Introduction
Urothelial carcinoma of the bladder (UCB) is the
second most common genitourinary tumor in men, significantly
increasing worldwide cancer statistics. The overall survival rate
for bladder cancer recorded for a five-year follow-up period is
~78% (1). It is a heterogeneous
neoplasm of unpredictable clinical course. Approximately 75% of
cases are classified as non-muscle invasive bladder cancer (NMIBC;
pTa/pT1 stage) at initial diagnosis. The remaining 25% of cases
consist of tumors displaying the ability of muscular invasion and
are classified as muscle invasive bladder cancer (MIBC; pT2–pT4
stage). Genetic studies suggest that the events of initiation,
promotion, and progression of UCB can be described by a model based
on two distinct molecular alteration patterns showing divergent
clinical behavior (2–4). Tobacco smoking and occupational
exposure to aromatic amines are considered well-established
exogenous risk factors for UCB (5).
Furthermore, genetic and epigenetic alterations also contribute to
UCB risk (6–9). Thus, interaction between environmental
exposure and genetic susceptibility can greatly increase DNA damage
and thus trigger urothelial precancerous events; therefore, the
prompt elimination of such damage counteracts malignant
transformation (10).
DNA repair mechanisms are multistep processes
essential for maintaining genomic stability. Distinct DNA repair
pathways recognize different types of DNA damage (11,12).
The base excision repair (BER) pathway plays a critical role by
removing nucleobase modifications by oxidation, alkylation,
deamination, and also backbone single-strand DNA (ssDNA) breaks
(13). A BER malfunction could thus
lead to accumulation of DNA lesions with a direct impact on the
risk of tumorigenesis, including UCB (14). The enzyme APE1 has a mainframe role
in BER by correcting apurinic/apyrimidinic (AP) sites, which are
pre-mutagenic lesions able to stall DNA replication forks. Besides
its role in DNA repair, APE1 is also a transcriptional regulator of
gene expression (15). The protein
XRCC1 plays a crucial role in BER. Although no direct catalytic
activity has been observed for XRCC1, it operates as a scaffold
protein for other BER enzymes (16,17).
DNA polymerase β (Pol β), a member of the X family DNA polymerases,
is the major polymerase in BER due to its dRP-lyase activity and
the main gap-filling enzyme (18).
Several molecular markers have been investigated to
improve UCB diagnosis and to provide potential therapeutic targets.
Gene expression profiling has been used as a strategy to identify
molecular subtypes, which could refine the classification of
bladder tumors (19–21). However, to date, no analyzed marker
has appeared conclusive for clinical practice. The aim of the
present study was to investigate whether variations in the levels
of transcripts APE1, XRCC1 and POLB could
influence the clinical outcomes of patients diagnosed with primary
UCB. The analyses were performed by quantitative real-time PCR
(qPCR) methodology (22). Our
findings suggest a significant correlation between the reduced
levels of APE1 transcripts and the poor survival of UCB
patients.
Materials and methods
Patient database
Fifty-two patients diagnosed with primary UCB
submitted to transurethral resection (TUR) at the Department of
Urology, Brazilian National Cancer Institute, Rio de Janeiro,
between 2004 and 2009 were eligible for this study. After TUR,
fresh tissue samples were immediately stored in liquid nitrogen
until further analysis. This project was approved by the Ethics
Committee of the Brazilian National Cancer Institute. At
recruitment, informed consent was obtained from each eligible
subject. Patient records were accessed to collect the following
information: date of registration at the hospital, age at
diagnosis, gender, pathological grade and stage, recurrence, and
disease-specific mortality (DSM).
The specimens obtained were classified as 25
low-grade papillary urothelial carcinoma and 27 high-grade
papillary urothelial carcinoma. Pathological staging consisted of
42 NMIBC specimens (Ta/T1 stage) and 10 MIBC specimens (T2–T4
stage). The study group included 43 men and 9 women with a mean age
of 68 years (range 48 to 92). Median follow-up was 55.5 months
(range 2 to 101 months). Disease recurred in 51.9% of all patients
(range 1 to 33 months) and 23.1% died (range 3 to 95 months) of
UCB. Table I shows the major
clinicopathologic characteristics of the UCB patients in this
study.
 | Table ICharacteristics of the investigated
UCB patients. |
Table I
Characteristics of the investigated
UCB patients.
|
Characteristics | Data |
|---|
| No. of patients
with primary tumor diagnosis | 52 |
| Mean age
(years) | 68 |
| Gender, n (%) |
| Female | 9 (17) |
| Male | 43 (83) |
| Grade, n (%) |
| Low-grade
(LG) | 25 (48) |
| High-grade
(HG) | 27 (52) |
| Stage, n (%) |
| Ta/T1 | 42 (81) |
| T2–T4 | 10 (19) |
| Follow-up
(months) | 55.5 |
| Disease-specific
mortality (DSM), n (%) |
| Low-grade
(LG) | 5 (20) |
| High-grade
(HG) | 7 (26) |
| Ta/T1 | 7 (17) |
| T2–T4 | 5 (50) |
| Recurrence, n
(%) |
| Low-grade
(LG) | 12 (48) |
| High-grade
(HG) | 15 (55.6) |
| Ta/T1 | 19 (45.2) |
| T2–T4 | 8 (80) |
RNA isolation and cDNA synthesis
Total RNA was isolated from tissue samples using
TRIzol® reagent according to the manufacturer's
instructions (Invitrogen, Carlsbad, CA, USA). After the extraction
process, all samples were treated with DNase I (RNase-free)
[Uniscience, New England Biolabs (NEB), Ipswich, MA, USA] to remove
any residual DNA from the previous step. RNA quality was evaluated
using 1% agarose gel electrophoresis in 1X TBE buffer
(Tris-borate-EDTA in DEPC water). Subsequently, RNA quantification
was performed on the NanoDrop spectrophotometer (Thermo Scientific,
Waltham, MA, USA) and the 260/280 nm ratio was measured to obtain
RNA purity. Total RNA (1 µg) was converted by reverse transcription
to cDNA using the SuperScript™ II reverse transcriptase
and random primers (both from Invitrogen), according to the
manufacturer's instructions. Following cDNA synthesis, the samples
were stored at −20°C.
Quantitative real-time PCR analysis
The relative expression of target transcripts was
compared with cDNA pools from non-cancerous tissues adjacent to the
tumor, which were used as reference samples. Quantitative real-time
PCR (qPCR) reactions were performed with 100 ng cDNA samples in
uniplex reactions by using a mix of Platinum®
SYBR®-Green qPCR SuperMix-UDG (Invitrogen) in Platform
Applied Biosystems (ABI PRISM® 7000 sequence detection
system; Applied Biosystems, Foster City, CA, USA). Details
regarding GenBank accession numbers, primer sequences, Tm values,
and amplicon length are listed in Table II. The GAPDH gene was used
as reference and its expression level was measured in all samples
individually to normalize APE1, XRCC1 and POLB
expression. The relative quantification of the results obtained by
qPCR was based on the comparative Ct method
(2−ΔΔCt).
 | Table IIGenBank accession no., primer
sequences, amplicon length and the expected Tm value for the qPCR
assays. |
Table II
GenBank accession no., primer
sequences, amplicon length and the expected Tm value for the qPCR
assays.
| Gene | GenBank accession
no. | Forward primer
(5′→3′) | Reverse primer
(5′→3′) | Amplicon length
(bp) | TM value(°C) |
|---|
| APE1 | NM_001641 |
CAATACTGGTCAGCTCCTTCGG |
TCATGCTCCTCATCGCCTATG | 127 | 83 |
| XRCC1 | NM_006297 |
GACACTTACCGAAAATGGCGG |
GCCATCATTCCCAATGTCCA | 111 | 83 |
| POLB | NM_002690 |
CAATGAGTACACCATCCGTCCC |
GTTCCCGGTATTTCCACTGGA | 110 | 81 |
| GAPDH | NM_001256799 |
GCAAATTCCATGGCACCGT |
TCGCCCCACTTGATTTTGG | 106 | 82 |
The qPCR conditions were conducted as follows: 1
cycle at 50°C (5 min) and 94°C (3 min) for enzyme activation,
followed by 40 cycles at 94°C (45 sec), 50°C (30 sec), 72°C (30
sec), and a subsequent final extension at 72°C (5 min). Net gene
expression for each patient was then compared, as a panel, for low-
and high-grade bladder tumors.
Genotyping
Genotyping assays to evaluate the APE1 T1349G
polymorphism were performed by sequencing cDNA samples. The PCR
amplification was carried out in the Veriti® Thermal
Cycler (Applied Biosystems) by using the primers
5′-GCTTCGAGCCTGGATTAAGAA-3′ (forward) and
5′-GGCCTGCATTAGGTACATATGCT-3′ (reverse) to amplify the target
fragment of APE1. PCR conditions were 94°C (10 min), followed by 40
cycles of 94°C (45 sec), 50°C (30 sec), and 72°C (30 sec), and
subsequent final extension at 72°C (5 min). PCR products were
purified by using GFX PCR DNA and Gel Band Purification kit (GE
Healthcare Life Sciences, Chalfont, UK), according to the
manufacturer's instructions. The cDNA sequencing was carried out on
the Sequencer 3130 Genetic Analyzer (Applied Biosystems) and the
results were analyzed with the Sequencher Program (Gene Codes
Corporation, Ann Arbor, MI, USA) using the reference sequence from
NCBI (NM_001244249.1).
Statistical analyses
Contingency tables were used to associate UCB tumor
grade (low-grade vs. high-grade) and stage (pTa/pT1 vs. pT2-pT4)
with clinical outcomes of recurrence and DSM. The Fisher's exact
test was adopted to test the statistical significance of the
association between these parameters. Statistical analysis was
carried out with statistical program R (v.2.15.1). The association
between tumor grade and stage with expression profile was evaluated
by non-parametric Mann-Whitney test with SPSS software (v.17.0).
The optimal cut-off point for sensitivity and specificity of the
transcripts was estimated by receiver operating characteristic
(ROC) curves. Univariable recurrence and survival probabilities
were estimated by using the Kaplan-Meier method. Univariable and
multivariable Cox regression models addressed time to recurrence
and mortality. For all statistical tests, p<0.05 was considered
to indicate a statistically significant result.
Results
Association of APE1, XRCC1 and POLB
transcript levels and clinicopathological parameters
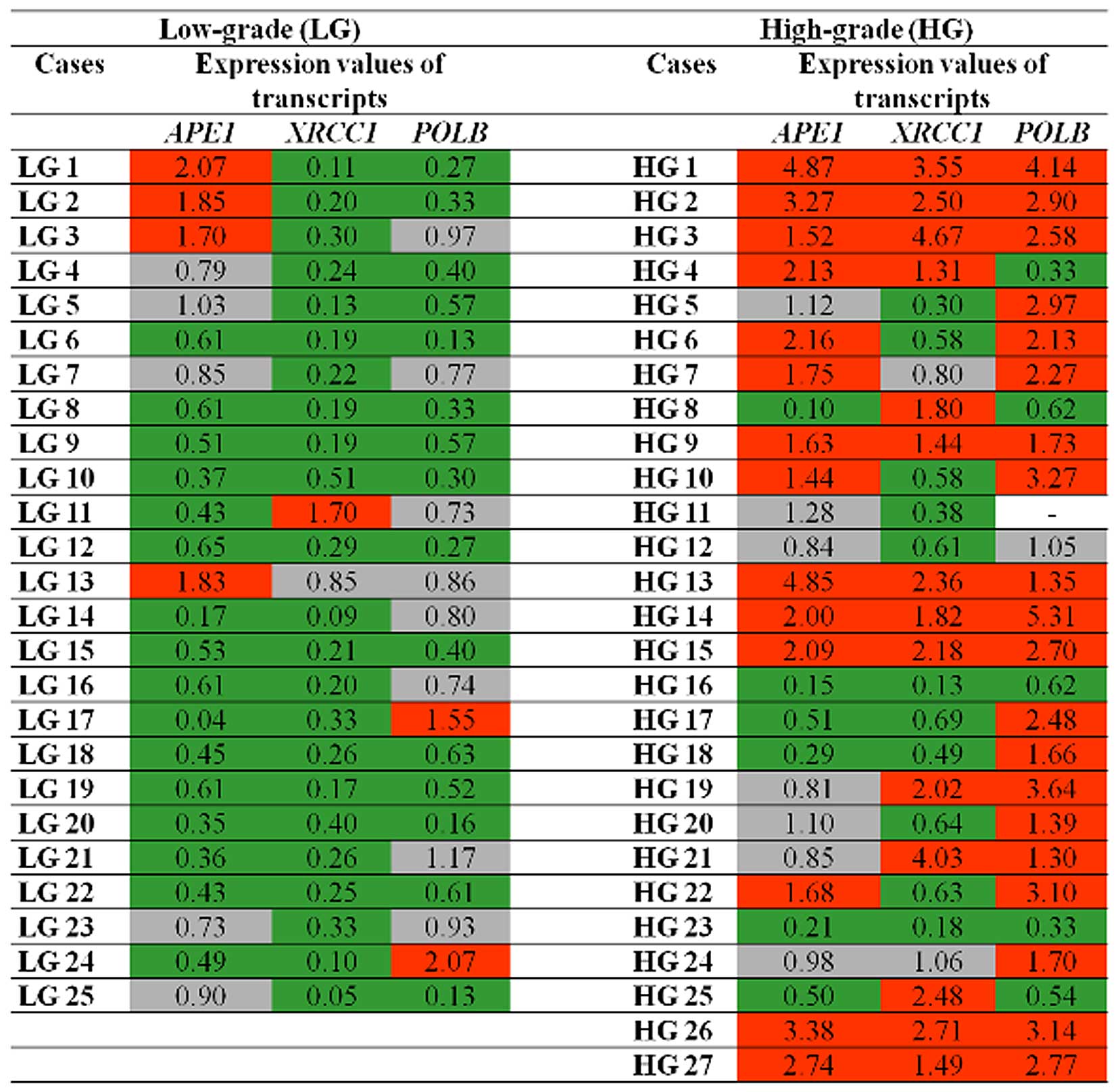
All samples from MIBC patients (T2-T4 stage) were
also classified as high-grade tumors and are referred to in
Fig. 1 as HG5, HG8, HG9, HG10,
HG11, HG13, HG22, HG23, HG24, and HG25. As shown in Table III, APE1, XRCC1 and
POLB transcript levels were similar between the NMIBC
(Ta/T1) and MIBC (T2–T4) groups and therefore were not associated
with stage (p>0.05).
 | Table IIIMedian variation in transcript levels
in each UCB group. |
Table III
Median variation in transcript levels
in each UCB group.
| Transcripts | Median value of
transcript expression
| P-value | Median value of
transcript expression
| P-value |
|---|
| Low-grade
(n=25) | High-grade
(n=27) | Ta/T1 stage
(n=10) | T2–T4 stage
(n=42) |
|---|
| APE1 | 0.61 | 1.44 | 0.005a | 1.18 | 1.38 | 0.745 |
| XRCC1 | 0.22 | 1.31 | 0.000a | 0.90 | 1.12 | 0.150 |
| POLB | 0.57 | 2.20 | 0.000a | 1.35 | 1.73 | 0.235 |
However, as summarized in Table III, statistical analysis revealed
highly significant differences in BER transcript levels between
low- and high-grade groups (p<0.01). In particular, increased
levels of APE1, XRCC1 and POLB transcripts
were detected in high-grade tumors (Fig. 1 and Table III).
According to Fig. 1,
reduced levels of XRCC1 transcripts were observed in 92% of
low-grade tumors, followed by APE1 (64%) and POLB
(60%). In addition, the triple combination of reduced levels of
APE1/XRCC1/POLB transcripts was observed in
40% of this tumor group. With regard to high-grade tumors, high
levels of POLB transcripts were detected in nearly 77% of
these tumors, followed by APE1 (52%) and XRCC1 (52%).
Likewise, our data revealed that 33.3% of high-grade tumors
exhibited combined increased levels of the
APE1/XRCC1/POLB transcripts.
By observing the panel displayed in Fig. 1, it can be seen that expression of
at least one of the BER genes varied among tumor samples of all UCB
patients investigated in the present study, appearing different
from the levels detected in the noncancerous samples. These
findings place BER gene transcriptional activity as a common
genetic alteration in UCB pathogenesis.
Correlation of clinicopathological
characteristics with clinical outcomes
To assess whether tumor grade or stage has an
influence on clinical outcomes, we investigated the relationship
between these parameters. The presence of the first recurrence
episode was associated with neither tumor stage nor tumor grade
(Table IV). Similarly, there was
no association between tumor grade and DSM (p=0.746). However, the
mortality rate was higher and positively associated with advanced
pathological stage (p=0.039).
 | Table IVCorrelation of the pathological grade
and stage with overall survival and recurrence-free survival in the
UCB patients. |
Table IV
Correlation of the pathological grade
and stage with overall survival and recurrence-free survival in the
UCB patients.
| Parameters | Grade
| P-value | Parameters | Stage
| P-value |
|---|
| Low-grade
(n=25) | High-grade
(n=27) | Ta/T1 (n=42) | T2–T4 (n=10) |
|---|
| First recurrence, n
(%)episodea | 12 (48) | 15 (55.6) | 0.786 | First recurrence, n
(%)episodea | 19 (45.2) | 8 (80) | 0.078 |
| DSMa, n (%) | 5 (20) | 7 (25.9) | 0.746 | DSMa, n (%) | 7 (16.7) | 5 (50) |
0.039c |
| Overall survival
rateb | | | | Overall survival
rateb | | | |
| 2-year | 95.2% | 77.3% | 0.596 | 2-year | 92.3% | 57.1% |
0.013c |
| 5-year | 78.1% | 73.2% | | 5-year | 82.9% | 45.7% | |
| Recurrence-free
survivalb | | | | Recurrence-free
survivalb | | | |
| 2-year | 95.2% | 85% | 0.457 | 1-year | 31.6% | 25% | 0.585 |
| 5-year | 65.1% | 63.6% | | 2-year | 10.5% | 12.5% | |
Regarding the overall survival rate recorded at two
and five years after diagnosis, no significant difference between
low- and high-grade groups was observed (p=0.596, Table IV). However, MIBC patients had
significantly shorter overall survival compared with the NMIBC
patients (p=0.013). In relation to recurrence-free survival, we did
not observe a significant difference between tumor grade or stage
(p>0.05).
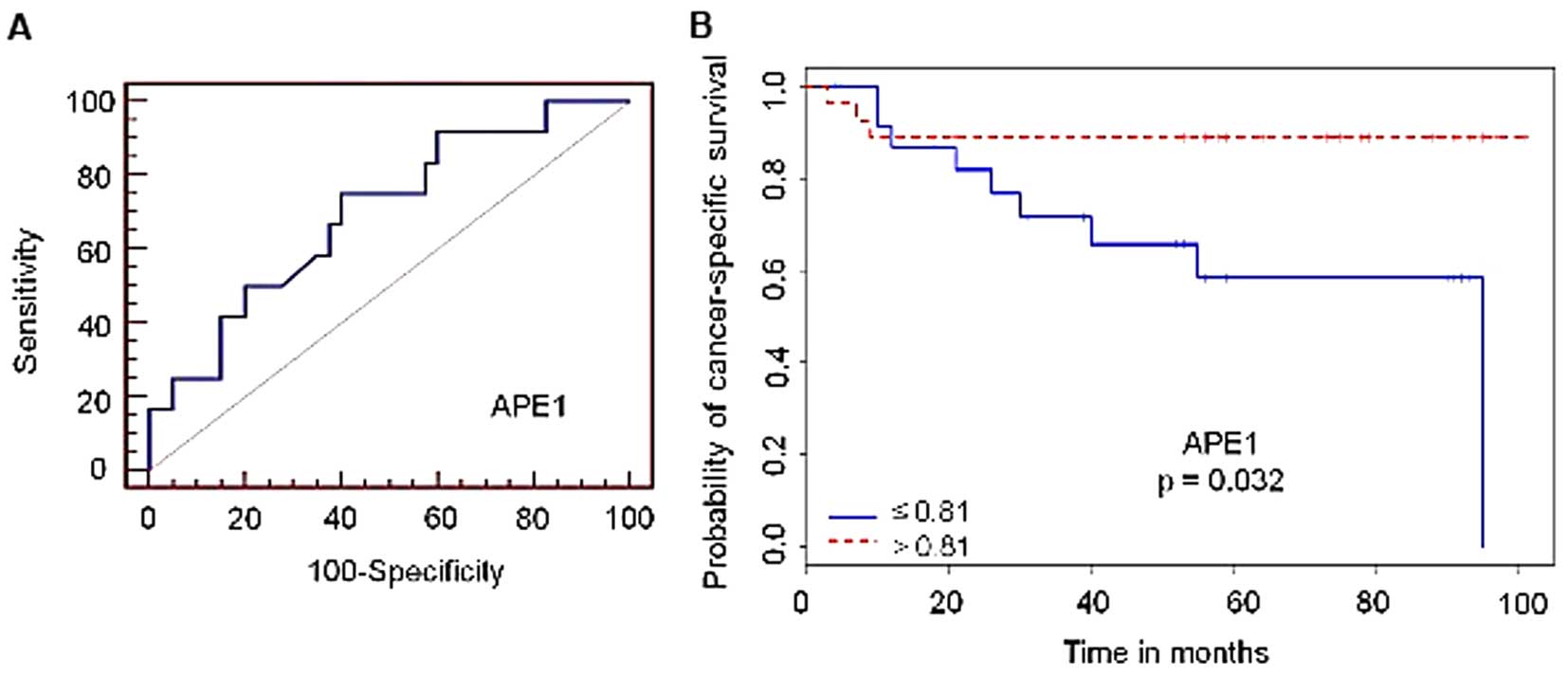
Association of reduced levels of APE1
with cancer-specific mortality
The levels of APE1, XRCC1 and
POLB transcripts were compared with the selected clinical
outcomes (DSM or recurrence) in each tumor grade and stage group to
assess predictive value. Overall, no significant correlation was
observed between transcript levels and DSM or recurrence, in either
pathological grade or stage (data not shown). Despite that, by
analyzing all patients in the study regardless of tumor stage or
grade, univariable analysis revealed that reduced levels of
APE1 transcripts were significantly associated with DSM
(p=0.032, Fig. 2). According to the
ROC plot, specificity and sensitivity for the APE1 gene
were, respectively, 60% and 75%. Therefore, these results revealed
that APE1 expression may be an indicator of poor prognosis
in UCB patients.
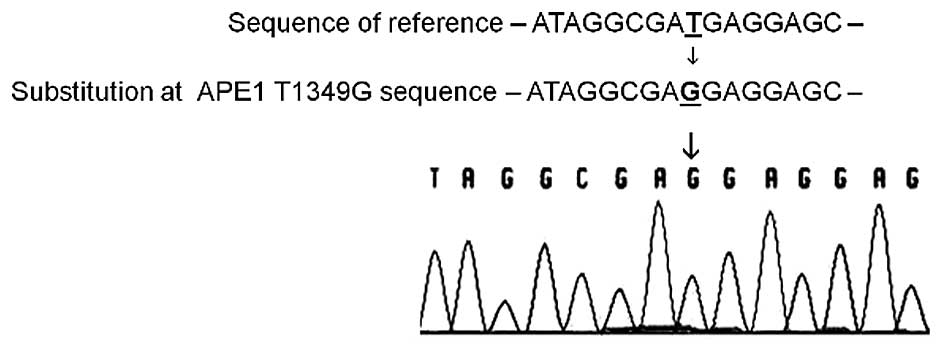
APE1 T1349G polymorphism
Genotyping assays to evaluate the APE1 T1349G
polymorphism were performed by preferentially sequencing the cDNA
samples from a subset of patients who concomitantly presented
reduced levels of APE1 transcripts and death and/or
recurrence events (Fig. 3). As
summarized in Table V, the G allele
variant was observed in 75% (9/12) of this subset, being present in
67% (6/9) among the low-grade cases analyzed. In addition, the
variant genotype (TG/GG) was found in 80% (4/5) of the low-grade
cases, whose outcome was either death and/or recurrence. All
high-grade patients analyzed had variant genotype (TG/GG), while
only individuals with the variant genotypes TG (T2–T4 stage)
succumbed to the disease.
 | Table VGenotype frequencies of the APE1
T1349G polymorphism among the UCB patients. |
Table V
Genotype frequencies of the APE1
T1349G polymorphism among the UCB patients.
| Genotype | Low-grade (LG), n=9
| High-grade (HG),
n=3
|
|---|
| n (%) | Death | Recurrence | n (%) | Death | Recurrence |
|---|
| TT | 3 (33) | LG17 | LG17 | 0 (0) | – | – |
| TG | 3 (33) | LG23 | LG23 | 2 (67) | HG8; HG23 | HG8 |
| GG | 3 (33) | LG11; LG19;
LG22 | LG11; LG19;
LG22 | 1 (33) | – | HG17 |
Discussion
Urothelial carcinoma of the bladder (UCB) is a
heterogeneous disease comprising multiple and complex molecular
changes associated with initiation and tumor progression. Previous
indications that pro-oxidant status appears to play a role in
genitourinary carcinogenesis and its correlation with tumor
invasion and metastasis encouraged our team to screen for molecular
markers of genetic stability as an attempt to improve UCB prognosis
(23,24). DNA repair mechanisms act as a
barrier to prevent genetic instability and cancer susceptibility. A
few studies have evaluated the association between
dysfunction/dysregulation in particular DNA repair genes and UCB
risk (25,26). Since APE1, XRCC1 and POLB proteins
operate downstream the same DNA repair pathway to eliminate DNA
oxidative damage, their expression above basal levels found in our
study could reveal excessive oxidative stress in high-grade bladder
tumors.
XRCC1 protein is required for efficient DNA repair
of other BER-involved enzymes. Notwithstanding its core interplay
in BER, a recent meta-analysis did not associate a dysfunctional
XRCC1 polymorphism to UCB risk (27). Increased levels of XRCC1
transcripts were also associated with chemoresistance to cisplatin
in non-small cell lung cancer (28). The set of results presented in this
study revealed that XRCC1 transcripts were increased in 52%
of high-grade tumors, whereas 92% of low-grade tumors displayed
reduced levels of XRCC1. Since single nucleotide
polymorphisms (SNPs) cannot be ascribed to such status,
transcriptional alterations for XRCC1 were revealed in this
study to better characterize the histopathological grade.
In higher eukaryotes, DNA polymerases participate in
replication, repair and recombination. Pol β is the major BER DNA
polymerase. Pol β is minimally expressed in normal cells but has
been shown to be overexpressed in different tumors both at the
transcriptional and protein levels, including bladder tumors
(29). Approximately 30% of all
human tumors express variant Pol β (30). As previously described in other
studies, Pol β destabilization plays an important role in
disrupting cell cycle control and increasing mutation rates. Being
an error-prone DNA polymerase, excess Pol β could disturb the
processivity of replication forks and increase genetic instability
and tumorigenesis by curtailing the activity of error-free DNA
polymerases during replication (31,32).
Furthermore, Pol β can also enhance mutagenesis by incorporation of
ribonucleotides into DNA and by competition with replicative DNA
polymerases (33,34). Our results revealed that the
POLB transcripts had increased levels in ~77% of high-grade
tumors, suggesting a strong correlation of POLB
dysregulation with tumorigenesis and progression events.
APE1 is a multifunctional protein essential in BER
and a redox transcriptional co-activator. APE1 participates in the
initial steps of BER by recognizing and nicking potentially
genotoxic and mutagenic abasic (AP) sites in different substrates,
such as double-strand DNA, hybrid DNA/RNA and RNA molecule
(35). APE1 elevated levels have
been reported to occur in a group of tumors (36–38).
In general, its increased levels provide resistance to
chemotherapeutic drugs and ionizing radiation and, as expected,
marked cisplatin sensitivity was observed to occur in vitro
in tumor cells knocked out for APE1 (39,40).
Nonetheless, Sak et al reported that increased APE1 protein
levels were correlated with improved cancer-specific survival in
patients with muscle-invasive bladder cancer following radical
radiotherapy (41). In the present
study, reduced APE1 levels at initial diagnosis were
significantly associated with poor clinical outcome, regardless of
the chosen therapeutical approach, pathological grade, or stage.
Moreover, reduced levels of APE1 also compromise the
regulation of p53 target genes through its redox activity and may
result in extensive genomic instability. All of these factors could
explain why reduced levels of APE1 transcripts resulted in
poorer UCB patient survival, causing it to be an important gene for
clinical cancer-specific survival prediction.
In our study, differences in clinical outcomes such
as recurrence and DSM were not correlated with pathological grade.
However, we identified higher mortality rates and reduced overall
survival in patients with advanced tumor stage. Since all MIBC
patients analyzed were also designated as high-grade tumors, the
characteristic highly aggressive profile of these tumors could
explain the increased mortality and poor survival in this subset of
UCB patients. The comprehensive profiling of BER transcripts in
each pathological grade group highlights meaningful differences in
the molecular signature of low- and high-grade bladder tumors that
could be attributed to differential transcriptional regulation
ensued by tumorigenesis and oxidative stress. Therefore, these
differences in BER transcript levels able to characterize two
distinct histologic UCB groups found in our study could complement
molecular data from previous studies associating low- and
high-grade UCB groups with distinct genetic alteration patterns
(2–4). Moreover, APE1 analyses
presented good specificity and sensitivity parameters (60%
specificity and 75% sensitivity) which represent important
information for a suitable clinical test.
In carcinogenesis events, the most extensively
studied polymorphism in the APE1 gene is the T to G
transversion (T1349G, also known as Asp148Glu, rs3136820) (42). Functional studies on this
polymorphism have shown that the G variant allele may have an
impact on APE1 endonuclease and reduce the ability to communicate
with other BER proteins (43). In
our study, some patients with reduced levels of APE1
transcripts also experienced death and/or recurrence events. In
this context, we performed an additional analysis to investigate
the presence of the APE1 T1349G polymorphism from this group
of patients. Our data pointed out that all high-grade patients
analyzed had variant genotypes (TG/GG), but only individuals with
genotype TG (T2–T4 stage) succumbed to the disease. With regard to
the low-grade group, the G allele variant occurred in 80% (4/5) of
the cases, whose outcome was mortality and/or recurrence events. In
a study for functional characterization of polymorphisms in DNA
repair genes and chromosome aberrations by using X-rays or
ultraviolet (UV) light to irradiate blood lymphocytes, samples from
individuals with the TG or GG genotype showed higher levels of
damage, including aberrant cells, chromatid breaks, chromatid
exchanges, deletions, and dicentrics (44). A meta-analysis study showed that
individuals carrying at least one G allele were associated with a
higher cancer risk than subjects with the TT genotype (45). However, a meta-analysis study
performed in 2013 suggested that the APE1 T1349G
polymorphism was not associated with bladder cancer risk among
Asians or non-Asians (46).
Consistent with these observations, our data suggest that besides
interfering with APE1 enzymatic activity, as shown in other
studies, the G allele may also modify the APE1
transcriptional levels and result in worse clinical outcome for
bladder cancer patients. Our small sample size for this
polymorphism analysis (n=9 for low-grade and n=3 for high-grade
groups) should be interpreted with caution, as more detailed data
are needed to verify these findings. However, taken together, our
results indicate that APE1 is an attractive candidate gene
to open new perspective studies for evaluating clinical outcomes of
UCB patients. Further analyses are required to validate APE1
as a reliable predictive molecular marker for UCB.
Finally, the present study thus reinforces the
notion that DNA repair gene expression must be finely tuned in
order to avoid genetic instability and the process of tumorigenesis
(47). Despite current effective
treatments to ensure better results for UCB patients, diagnostic
and prognostic exams are still partially accurate and should be
complemented with auxiliary tests. Therefore, more clinical
research on UCB should be encouraged to allow a significant
increase in clinical data, better comprehension of the biology of
the disease, and optimization of diagnostic and prognostic tests
(48).
In conclusion, evaluation of DNA repair transcripts
revealed that high-grade tumors exhibited elevated levels of
APE1, XRCC1, and POLB as compared with
low-grade tumors. Taken together, this panel adds information on
the histopathology of bladder tumors. By analyzing all patients,
only APE1 reduced levels were an independent predictor of
cancer-specific mortality in primary UCB regardless of pathological
grade or stage, and APE1 may represent a possible candidate
gene for evaluating clinical outcomes in UCB. Despite this,
evidence suggests that dysregulated BER transcription may promote
bladder carcinogenesis.
Acknowledgments
The authors acknowledge all participing clinicians,
investigators, patients, and everyone who encouraged this study.
This study was supported by the Brazilian Development Agencies:
CAPES, FAPERJ and Prog. Oncobiologia FAF/Onco III.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Knowles MA: Molecular subtypes of bladder
cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis.
27:361–373. 2006. View Article : Google Scholar
|
|
3
|
DeGraff DJ, Cates JM, Mauney JR, Clark PE,
Matusik RJ and Adam RM: When urothelial differentiation pathways go
wrong: Implications for bladder cancer development and progression.
Urol Oncol. 31:802–811. 2013. View Article : Google Scholar
|
|
4
|
Castillo-Martin M, Domingo-Domenech J,
Karni-Schmidt O, Matos T and Cordon-Cardo C: Molecular pathways of
urothelial development and bladder tumorigenesis. Urol Oncol.
28:401–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burger M, Catto JW, Dalbagni G, Grossman
HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C,
Shariat S, et al: Epidemiology and risk factors of urothelial
bladder cancer. Eur Urol. 63:234–241. 2013. View Article : Google Scholar
|
|
6
|
Kim WJ and Bae SC: Molecular biomarkers in
urothelial bladder cancer. Cancer Sci. 99:646–652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shariat SF, Tokunaga H, Zhou J, Kim J,
Ayala GE, Benedict WF and Lerner SP: p53, p21, pRB, and p16
expression predict clinical outcome in cystectomy with bladder
cancer. J Clin Oncol. 22:1014–1024. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang J, Xu Z, Li J, Zhang R, Zhang G, Ji
H, Song B and Chen Z: XPC epigenetic silence coupled with p53
alteration has a significant impact on bladder cancer outcome. J
Urol. 184:336–343. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan C, Kim YW, Ha YS, Kim IY, Kim YJ, Yun
SJ, Moon SK, Bae SC and Kim WJ: RUNX3 methylation as a predictor
for disease progression in patients with non-muscle-invasive
bladder cancer. J Surg Oncol. 105:425–430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hoeijmakers JH: Genome maintenance
mechanisms for preventing cancer. Nature. 411:366–374. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Christmann M, Tomicic MT, Roos WP and
Kaina B: Mechanisms of human DNA repair: An update. Toxicology.
193:3–34. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wood RD, Mitchell M and Lindahl T: Human
DNA repair genes, 2005. Mutat Res. 577:275–283. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zharkov DO: Base excision DNA repair. Cell
Mol Life Sci. 65:1544–1565. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Figueroa JD, Malats N, Real FX, Silverman
D, Kogevinas M, Chanock S, Welch R, Dosemeci M, Tardón A, Serra C,
et al: Genetic variation in the base excision repair pathway and
bladder cancer risk. Hum Genet. 121:233–242. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fritz G: Human APE/Ref-1 protein. Int J
Biochem Cell Biol. 32:925–929. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kubota Y, Nash RA, Klungland A, Schär P,
Barnes DE and Lindahl T: Reconstitution of DNA base excision-repair
with purified human proteins: Interaction between DNA polymerase
beta and the XRCC1 protein. EMBO J. 15:6662–6670. 1996.PubMed/NCBI
|
|
17
|
Campalans A, Marsin S, Nakabeppu Y,
O'connor TR, Boiteux S and Radicella JP: XRCC1 interactions with
multiple DNA glycosylases: A model for its recruitment to base
excision repair. DNA Repair (Amst). 4:826–835. 2005. View Article : Google Scholar
|
|
18
|
Allinson SL, Dianova II and Dianov GL: DNA
polymerase beta is the major dRP lyase involved in repair of
oxidative base lesions in DNA by mammalian cell extracts. EMBO J.
20:6919–6926. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dyrskjøt L, Thykjaer T, Kruhøffer M,
Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H and Orntoft TF:
Identifying distinct classes of bladder carcinoma using
microarrays. Nat Genet. 33:90–96. 2003. View Article : Google Scholar
|
|
20
|
Mengual L, Burset M, Ars E, Lozano JJ,
Villavicencio H, Ribal MJ and Alcaraz A: DNA microarray expression
profiling of bladder cancer allows identification of noninvasive
diagnostic markers. J Urol. 182:741–748. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Damrauer JS, Hoadley KA, Chism DD, Fan C,
Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS, et
al: Intrinsic subtypes of high-grade bladder cancer reflect the
hallmarks of breast cancer biology. Proc Natl Acad Sci USA.
111:3110–3115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gecit I, Aslan M, Gunes M, Pirincci N,
Esen R, Demir H and Ceylan K: Serum prolidase activity, oxidative
stress, and nitric oxide levels in patients with bladder cancer. J
Cancer Res Clin Oncol. 138:739–743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kumar B, Koul S, Khandrika L, Meacham RB
and Koul HK: Oxidative stress is inherent in prostate cancer cells
and is required for aggressive phenotype. Cancer Res. 68:1777–1785.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Michiels S, Laplanche A, Boulet T, Dessen
P, Guillonneau B, Méjean A, Desgrandchamps F, Lathrop M, Sarasin A
and Benhamou S: Genetic polymorphisms in 85 DNA repair genes and
bladder cancer risk. Carcinogenesis. 30:763–768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Narter KF, Ergen A, Agaçhan B, Görmüs U,
Timirci O and Isbir T: Bladder cancer and polymorphisms of DNA
repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1). Anticancer Res.
29:1389–1393. 2009.PubMed/NCBI
|
|
27
|
Zhuo W, Zhang L, Cai L, Zhu B and Chen Z:
XRCC1 Arg399Gln polymorphism and bladder cancer risk: Updated
meta-analyses based on 5767 cases and 6919 controls. Exp Biol Med
(Maywood). 238:66–76. 2013. View Article : Google Scholar
|
|
28
|
Weaver DA, Crawford EL, Warner KA,
Elkhairi F, Khuder SA and Willey JC: ABCC5, ERCC2, XPA and XRCC1
transcript abundance levels correlate with cisplatin
chemoresistance in non-small cell lung cancer cell lines. Mol
Cancer. 4:182005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Albertella MR, Lau A and O'Connor MJ: The
overexpression of specialized DNA polymerases in cancer. DNA Repair
(Amst). 4:583–593. 2005. View Article : Google Scholar
|
|
30
|
Starcevic D, Dalal S and Sweasy JB: Is
there a link between DNA polymerase beta and cancer? Cell Cycle.
3:998–1001. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Osheroff WP, Jung HK, Beard WA, Wilson SH
and Kunkel TA: The fidelity of DNA polymerase beta during
distributive and processive DNA synthesis. J Biol Chem.
274:3642–3650. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bergoglio V, Pillaire MJ, Lacroix-Triki M,
Raynaud-Messina B, Canitrot Y, Bieth A, Garès M, Wright M, Delsol
G, Loeb LA, et al: Deregulated DNA polymerase beta induces
chromosome instability and tumorigenesis. Cancer Res. 62:3511–3514.
2002.PubMed/NCBI
|
|
33
|
Bergoglio V, Ferrari E, Hübscher U, Cazaux
C and Hoffmann JS: DNA polymerase beta can incorporate
ribonucleotides during DNA synthesis of undamaged and CPD-damaged
DNA. J Mol Biol. 331:1017–1023. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pillaire MJ, Betous R, Conti C, Czaplicki
J, Pasero P, Bensimon A, Cazaux C and Hoffmann JS: Upregulation of
error-prone DNA polymerases beta and kappa slows down fork
progression without activating the replication checkpoint. Cell
Cycle. 6:471–477. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Berquist BR, McNeill DR and Wilson DM III:
Characterization of abasic endonuclease activity of human Ape1 on
alternative substrates, as well as effects of ATP and sequence
context on AP site incision. J Mol Biol. 379:17–27. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bobola MS, Blank A, Berger MS, Stevens BA
and Silber JR: Apurinic/apyrimidinic endonuclease activity is
elevated in human adult gliomas. Clin Cancer Res. 7:3510–3518.
2001.PubMed/NCBI
|
|
37
|
Kelley MR, Cheng L, Foster R, Tritt R,
Jiang J, Broshears J and Koch M: Elevated and altered expression of
the multifunctional DNA base excision repair and redox enzyme
Ape1/ref-1 in prostate cancer. Clin Cancer Res. 7:824–830.
2001.PubMed/NCBI
|
|
38
|
Moore DH, Michael H, Tritt R, Parsons SH
and Kelley MR: Alterations in the expression of the DNA
repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin
Cancer Res. 6:602–609. 2000.PubMed/NCBI
|
|
39
|
Robertson KA, Bullock HA, Xu Y, Tritt R,
Zimmerman E, Ulbright TM, Foster RS, Einhorn LH and Kelley MR:
Altered expression of Ape1/ref-1 in germ cell tumors and
overexpression in NT2 cells confers resistance to bleomycin and
radiation. Cancer Res. 61:2220–2225. 2001.PubMed/NCBI
|
|
40
|
Zhang Y, Wang J, Xiang D, Wang D and Xin
X: Alterations in the expression of the apurinic/apyrimidinic
endonuclease-1/redox factor-1 (APE1/Ref-1) in human ovarian cancer
and indentification of the therapeutic potential of APE1/Ref-1
inhibitor. Int J Oncol. 35:1069–1079. 2009.PubMed/NCBI
|
|
41
|
Sak SC, Harnden P, Johnston CF, Paul AB
and Kiltie AE: APE1 and XRCC1 protein expression levels predict
cancer-specific survival following radical radiotherapy in bladder
cancer. Clin Cancer Res. 11:6205–6211. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gu D, Wang M, Wang S, Zhang Z and Chen J:
The DNA repair gene APE1 T1349G polymorphism and risk of gastric
cancer in a Chinese population. PLoS One. 6:e289712011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hadi MZ, Coleman MA, Fidelis K,
Mohrenweiser HW and Wilson DM III: Functional characterization of
Ape1 variants identified in the human population. Nucleic Acids
Res. 28:3871–3879. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Au WW, Salama SA and Sierra-Torres CH:
Functional characterization of polymorphisms in DNA repair genes
using cytogenetic challenge assays. Environ Health Perspect.
111:1843–1850. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gu D, Wang M, Wang M, Zhang Z and Chen J:
The DNA repair gene APE1 T1349G polymorphism and cancer risk: A
meta-analysis of 27 case-control studies. Mutagenesis. 24:507–512.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu C, Yin Q, Li L, Zhuang YZ, Zu X and
Wang Y: APE1 Asp148Glu gene polymorphism and bladder cancer risk: A
meta-analysis. Mol Biol Rep. 40:171–176. 2013. View Article : Google Scholar
|
|
47
|
Sweasy JB, Lang T and DiMaio D: Is base
excision repair a tumor suppressor mechanism? Cell Cycle.
5:250–259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kunath F, Krause SF, Wullich B, Goebell
PJ, Engehausen DG, Burger M, Meerpohl JJ and Keck B: Bladder cancer
- the neglected tumor: A descriptive analysis of publications
referenced in MEDLINE and data from the register
ClinicalTrials.gov. BMC Urol. 13:562013. View Article : Google Scholar
|