Introduction
Liver cancer is one of the most prevalent malignant
tumors, and is the third leading cause of cancer-related mortality
worldwide, especially in parts of Asia and Africa (1). There are ~750,000 new cases of liver
cancer, in which 85–90% are hepatocellular carcinoma (HCC),
reported globally per year (2).
Current standard practices for the treatment of liver cancer,
surgical resection and liver transplantation, are less than
satisfactory due to intra- and extra-hepatic metastasis and
post-surgical recurrence (3,4).
Numerous studies have determined the pathogenesis of liver cancer;
however, the accurate molecular mechanisms underlying the
pathogenesis and progression of liver cancer remain poorly
understood.
Galectin-9 (Gal-9), a β-galactoside-binding lectin
with two carbohydrate-recognition domains, was first identified as
an eosinophil chemoattractant and activation factor (5,6). An
increasing amount of evidence suggests that Gal-9 may modulate a
variety of biological functions and play an important role in both
immune response and tumor progression (7,8). As
one of the specific ligands of T-cell immunoglobulin and mucin
domain 3 (Tim-3), a Th1-specific type 1 membrane protein, Gal-9 is
an important inhibitory immune molecule (9). Binding of Gal-9 to Tim-3 causes an
inhibitory signal that results in the apoptosis of effector cells,
negatively regulates Th1-type immunity and induces tumor immune
tolerance and immune evasion (10,11).
Subsequently, Gal-9 was identified to be ubiquitously expressed in
a variety of tumor tissues and cell lines and its expression level
was closely related with malignant tumor prognosis (12).
microRNAs (miRNAs) are a class of non-coding
single-stranded RNA molecules with ~22–24 nucleotides, which bind
to the 3′-untranslated region (3′UTR) of target mRNAs by base
pairing. miRNAs regulate the expression of target genes through
post-transcriptional inhibition or induced degradation (13–15).
By inhibiting the protein synthesis of multiple targets, miRNAs
play an important role in many cellular pathways (16,17).
Moreover, recent evidence shows that miRNAs act as tumor-suppressor
genes or oncogenes in human cancers (18). The abnormal expression of miRNAs may
affect the development and progression of liver cancer.
However, to the best of our knowledge, no previous
study has aimed to determine the mechanism of Gal-9 and miRNAs
involved in liver cancer. We hypothesized that there might be
miRNAs that regulate the expression of Gal-9 and affect the
Tim3/Gal-9 pathway influencing the immune response and tumor
progression in liver cancer. In the present study, we found that 4
miRNAs (miR-22, 296-3p, 455-5p and 491-5p) were potential
regulators of Gal-9 and confirmed that miR-22 may directly inhibit
Gal-9 expression and cause lymphocyte apoptosis and tumor cell
proliferation. Our results suggest that miR-22 has potential
therapeutic value for the treatment of liver cancer.
Materials and methods
Tissue collection
Primary human liver cancer tissue and corresponding
adjacent tissue (<2 cm away from the tumor area) samples were
obtained from 10 patients who underwent primary surgical resection
of liver cancer at the Department of Hepatobiliary Surgery, Union
Hospital, Tongji Medical College, Huazhong University of Science
and Technology. All participants provided written informed consent,
and the study protocols were approved by the hospital ethics
committee. Medical records of the patients, including age and
gender, tumor staging, pathological diagnosis and surgical records
were collected. The tissues were immediately snap-frozen in liquid
nitrogen and stored at −80°C until total RNA was extracted.
Cell culture
Normal hepatocyte cell line Lo2 and human
hepatocellular carcinoma cell lines HepG2 and SMMC7721 were
purchased from the Chinese Institute of Biochemistry and Cell
Biology and cultured in RPMI-1640 medium (Hyclone, Logan, Utah,
USA) containing 10% fetal bovine serum (FBS) (Gibco, Grand Island,
NY, USA), 1 U/ml penicillin and 1 µg/ml streptomycin
(Gibco). Cells were grown in a humidified atmosphere of 5% carbon
dioxide at 37°C. All cell lines were authenticated and
characterized by the cell bank. The cells were expanded immediately
and multiple aliquots were cryopreserved. Cells were used within 6
months of resuscitation
Quantitative real-time PCR (qRT-PCR)
Total RNA and small RNA were extracted from the cell
lines and tissues using TRIzol® reagent (Invitrogen,
Carlsbad, CA, USA). The purity and integrity of the RNA sample were
assessed using a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo
Scientific, Waltham, MA, USA), and 500 ng RNA was transcribed into
cDNA using the PrimeScript™ RT reagent kit (Takara, Dalian, China)
according to the manufacturer's instructions. The obtained cDNA was
used as a template to perform PCR amplification using the
SYBR® Premix Ex Taq™ II kit (Takara). Human Gal-9,
GAPDH, miRNAs and U6 mRNA levels were analyzed using qRT-PCR with
the Mx3000P system (Agilent Technologies, Santa Clara, CA, USA).
GAPDH was used as an internal control for Gal-9 detection and U6
for miRNA detection. Each 20 µl reaction system consisted of
2 µl of cDNA, 10 µl SYBR® Premix Ex Taq™
II, 10 µmol/l of both sense and antisense primers. All PCR
reactions were carried out as follows: initial denaturation at 95°C
for 30 sec, followed by 40 cycles of denaturation at 95°C for 5
sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec.
Each experiment contained at least three replicates, and the
results were calculated according to the method 2−ΔΔCT.
The PCR primers used in this study are available upon request and
are listed in Table I.
 | Table IPCR primers used in the present
study. |
Table I
PCR primers used in the present
study.
| Genes | Primer sequences
(5′ to 3′) |
|---|
| Gal-9 | F:
CGCCCCTGGACAGATGTT
R: GACAGGAGGATGGACTTGGA |
| GAPDH | F:
GAAGGTGAAGGTCGGAGTC
R: GAAGATGGTGATGGGATTTC |
| miR-22 | RT:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGTTC
F: GCCGAGGGTTGGGTGGAG |
| miR-296-3p | RT:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGAGAGC
F: GCCGAGGGTTGGGTGGAG |
| miR-455-5p | RT:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGATGTA
F: GCCGTATGTGCCTTTGGACT |
| miR-491-5p | RT:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCTCATG
F: GCCGAGTGGGGAACCCTT |
| U6 | RT:
AACGCTTCACGAATTTGCGT
F: CTCGCTTCGGCAGCACA
R: AACGCTTCACGAATTTGCGT
aR: GTGCAGGGTCCGAGGT |
Western blot analysis
Protein was extracted from the cells using RIPA
lysis buffer containing EDTA-free Complete Protease Inhibitors
(Roche, Mannheim, Germany). After the protein concentration was
measured, it was mixed with SDS loading buffer, separated by 10%
SDS-PAGE and transferred onto nitrocellulose membranes. To block
nonspecific binding, the membranes were incubated at room
temperature for 1 h with 5% skim milk powder, followed by an
overnight incubation at 4°C with the primary antibodies anti-Gal-9
(ImmunoWay Biotechnology Company, Newark, DE, USA) or anti-β-actin
(Cell Signaling Technology, Boston, MA, USA) and the blots were
incubated with horseradish peroxidase-labeled secondary antibodies
(Cell Signaling Technology). Signals were visualized by ECL
chemiluminescence. Changes in protein expression were quantified
using ImageJ software (National Institutes of Health, Bethesda, MD,
USA). Equal protein loading was assessed by expression of
β-actin.
Target prediction
Bioinformatic analysis was carried out using
specific programs: miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/).
Plasmid construction, miRNA synthesis and
transfection
The full-length Gal-9 3′UTR containing a putative
miR-22 binding site was amplified by PCR from genomic DNA and
cloned at the EcoRI and HindIII sites into the pCMV
vector. The specific primers for the wild-type Gal-9 3′UTR vector
were as follows: 5′-ATAGAATTCGCGGCTTCCTGGCCCTG-3′ and
5′-CGCAAGCTTTGAATGTGCCAACAAGCA-3′. The specific primers for the
mutant-type Gal-9 3′UTR vector were as follows:
5′-ATAGAATTCGCGGCTTCCTGGCGGAC-3′ and
5′-CGCAAGCTTTGAATGTGCCAACAGCGC-3′. Wild-type and mutant-type
inserts were confirmed by sequencing. For the expression of Gal-9,
the genomic fragment of Homo sapiens Gal-9 precursor was
amplified by PCR using the primer pairs:
5′-GGAGAATTCGAGATGGCCTTCAGCGGTTCCCAG-3 and
5′-CCACTCGAGCGCCTATGTCTGCACATGGGTCAG-3′. The PCR product was cloned
into pcDNA3.1 at the EcoRI and XhoI sites and named
the Gal-9/pcDNA3.1 vector. miR-22-mimic, miR-491-mimic and
miR-control mimic (negative control) were synthesized by Guangzhou
RiboBio Co., Ltd. (Guangzhou, China). Cells were transiently
transfected by use of Lipofectamine 2000 transfection reagent
(Invitrogen, Carlsbad, CA, USA) for 24 h, according to the
manufacturer's protocol.
Luciferase reporter gene assay
HepG2 cells were plated in a 96-well plate and
co-transfected with miRNA mimics or control mimics with pCMV-Gal-9
3′UTR-WT or pCMV-Gal-9 3′UTR-MU and PRL-TK (Promega, Beijing,
China), using Lipofectamine 2000. Cells were collected 24 h after
transfection and luciferase activity was analyzed using the
Dual-Luciferase Reporter Assay System Modulus™ Single Tube
Multimode Reader (Turner BioSystems, Madison, WI, USA). The pRL-TK
vector that provided the constitutive expression of Renilla
luciferase was co-transfected as an internal control to correct for
differences in both transfection and harvest efficiencies.
Transfections were carried out in duplicates and repeated at least
twice in independent experiments.
Isolation of peripheral blood mononuclear
cells (PBMCs) and co-culture experiments of PBMCs and liver cancer
cells
PBMCs were isolated from the venous blood of healthy
blood donors. Aliquots of 3 ml of blood were diluted in 3 ml PBS
and layered on 6 ml Lymphocyte Separation Media (MD Pacific,
Tianjin, China). After centrifugation at 2,000 rpm for 20 min at
room temperature, the PBMCs were recovered from the interphase and
washed twice with serum-free RPMI-1640 medium (Hyclone, Logan,
Utah, USA) and centrifuged at 1,000 rpm for 10 min. PBMCs were
resuspended in complete culture medium and adjusted to
3×106 cells/ml.
For the co-culture experiments, HepG2 liver cancer
cells were washed with PBS, diluted in complete medium and
transferred to the wells. Transfections were performed using
Lipofectamine 2000. After a 24-h transfection, PBMCs were plated in
each well for co-culture. After 48 h, the cancer cells and the
PBMCs were separated for further analysis.
Cell proliferation assay
The WST-1 assay was used to evaluate cell
proliferation. After a 48-h co-culture, 10 µl/well cell
proliferation reagent WST-1 (Roche, Mannheim, Germany) was added
and incubated for 2 h. The cells were shaken thoroughly for 1 min
on a shaker and the absorbance was measured using an ELX808
Universal Microplate Reader (Bio Tek, Winooski, VT, USA). The
wavelength for measuring the absorption of the formazan product was
450 nm. All samples were performed in triplicate.
Flow cytometric analysis of cell
apoptosis
A cell apoptosis assay was performed using an
Annexin-V-Fluos staining kit (Roche, Mannheim, Germany). After
co-culture of the PBMCs and HepG2 cells for 48 h, PBMCs were
collected and centrifuged at 1,000 rpm for 5 min and washed with
PBS twice. The cell pellet was resuspended in 100 µl of
Annexin-V-Fluos labeling solution (Roche) and incubated 10–15 min
at 15–25°C. All specimens were assessed on a FACS Calibur flow
cytometer (BD, Franklin Lake, NJ, USA), and data were analyzed with
FlowJo software.
Statistical analysis
The statistical significance was determined using
the Student's t-test. All data are expressed as the mean ± SD. A
P-value <0.05 was considered to indicate a statistically
significant result. All experiments were repeated more than three
times and each experiment consisted of triple wells. One
representative experiment was selected to construct diagrams and
for data analysis.
Results
Expression of Gal-9 in the liver cancer
cell lines
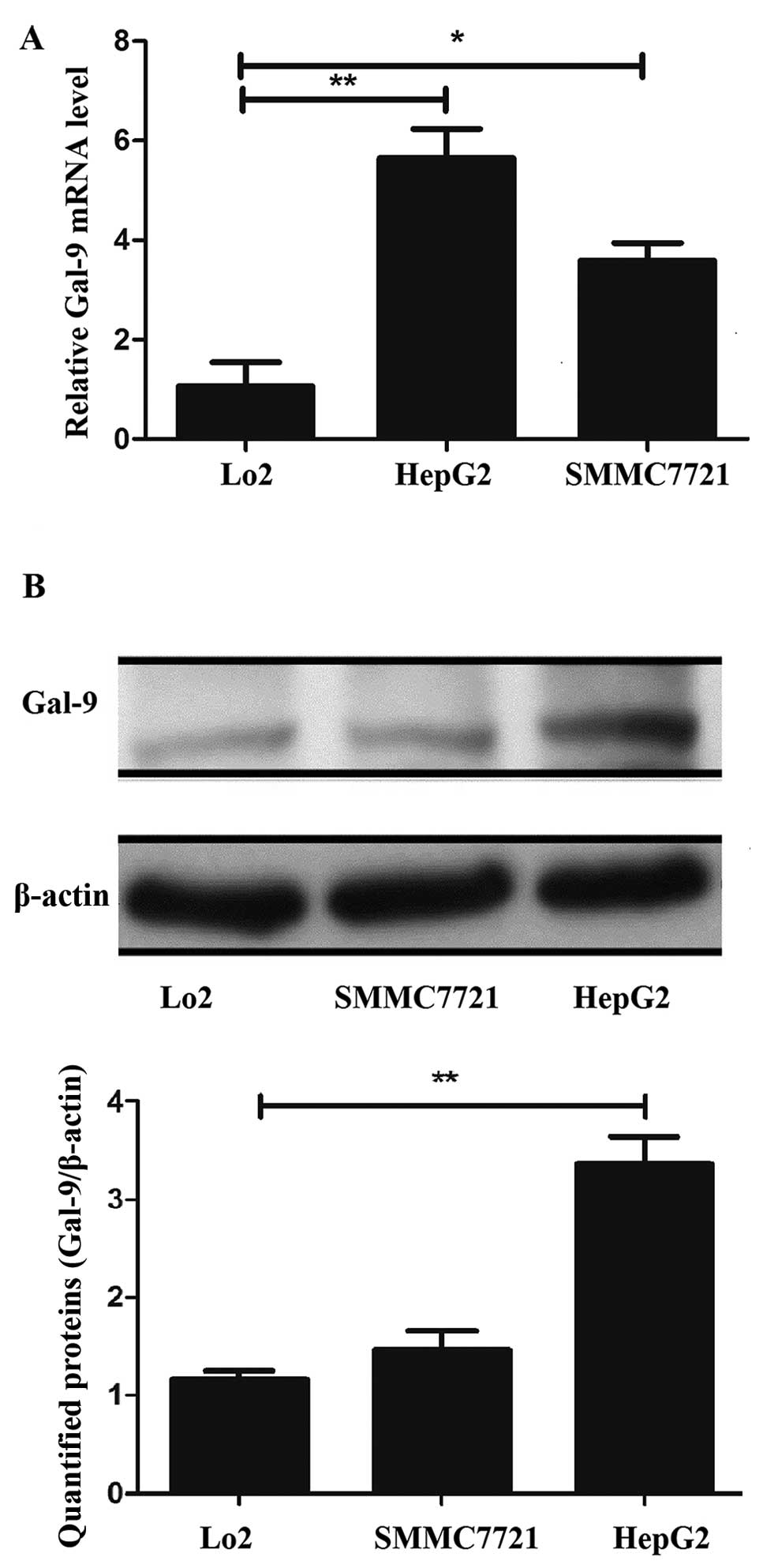
To identify the expression levels of the
Gal-9 mRNA and protein in normal liver and liver cancer
cells, we performed qRT-PCR and western blot analysis in Lo2 cells
as well as in HepG2 and SMMC7721 cells. The Gal-9 mRNA level
in HepG2 cells was ~5.3-fold higher and the level in the SMMC7721
was 2.4-fold higher than that in the Lo2 cells. Furthermore, the
Gal-9 protein level in the HepG2 cells was ~2-fold higher
than that in the Lo2 cells. The results revealed that Gal-9 was
significantly upregulated in the liver cancer cells at the mRNA and
protein levels compared to these levels in the normal liver cells
(p<0.05, Fig. 1A and B),
particularly in the HepG2 cells.
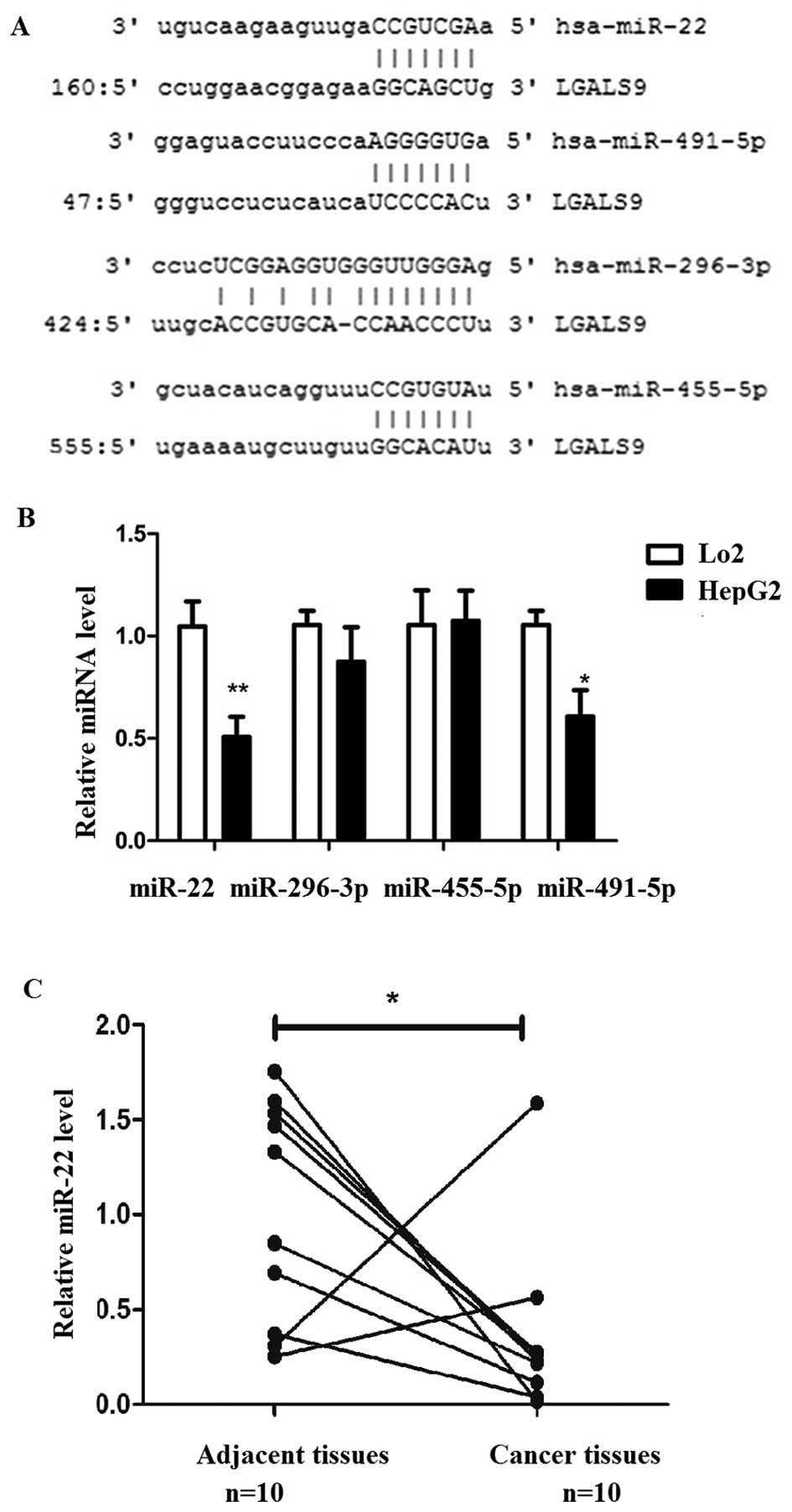
Identification of candidate miRNAs
targeting Gal-9 and investigation of their expression levels in
human liver cancer tissues and cell lines
Target prediction programs miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/) were used to predict and
identify miRNAs that possibly target Gal-9 in liver cancer. The
overlapping prediction analysis suggested that 4 miRNAs (miR-22,
296-3p, 455-5p and 491-5p) were potential regulators of
Gal-9 (Fig. 2A).
We determined the expression of these 4 miRNAs in
normal liver (Lo2) and liver cancer (HepG2) cells. The results
showed that miR-22 and miR-491 were significantly down-regulated in
liver cancer cells than these miRNAs in normal liver cells,
particularly miR-22 (Fig. 2B).
miR-22 was focused on and its expression level was determined in
liver cancer and corresponding adjacent tissues. miR-22 was
downregulated in liver cancer tissues compared to that in the
corresponding adjacent tissues (Fig.
2C).
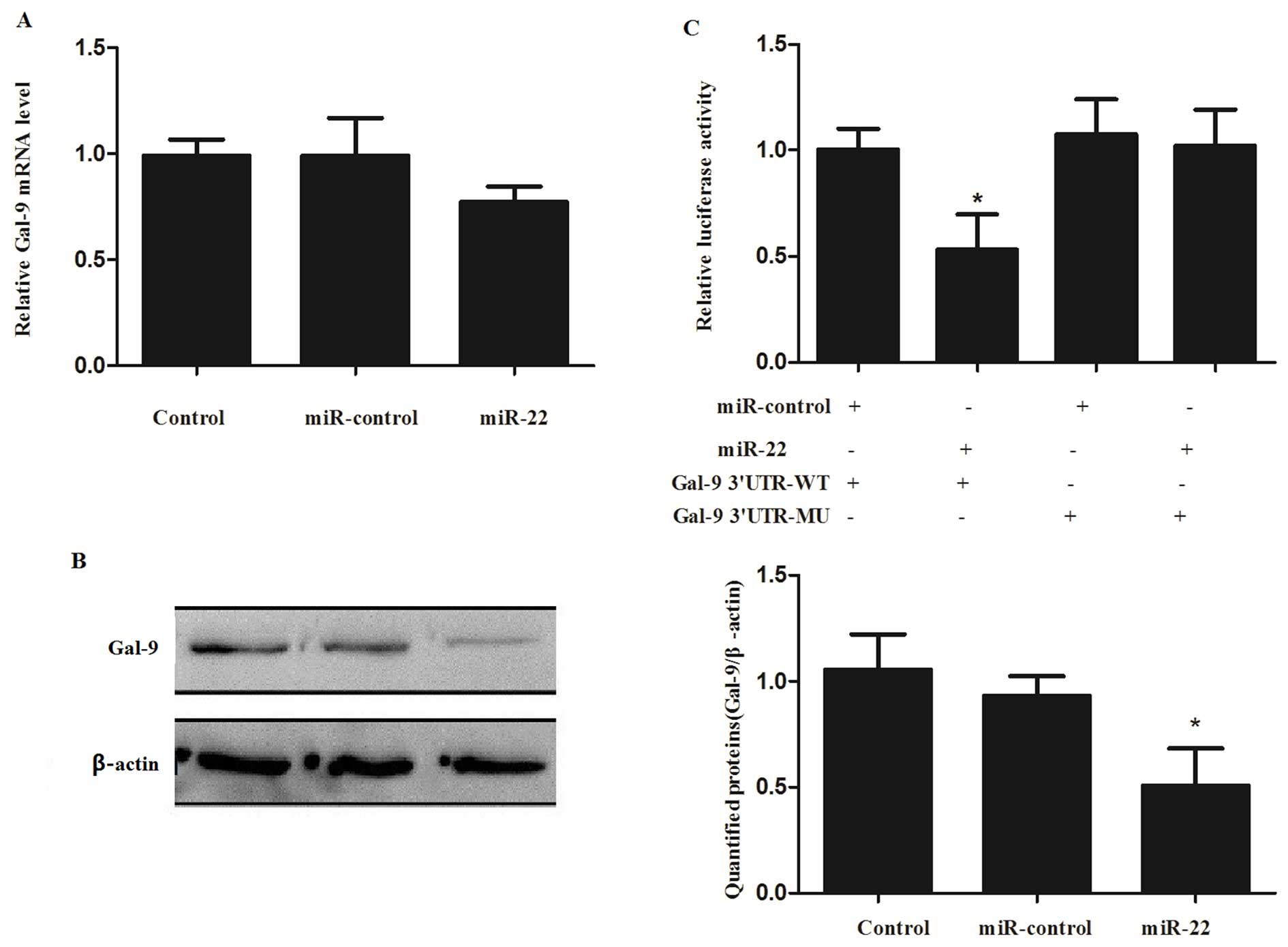
Direct inhibition of miR-22 on the
expression of Gal-9 via binding to the specific target site in
Gal-9 3′UTR
To investigate whether the expression of
Gal-9 is regulated by miR-22, the miR-control or miR-22
mimic were transfected into HepG2 cells and the expression levels
of Gal-9 mRNA and protein were detected using qRT-PCR and
western blot analysis, respectively. We found that the expression
of Gal-9 mRNA was slightly decreased, but did not reach a
significant level (Fig. 3A).
However, the expression of Gal-9 protein was significantly
downregulated after transfection of the miR-22 mimics (Fig. 3B) compared with the negative control
and miR-control.
To further investigate the mechanisms underlying the
regulation of Gal-9 by miR-22, the 3′UTR of Gal-9 was
cloned into a reporter vector linking the luciferase open reading
frame downstream to generate pCMV-Gal-9-3′UTR wild-type (Gal-9
3′UTR-WT) and pCMV-Gal-9-3′UTR mutant-type (Gal-9 3′UTR-MU). The
two plasmids were transfected into HepG2 cells, respectively,
together with the miR-control or miR-22 mimic, respectively. The
luciferase activity was measured after a 24-h transfection. The
luciferase activity in the Gal-9 3′UTR-WT group, but not in the
Gal-9 3′UTR-MU, was significantly reduced in the cells
co-transfected with the miR-22 mimic when compared with the
miR-control (P=0.0268, Fig.
3C).
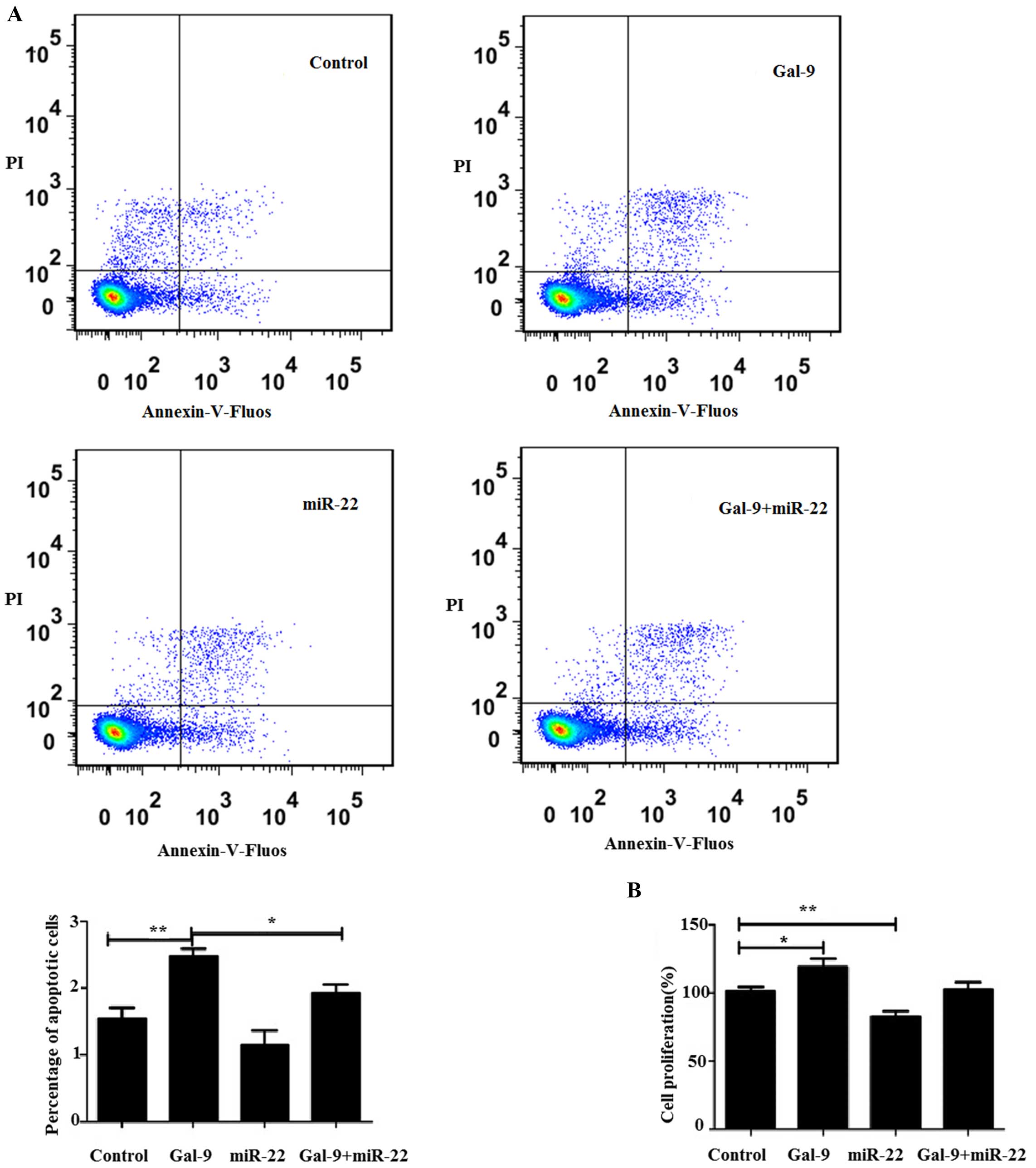
Influence of the miR-22/Gal-9 axis on
tumor cell proliferation and lymphocyte apoptosis
To determine whether the miR-22/Gal-9 axis affects
lymphocyte and tumor cell apoptosis, HepG2 cells were divided into
four groups: negative control, cells transfected with Gal-9
overexpression vector only, cells transfected with the miR-22
mimics only and cells co-transfected with the Gal-9
overexpression vector and miR-22 mimics. After a 24-h transfection,
the liver cancer cells were co-cultured with PBMCs for 48 h and the
apoptosis of PBMCs was detected. Our results showed that the
apoptosis rate of the PBMCs co-cultured with the HepG2 cells
transfected with the Gal-9 overexpression vector was higher
than that of the negative control. The apoptosis rate of the PBMCs
co-cultured with the HepG2 cells co-transfected with miR-22 mimics
and the Gal-9 overexpression vector was lower than that of
the cells transfected with Gal-9 only (Fig. 4A).
In order to further examine the role of the
miR-22/Gal-9 axis in tumor cell proliferation, a WST-1 assay
was used. The results showed that HepG2 cells transfected with the
Gal-9 overexpression vector exhibited higher proliferation
compared with the negative control. However, HepG2 cells
transfected with the miR-22 mimics only exhibited slower
proliferation when compared with the negative control. While HepG2
cells co-transfected with the miR-22 mimics and the Gal-9
overexpression vector showed no significantly different cell
proliferation compared to the negative control (Fig. 4B).
Discussion
The interaction between tumor cells and the tumor
microen-vironment contributes to the development and progression of
cancers (19). In the liver cancer
microenvironment, accompanied by extensive lymphocyte infiltration,
there is also a large amount of inhibitory factors, which cause the
effector T cells to dysfunction and finally leading to immune
escape of hepatic carcinoma cells (5,20,21).
Tim-3, an important inhibitory receptor, plays an important role in
these processes (22,23).
Gal-9 is one of the specific ligands of Tim-3 and
has been found extracellularly as well as intracellularly, both in
the nucleus and in the cytoplasm (8,24).
Gal-9 expression is widely distributed in tissues involved in the
immune system, such as the spleen, thymus and peripheral blood
lymphocytes, and in tissues of endodermal origin, such as the
liver, intestine, stomach and lung (25,26).
Until recently several studies have shown that expression of
Gal-9 varies in tumor cells when compared with their normal
counterparts. For example, breast, lung and melanoma cancer cell
lines showed low or absent Gal-9 expression (27,28),
while leukemia and colon cancer cell lines showed high expression
of Gal-9 (29). However, little
research has been performed to study Gal-9 expression in liver
cancer and normal liver cells. In the present study, we first
determined the expression of Gal-9 in human hepatocellular
carcinoma cell lines (HepG2 and SMMC7721) and in a normal
hepatocyte cell line (Lo2). The results showed that HepG2, SMMC7721
and Lo2 all expressed Gal-9 and Gal-9 was also found
to have high expression in human liver cancer cell lines.
In recent years, an important role has been
discovered for Gal-9 in health and disease (7). Subsequent studies have revealed that
Gal-9 modulates a variety of biological functions, such as
cell aggregation and adhesion, apoptosis of tumor cells and others
(8,30). The interaction between Gal-9 and
Tim-3 expressed on Th1 cells negatively regulates Th1-mediated
immune responses (31,32). Furthermore, some studies have
revealed that Gal-9 is implicated in the immune escape of tumors
through the induction of tumor-specific Tim3+ T-cell
death (33). In the present study,
we investigated the effect of Gal-9 on lymphocyte apoptosis
and tumor cell proliferation when they were co-cultured in liver
cancer cells. Our results showed that the apoptosis rate of PBMCs
co-cultured with HepG2 cells transfected with the Gal-9
overexpression vector was higher than that of the negative control,
while HepG2 cells transfected with the Gal-9 overexpression vector
only exhibited higher proliferation compared with the negative
control. These results suggest that in liver cancer, the binding of
Gal-9 to Tim3 may induce lymphocyte apoptosis and tumor cell immune
escape.
To investigate whether the expression of
Gal-9 is regulated by miRNAs, we predicted four miRNAs
(miR-22,296-3p, 455-5p and 491-5p) as candidate regulators of
Gal-9 and examined their expression levels. miR-22 was
significantly downregulated in both liver cancer cells and tissues.
miR-22, originally identified in HeLa cells, is a type of highly
evolutionarily conserved miRNA. Recent studies have shown that
miR-22 is an important regulator of cancer oncogenesis and tumor
behavior. Previous literature has reported that miR-22 is
overexpressed in prostate cancer, but is downregulated in breast
cancer, cholangiocarcinoma, multiple myeloma, and HCC (34). Our results showed that miR-22 was
significantly downregulated in both liver cancer cells and tissues,
consistent with previous results. Luciferase reporter gene assay
and detection of Gal-9 mRNA and protein expression after
transfection with the miR-22-mimics confirmed that miR-22 directly
targets Gal-9 and inhibits the expression of Gal-9 via
binding to the specific target site in the Gal-9 3′UTR.
We also observed the effect of the miR-22/Gal-9 axis
on lymphocyte exhaustion and apoptosis, as well as tumor cell
proliferation and immune evasion. The results showed that the
apoptosis rate of PBMCs co-culture with HepG2 cells co-transfected
with the miR-22 mimics and the Gal-9 overexpression vector
was lower than that of the cells transfected with Gal-9 only, while
HepG2 cells transfected with the miR-22 mimics only exhibited
slower proliferation when compared with the negative control and
HepG2 cells co-transfected with the miR-22 mimics and Gal-9
overexpression vector showed no significant difference in cell
proliferation compared to the negative control. The explanation of
these phenomena are as follows. Following transfection with miR-22,
the expression level of Gal-9 is decreased, disrupting the
interaction between Tim-3 and Gal-9 and preventing lymphocytes from
apoptosis and partly recovering effector T-cell function and
tempering tumor immune response, reducing tumor cell proliferation
and immune escape. It may be a novel immune therapeutic target for
treating patients with liver cancer
In the present study, we revealed that miR-22
suppresses the expression of Gal-9 and influences T cell
function and liver cancer cell immune evasion by affecting the
Tim3/Gal-9 pathway. These results would be helpful to investigate
the molecular mechanisms underlying liver cancer development from a
new perspective and identify novel therapeutic targets for the
prevention and treatment of liver cancer.
References
|
1
|
Yi X, Luk JM, Lee NP, Peng J, Leng X, Guan
XY, Lau GK, Beretta L and Fan ST: Association of mortalin (HSPA9)
with liver cancer metastasis and prediction for early tumor
recurrence. Mol Cell Proteomics. 7:315–325. 2008. View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Uchino K, Tateishi R, Shiina S, Kanda M,
Masuzaki R, Kondo Y, Goto T, Omata M, Yoshida H and Koike K:
Hepatocellular carcinoma with extrahepatic metastasis: Clinical
features and prognostic factors. Cancer. 117:4475–4483. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagahara K, Arikawa T, Oomizu S, Kontani
K, Nobumoto A, Tateno H, Watanabe K, Niki T, Katoh S, Miyake M, et
al: Galectin-9 increases Tim-3+ dendritic cells and
CD8+ T cells and enhances antitumor immunity via
galectin-9-Tim-3 interactions. J Immunol. 181:7660–7669. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fujihara S, Mori H, Kobara H, Rafiq K,
Niki T, Hirashima M and Masaki T: Galectin-9 in cancer therapy.
Recent Pat Endocr Metab Immune Drug Discov. 7:130–137. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wiersma VR, de Bruyn M, Helfrich W and
Bremer E: Therapeutic potential of Galectin-9 in human disease. Med
Res Rev. 33(Suppl 1): E102–E126. 2013. View Article : Google Scholar
|
|
8
|
Hirashima M, Kashio Y, Nishi N, Yamauchi
A, Imaizumi TA, Kageshita T, Saita N and Nakamura T: Galectin-9 in
physiological and pathological conditions. Glycoconj J. 19:593–600.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, He W, Zhou H, Yuan J, Wu K, Xu L
and Chen ZK: The Tim-3 ligand galectin-9 negatively regulates
CD8+ alloreactive T cell and prolongs survival of skin
graft. Cell Immunol. 250:68–74. 2007. View Article : Google Scholar
|
|
10
|
Freeman GJ, Casasnovas JM, Umetsu DT and
DeKruyff RH: TIM genes: A family of cell surface phosphatidylserine
receptors that regulate innate and adaptive immunity. Immunol Rev.
235:172–189. 2010.PubMed/NCBI
|
|
11
|
Sharma S, Sundararajan A, Suryawanshi A,
Kumar N, Veiga-Parga T, Kuchroo VK, Thomas PG, Sangster MY and
Rouse BT: T cell immunoglobulin and mucin protein-3
(Tim-3)/Galectin-9 interaction regulates influenza A virus-specific
humoral and CD8 T-cell responses. Proc Natl Acad Sci USA.
108:19001–19006. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Heusschen R, Griffioen AW and Thijssen VL:
Galectin-9 in tumor biology: A jack of multiple trades. Biochim
Biophys Acta. 1836:177–185. 2013.PubMed/NCBI
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorige-nicity, and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.
|
|
19
|
Cosse JP and Michiels C: Tumour hypoxia
affects the responsiveness of cancer cells to chemotherapy and
promotes cancer progression. Anticancer Agents Med Chem. 8:790–797.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fourcade J, Sun Z, Benallaoua M, Guillaume
P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V and Zarour HM:
Upregulation of Tim-3 and PD-1 expression is associated with tumor
antigen-specific CD8+ T cell dysfunction in melanoma
patients. J Exp Med. 207:2175–2186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fourcade J, Kudela P, Sun Z, Shen H, Land
SR, Lenzner D, Guillaume P, Luescher IF, Sander C, Ferrone S, et
al: PD-1 is a regulator of NY-ESO-1-specific CD8+ T cell
expansion in melanoma patients. J Immunol. 182:5240–5249. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hastings WD, Anderson DE, Kassam N,
Koguchi K, Greenfield EA, Kent SC, Zheng XX, Strom TB, Hafler DA
and Kuchroo VK: TIM-3 is expressed on activated human
CD4+ T cells and regulates Th1 and Th17 cytokines. Eur J
Immunol. 39:2492–2501. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kikushige Y, Shima T, Takayanagi S, Urata
S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki
Y, et al: TIM-3 is a promising target to selectively kill acute
myeloid leukemia stem cells. Cell Stem Cell. 7:708–717. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thijssen VL, Hulsmans S and Griffioen AW:
The galectin profile of the endothelium: Altered expression and
localization in activated and tumor endothelial cells. Am J Pathol.
172:545–553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Türeci O, Schmitt H, Fadle N, Pfreundschuh
M and Sahin U: Molecular definition of a novel human galectin which
is immu-nogenic in patients with Hodgkin's disease. J Biol Chem.
272:6416–6422. 1997. View Article : Google Scholar
|
|
26
|
Wada J, Ota K, Kumar A, Wallner EI and
Kanwar YS: Developmental regulation, expression, and apoptotic
potential of galectin-9, a beta-galactoside binding lectin. J Clin
Invest. 99:2452–2461. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Irie A, Yamauchi A, Kontani K, Kihara M,
Liu D, Shirato Y, Seki M, Nishi N, Nakamura T, Yokomise H, et al:
Galectin-9 as a prognostic factor with antimetastatic potential in
breast cancer. Clin Cancer Res. 11:2962–2968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kageshita T, Kashio Y, Yamauchi A, Seki M,
Abedin MJ, Nishi N, Shoji H, Nakamura T, Ono T and Hirashima M:
Possible role of galectin-9 in cell aggregation and apoptosis of
human melanoma cell lines and its clinical significance. Int J
Cancer. 99:809–816. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lahm H, André S, Hoeflich A, Fischer JR,
Sordat B, Kaltner H, Wolf E and Gabius HJ: Comprehensive galectin
fingerprinting in a panel of 61 human tumor cell lines by RT-PCR
and its implications for diagnostic and therapeutic procedures. J
Cancer Res Clin Oncol. 127:375–386. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Asakura H, Kashio Y, Nakamura K, Seki M,
Dai S, Shirato Y, Abedin MJ, Yoshida N, Nishi N, Imaizumi T, et al:
Selective eosinophil adhesion to fibroblast via IFN-gamma-induced
galectin-9. J Immunol. 169:5912–5918. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu C, Anderson AC, Schubart A, Xiong H,
Imitola J, Khoury SJ, Zheng XX, Strom TB and Kuchroo VK: The Tim-3
ligand galectin-9 negatively regulates T helper type 1 immunity.
Nat Immunol. 6:1245–1252. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sánchez-Fueyo A, Tian J, Picarella D,
Domenig C, Zheng XX, Sabatos CA, Manlongat N, Bender O, Kamradt T,
Kuchroo VK, et al: Tim-3 inhibits T helper type 1-mediated auto-
and allo-immune responses and promotes immunological tolerance. Nat
Immunol. 4:1093–1101. 2003. View
Article : Google Scholar
|
|
33
|
Li H, Wu K, Tao K, Chen L, Zheng Q, Lu X,
Liu J, Shi L, Liu C, Wang G, et al: Tim-3/galectin-9 signaling
pathway mediates T-cell dysfunction and predicts poor prognosis in
patients with hepatitis B virus-associated hepatocellular
carcinoma. Hepatology. 56:1342–1351. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu
S, Wu M, Pan Z and Zhou W: microRNA-22, downregulated in
hepatocellular carcinoma and correlated with prognosis, suppresses
cell proliferation and tumourigenicity. Br J Cancer. 103:1215–1220.
2010. View Article : Google Scholar : PubMed/NCBI
|