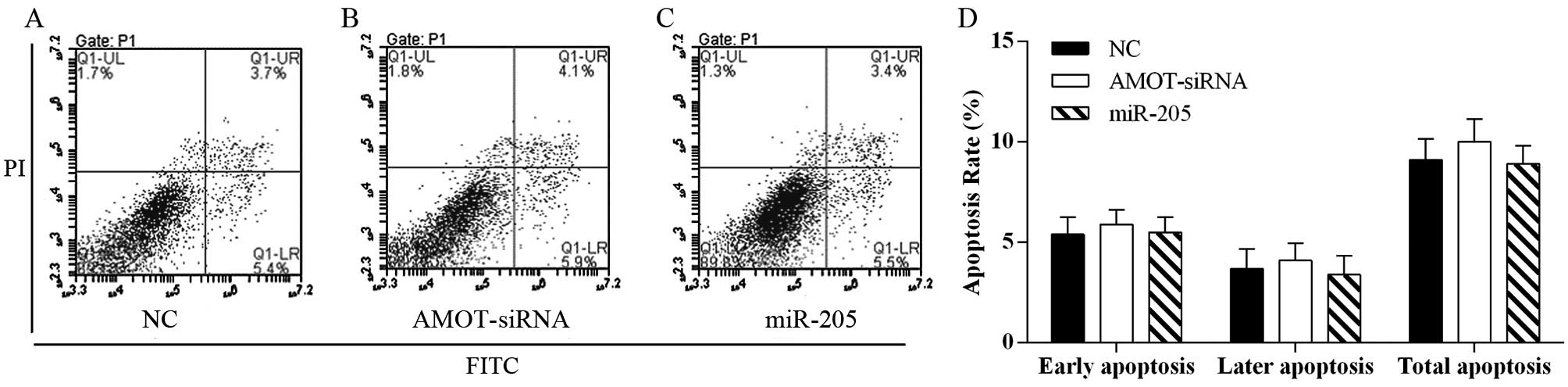
miR-205 does not affect breast cancer
cell apoptosis
In order to evaluate the effect of miR-205 on cell
apoptosis, the Annexin V-FITC/PI staining method was used to
perform the apoptosis assays. The data demonstrated that
overexpression of miR-205 as well as AMOT-siRNA transfection did
not affect cell apoptosis in the MCF-7 breast cancer cell line when
compared with the negative controls (Fig. 5).
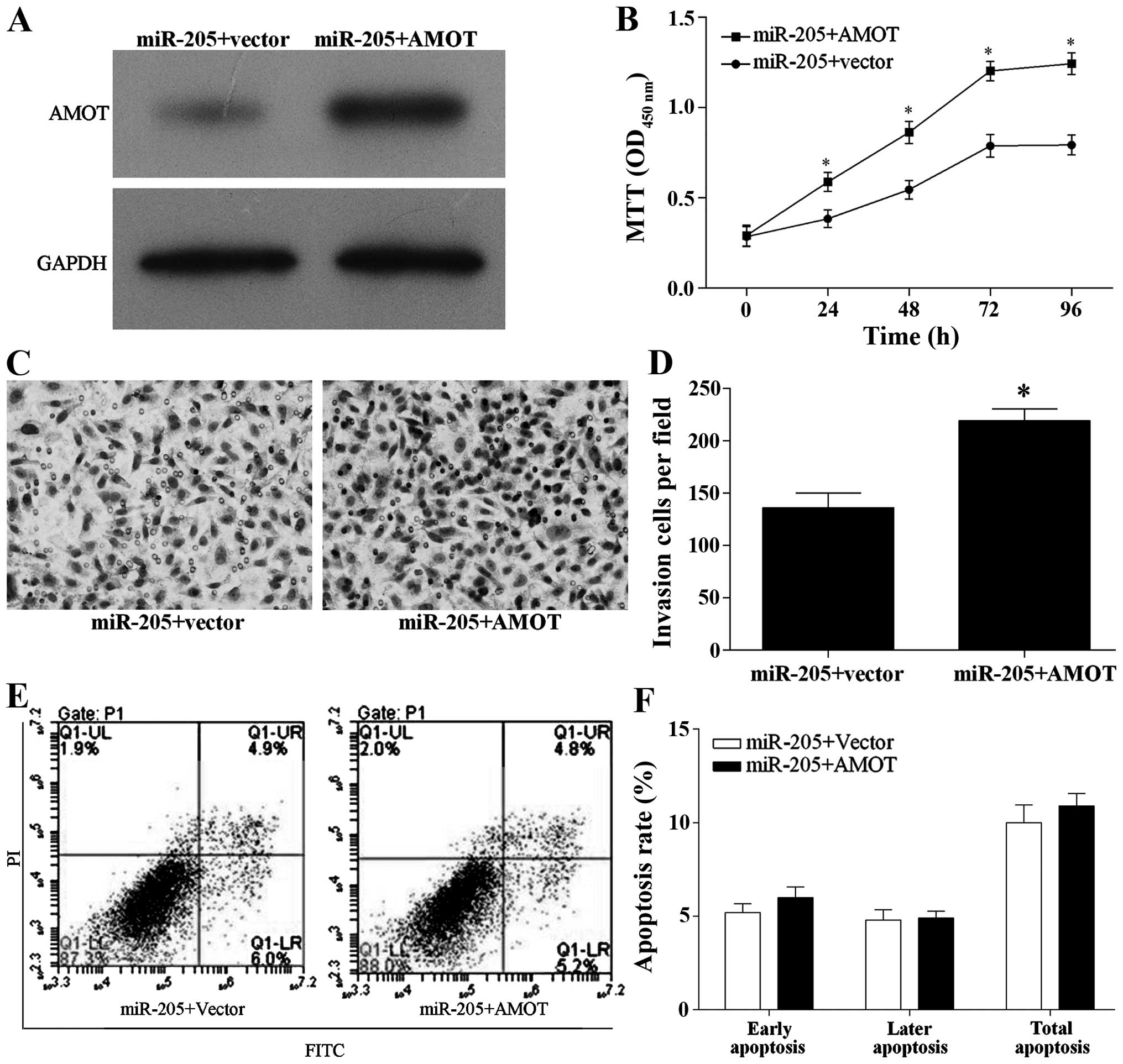
AMOT ameliorates the inhibitory effect of
miR-205 on cell proliferation and invasion
For further study, AMOT was over-expressed in
miR-205-overexpressing MCF-7 cells as shown by western blotting
(Fig. 6A). Moreover, overexpression
of AMOT in the MCF-7-overexpressing miR-205 cells significantly
decreased the inhibitory effect of miR-205 overexpression on breast
cancer cell proliferation and invasion (Fig. 6B–D), but did not affect cell
apoptosis (Fig. 6E and F).
Discussion
MicroRNAs (miRNAs) are endogenous, non-coding small
RNAs that target the 3′-untranslated regions (3′-UTR) of certain
messenger RNAs (mRNAs) to negatively regulate gene expression,
including mRNA degradation and translation inhibition (9). According to statistics, more than 30%
of all human genes as well as cellular processes are regulated or
controlled by miRNAs (20,21). More and more evidence has shown the
crucial impact of miRNAs on the occurrence and development of human
types of cancers (22–24). Dysregulation of miRNAs plays
important roles in cancer cell growth (25), invasion (26) and apoptosis (27).
Although miR-205 was identified many years ago, its
biological function has only recently been investigated. miR-205
acts as a tumor suppressor and is involved in many physiological
and pathological processes, such as hepatocellular carcinoma
(28), oxidative and endoplasmic
reticulum (ER) stress (29) and
endometrial cancer (30). A
previous study indicated that the expression of miR-205 is
frequently reduced in various types of cancer (31,32),
including breast cancer (17),
which suggests that miR-205 plays an important role in the
tumorigenesis and tumor progression of breast cancer. However, the
function of miR-205 in breast cancer is poorly understood.
In the present study, we showed that miR-205 was
significantly downregulated in breast tumor tissues, indicating a
potential role of miR-205 in breast cancer. In order to understand
whether miR-205 downregulation bears a biological role, the role of
miR-205 in breast cancer cell growth, invasion and apoptosis was
analyzed. Overexpression of miR-205 significantly inhibited cell
proliferation and reduced the migration and invasion of breast
cancer cells, and downregulated the expression of angiomotin
(AMOT). However, overexpression of miR-205 did not markedly affect
cell apoptosis. Overexpression of AMOT, a target gene of miR-205,
partially ameliorated the inhibitory effect on breast cancer cell
proliferation and invasion that was caused by miR-205
overexpression. These results suggest that miR-205 suppressed
breast cancer cell proliferation and invasion, which strongly
argues for the existence of a close correlation between miR-205 and
breast cancer occurrence and development.
AMOT is a membrane-associated protein that is
involved in controlling cell migration (33). Furthermore, it binds to the
endothelial cell surface of angiogenic tissues (6). p80-AMOT is an isoform of AMOT, which
enhances cell migration and stabilizes tubes in vitro
(34). A previous study showed that
the ERK1/2 pathway was activated through the Rac1 GTPase indirectly
by AMOT (35). In addition, the
expression of AMOT was upregulated in breast cancer (5), and expression of AMOT enhanced
ERK1/2-dependent proliferation of MCF-7 cells (36). AMOT also plays an important role in
altering tumor vessel permeability and hampering the growth of
tumors (37). In the present study,
we identified that AMOT is a direct target of miR-205 in breast
cancer through western blotting and luciferase assays. The results
showed that overexpression of miR-205 as well as AMOT knockdown by
AMOT-siRNA markedly reduced the expression of AMOT, and exhibited
significant suppression of proliferation and invasion. In addition,
overexpression of AMOT ameliorated the inhibitory effect of miR-205
overexpression on breast cancer cell growth and invasion in part.
All of the results indicate that downregulation of miR-205 promotes
the proliferation and invasive capacity of breast cancer through
the AMOT-mediated signal pathway.
In conclusion, we demonstrated that miR-205 is
downregulated in breast cancer tissues, and targets AMOT.
Overexpression of miR-205 downregulated the expression of AMOT and
inhibited the proliferative and the invasive capacities of breast
cancer cells. Collectively, the present study may lead to new
diagnostic and therapeutic approaches for breast cancer, and
enhances our understanding of the post-transcriptional regulation
of AMOT.
Acknowledgments
We thank the Department of General Surgery, Sun
Yat-Sen University Cancer Center for providing the tumor tissues
and matched adjacent normal tissues for the present study. We also
thank Guangzhou Vipotion Biotechnology Co., Ltd. for the assistance
in the vector construction of the luciferase reporter assay.
References
|
1
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Connolly R and Stearns V: Epigenetics as a
therapeutic target in breast cancer. J Mammary Gland Biol
Neoplasia. 17:191–204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sandhu R, Roll JD, Rivenbark AG and
Coleman WB: Dysregulation of the epigenome in human breast cancer:
Contributions of gene-specific DNA hypermethylation to breast
cancer pathobiology and targeting the breast cancer methylome for
improved therapy. Am J Pathol. 185:282–292. 2015. View Article : Google Scholar
|
|
4
|
Karsli-Ceppioglu S, Dagdemir A, Judes G,
Ngollo M, Penault-Llorca F, Pajon A, Bignon YJ and Bernard-Gallon
D: Epigenetic mechanisms of breast cancer: An update of the current
knowledge. Epigenomics. 6:651–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang WG, Watkins G, Douglas-Jones A,
Holmgren L and Mansel RE: Angiomotin and angiomotin like proteins,
their expression and correlation with angiogenesis and clinical
outcome in human breast cancer. BMC Cancer. 6:162006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Holmgren L, Ambrosino E, Birot O, Tullus
C, Veitonmäki N, Levchenko T, Carlson LM, Musiani P, Iezzi M,
Curcio C, et al: A DNA vaccine targeting angiomotin inhibits
angiogenesis and suppresses tumor growth. Proc Natl Acad Sci USA.
103:9208–9213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Troyanovsky B, Levchenko T, Månsson G,
Matvijenko O and Holmgren L: Angiomotin: An angiostatin binding
protein that regulates endothelial cell migration and tube
formation. J Cell Biol. 152:1247–1254. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bratt A, Birot O, Sinha I, Veitonmäki N,
Aase K, Ernkvist M and Holmgren L: Angiomotin regulates endothelial
cell-cell junctions and cell motility. J Biol Chem.
280:34859–34869. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu M, Roth A, Yu M, Morris R, Bersani F,
Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, et al: The
IGF2 intronic miR-483 selectively enhances transcription from IGF2
fetal promoters and enhances tumorigenesis. Genes Dev.
27:2543–2548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schetter AJ, Okayama H and Harris CC: The
role of microRNAs in colorectal cancer. Cancer J. 18:244–252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu H, Zhu S and Mo YY: Suppression of cell
growth and invasion by miR-205 in breast cancer. Cell Res.
19:439–448. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sempere LF, Christensen M, Silahtaroglu A,
Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S and Cole CN:
Altered microRNA expression confined to specific epithelial cell
subpopulations in breast cancer. Cancer Res. 67:11612–11620. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio MV, Casalini P, Piovan C, Di Leva G,
Merlo A, Triulzi T, Ménard S, Croce CM and Tagliabue E:
microRNA-205 regulates HER3 in human breast cancer. Cancer Res.
69:2195–2200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagadia R, Pandit P, Coman WB,
Cooper-White J and Punyadeera C: miRNAs in head and neck cancer
revisited. Cell Oncol. 36:1–7. 2013. View Article : Google Scholar
|
|
22
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang YW, Shi DB, Chen X, Gao C and Gao P:
Clinicopathological significance of microRNA-214 in gastric cancer
and its effect on cell biological behaviour. PLoS One.
9:e913072014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Inoue T, Iinuma H, Ogawa E, Inaba T and
Fukushima R: Clinicopathological and prognostic significance of
microRNA-107 and its relationship to DICER1 mRNA expression in
gastric cancer. Oncol Rep. 27:1759–1764. 2012.PubMed/NCBI
|
|
25
|
Fei X, Qi M, Wu B, Song Y, Wang Y and Li
T: MicroRNA-195-5p suppresses glucose uptake and proliferation of
human bladder cancer T24 cells by regulating GLUT3 expression. FEBS
Lett. 586:392–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu F, Deng H, Yao H, Liu Q, Su F and Song
E: mir-30 reduction maintains self-renewal and inhibits apoptosis
in breast tumor-initiating cells. Oncogene. 29:4194–4204. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang T, Zhang J, Cui M, Liu F, You X, Du
Y, Gao Y, Zhang S, Lu Z, Ye L, et al: Hepatitis B virus X protein
inhibits tumor suppressor miR-205 through inducing hypermethylation
of miR-205 promoter to enhance carcinogenesis. Neoplasia.
15:1282–1291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Muratsu-Ikeda S, Nangaku M, Ikeda Y,
Tanaka T, Wada T and Inagi R: Downregulation of miR-205 modulates
cell susceptibility to oxidative and endoplasmic reticulum stresses
in renal tubular cells. PLoS One. 7:e414622012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang G, Hou X, Li Y and Zhao M: miR-205
inhibits cell apoptosis by targeting phosphatase and tensin homolog
deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC
Cancer. 14:440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang N, Li Q, Feng NH, Cheng G, Guan ZL,
Wang Y, Qin C, Yin CJ and Hua LX: miR-205 is frequently
downregulated in prostate cancer and acts as a tumor suppressor by
inhibiting tumor growth. Asian J Androl. 15:735–741. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kalogirou C, Spahn M, Krebs M, Joniau S,
Lerut E, Burger M, Scholz CJ, Kneitz S, Riedmiller H and Kneitz B:
MiR-205 is progressively down-regulated in lymph node metastasis
but fails as a prognostic biomarker in high-risk prostate cancer.
Int J Mol Sci. 14:21414–21434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Levchenko T, Aase K, Troyanovsky B, Bratt
A and Holmgren L: Loss of responsiveness to chemotactic factors by
deletion of the C-terminal protein interaction site of angiomotin.
J Cell Sci. 116:3803–3810. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Levchenko T, Bratt A, Arbiser JL and
Holmgren L: Angiomotin expression promotes hemangioendothelioma
invasion. Oncogene. 23:1469–1473. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yi C, Troutman S, Fera D,
Stemmer-Rachamimov A, Avila JL, Christian N, Persson NL, Shimono A,
Speicher DW, Marmorstein R, et al: A tight junction-associated
Merlin-angiomotin complex mediates Merlin's regulation of mitogenic
signaling and tumor suppressive functions. Cancer Cell. 19:527–540.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ranahan WP, Han Z, Smith-Kinnaman W,
Nabinger SC, Heller B, Herbert BS, Chan R and Wells CD: The adaptor
protein AMOT promotes the proliferation of mammary epithelial cells
via the prolonged activation of the extracellular signal-regulated
kinases. Cancer Res. 71:2203–2211. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Arigoni M, Barutello G, Lanzardo S, Longo
D, Aime S, Curcio C, Iezzi M, Zheng Y, Barkefors I, Holmgren L, et
al: A vaccine targeting angiomotin induces an antibody response
which alters tumor vessel permeability and hampers the growth of
established tumors. Angiogenesis. 15:305–316. 2012. View Article : Google Scholar : PubMed/NCBI
|