Introduction
Mesenchymal stem cells (MSCs) are defined as cells
possessing the ability of self-renewal, proliferation and
differentiation into multiple lineages, including chondrocytes,
osteoblasts and adipocytes (1).
MSCs are known to have homing properties and can repair injured
tissues; thus, MSCs demonstrate potential as therapy for organ
defects, regeneration or certain other conditions, including bone
defects (2,3), hair loss (4), liver fibrosis (5), pancreatic islet transplantation,
immunomodulation (6) and neuronal
regeneration (7). MSCs can be found
in various tissues, including bone marrow, umbilical cord blood,
placenta and fat (8). Among these
sites, adipose tissue is accessible and abundant for the isolation
of adult stem cells. Therefore, human adipose tissue-derived MSCs
(hATMSCs) obtained by relatively less invasive procedures are an
attractive source for cell therapy.
Our previous study indicated that systemic
transplantation of hATMSCs did not induce tumor development
(9). MacIsaac et al
(10) also showed that no
malignancies were detected in normal mice treated with hATMSCs.
However, in tumor models, MSCs have been reported to play a role in
the process of carcinogenesis, tumor growth and metastasis,
although numerous studies have reported contradicting results; some
investigators found that MSCs promote tumor growth and others
report that MSCs inhibit tumor growth (11). Maestroni et al (12) reported that mouse bone
marrow-derived mesenchymal stem cells (BM-MSCs) inhibited the
proliferation and growth of Lewis lung carcinoma and B16 melanoma.
It was also reported that hBM-MSCs showed a tumor-suppressive
effect in an experimental model of Kaposi's sarcoma (13). BM-MSCs also exhibited an inhibitory
effect on A549 tumor cells both in vivo and in vitro
experiments (14). In contrast, a
promoting effect of MSCs on tumor growth was observed in other
studies. Zhu et al (15)
subcutaneously injected hBM-MSCs along with colon cancer cells, and
promotion of angiogenesis and tumor growth were noted. Xu et
al (16) showed that hBM-MSCs
promoted the growth and metastasis of osteosarcoma. The
tumor-promoting effect of BM-MSCs was also demonstrated by Tsai
et al (17) in HT-29 cancer
cells.
Some mechanisms of MSC-related tumor supporting
effects have been reported, which include vascular (18) and tumor stromal support (19), immunosuppressive effects (20) and metastatic support (21). On the other hand, the tumor
inhibitory mechanisms of MSCs could be due to cell cycle arrest
(22), angiogenesis inhibition
(23) and regulation of certain
soluble factors, such as Wnt inhibitor, Dickkopf-related protein-1
(DKK-1), that may block tumor-related cell signaling pathways
(24). Despite these explanations
regarding the action of MSCs, the tumor modulation mechanisms of
MSCs are extremely complicated and difficult to demonstrate. Thus,
in order for the clinical use of MSCs as a novel cell therapy for
various conditions, it is critical to understand the interactions
between tumors and MSCs. In the present study, we investigated the
tumor-modulatory effects of hATMSCs in several types of human
cancer cells to clarify the roles of hATMSCs on human cancers and
to determine the underlying mechanisms.
Materials and methods
Tumor cell lines
Human malignant melanoma cell line A-375, human lung
adenocarcinoma cell line A549 and human colorectal adenocarcinoma
cell line HT-29 were purchased from ATCC (Manassas, VA, USA). Human
epidermoid carcinoma cell line A-431, human gastric carcinoma cell
line NCI-N87 and human pancreatic adenocarcinoma cell line Capan-1
were obtained from the Korean Cell Line Bank (Seoul, Korea). The
cell lines were cultured in RPMI-1640 supplemented with 10% fetal
bovine serum (FBS) and 0.03% antibiotic-antimycotic (all from
Gibco, Grand Island, NY, USA) at 37°C in a 5% CO2
humidified chamber.
Isolation and characterization of
hATMSCs
hATMSCs were prepared from surplus banked stem cells
under good manufacturing practice conditions (RNL Bio, Seoul,
Korea) and used as anonymized materials. In brief, human abdominal
subcutaneous fat tissue was obtained by simple liposuction
following informed consent from donors, digested with collagenase I
(1 mg/ml), and filtered through a 100-μm nylon sieve to
remove cellular debris and centrifuged at 470 g for 5 min. The
pellet was resuspended in RCME cell attachment medium (RNL Bio)
containing 10% FBS and cultured at 37°C. After 24 h, the cell
medium was changed to RKCM cell growth medium (RNL Bio) containing
5% FBS. When the cells reached 80–90% confluency, they were
subcultured and expanded in RKCM medium until passage 3. The
immunophenotypes of the hATMSCs were analyzed using flow cytometric
analysis. Trypsinized hATMSCs were suspended [1×106
hATMSCs/25 μl of phosphate-buffered saline (PBS)] and
stained with FITC-conjugated CD31, CD34 and CD45 antibodies and
PE-conjugated CD73 and CD90 antibodies (BD Pharmingen, San Diego,
CA, USA).
In vivo tumor xenograft models
Severe combined immunodeficient (SCID) and nude mice
can be used as human tumor xenograft models (25). Six-week old female BALB/c-nude mice
(Orient Bio Inc., Seongnam, Korea) and hairless SCID mice (Charles
River, Wilmington, MA, USA) were housed in an environmentally
controlled room (22±2°C, 40–60% humidity and a 12-h light cycle).
The present study was approved by the Institutional Review Board
(IRB) and the Institutional Animal Care and Use Committee (IACUC)
of the Biomedical Research Institute at Seoul National University
Hospital. Cancer cells (5×106) were subcutaneously
inoculated into the back of mice. The injection was made through
the subcutaneous layer of the cervicodorsal part of the animals.
When the tumor volume reached close to 100 mm3, the mice
were intratumorally injected with 1×105 hATMSCs
suspended in PBS or PBS once a week for 8 weeks (n=3–6/group). The
tumor size was measured twice a week using a caliper (Mitutoyo,
Tokyo, Japan), and the tumor volumes were calculated as π/6 ×
(volume × width × height) according to a previously described
method (26). After sacrifice at 8
weeks, the tumor masses were removed, weighed and fixed in 10%
neutral buffered formaldehyde solution for histological analysis or
preserved at −70°C.
In vitro co-culture and trypan blue
exclusion assay
A549 and HT-29 cancer cells (1×105) were
co-cultured with hATMSCs in RPMI-1640 medium with 10% FBS in 6-well
plates for 72 h. Indirect co-culture was conducted using Transwell
inserts (0.4-μm pore size, polycarbonate membrane; SPL,
Pocheon, Korea) in 6-well plates to prevent cell-to-cell contact.
All of the experiments were conducted in triplicate. Total cell
number was assessed by a trypan blue exclusion assay using a
hemocytometer under a microscope.
Western blot analysis
The proteins were extracted using RIPA lysis buffer
(Millipore, Bedford, MA, USA) containing 0.1% phosphatase inhibitor
cocktail (Sigma-Aldrich, St. Louis, MO, USA) and protease inhibitor
(Roche, Indianapolis, IN, USA). The protein concentrations were
determined using a BCA protein assay kit (Pierce Biotechnology,
Rockford, IL, USA). Protein samples were loaded for immunoblotting
using antibodies against phosphorylated nuclear factor κB (NF-κB)
p65, β-actin (Cell Signaling Technology, Beverly, MA, USA), p-JAK3,
p-STAT3 and β-catenin (Santa Cruz Biotechnology, Santa Cruz, CA,
USA). Densitometry of the target bands was analyzed using ImageJ
[National Center for Biotechnology Information (NCBI)].
RNA isolation and microarray
analysis
Total RNA from the tumors was isolated using Ambion
mirVana miRNA Isolation kit (Ambion, Austin, TX, USA). RNA quality
was assessed using an Agilent 2100 Bioanalyzer using the RNA 6000
Nano Chip (Agilent Technologies, Amstelveen, The Netherlands), and
the quantity was determined by the ND-1000 spectrophotometer
(Thermo Scientific, Wilmington, DE, USA). Affymetrix
GeneChip® Human Gene 2.0 ST Array (Affymetrix, Santa
Clara, CA, USA) was used to analyze differential gene expression
profiles as previously described (27). In order to determine whether genes
were differentially expressed among the groups, the genes that
showed >1.5-fold difference between the average signal values of
the control and test groups were selected. Additionally, genes with
a p-value <0.05 were extracted by Student's t-test (two-tailed,
unpaired). After significant genes were identified, the web-based
tool Database for Annotation, Visualization and Integrated
Discovery (DAVID) was used to perform the biological interpretation
of the differentially expressed genes. The genes were classified
based on the information of gene function in Gene Ontology (GO)
(http://david.abc.ncifcrf.gov/home.jsp).
Flow cytometric analysis
Cancer cells were analyzed by a flow cytometer
(FACSCalibur; Becton-Dickinson, San Jose, CA, USA) to assess levels
of proliferation, apoptosis and NF-κB expression. Apoptosis and
proliferation were evaluated using Annexin V-FITC Early Apoptosis
Detection kit (Cell Signaling Technology) and BrdU Flow kit (BD
Pharmingen), respectively, according to the manufacturer's
instructions. Harvested tumor cells were centrifuged, resuspended
in PBS and fixed in 2–4% formaldehyde. Phospho-NF-κB p65-Alexa
Fluor 647 conjugate (Cell Signaling Technology) antibody was used
for NF-κB analysis.
Statistical analysis
Statistical analysis of the results was performed
using one-way ANOVA (SPSS, Inc., Chicago, IL, USA). p-values of
<0.05 were considered to indicate statistically significant
results. If the variance was significant, the data were analyzed by
the multiple comparison procedure of Dunnett's test.
Results
Effects of the hATMSCs on the growth of
various types of tumors in vivo xenograft models
For immunophenotypic characterization of the
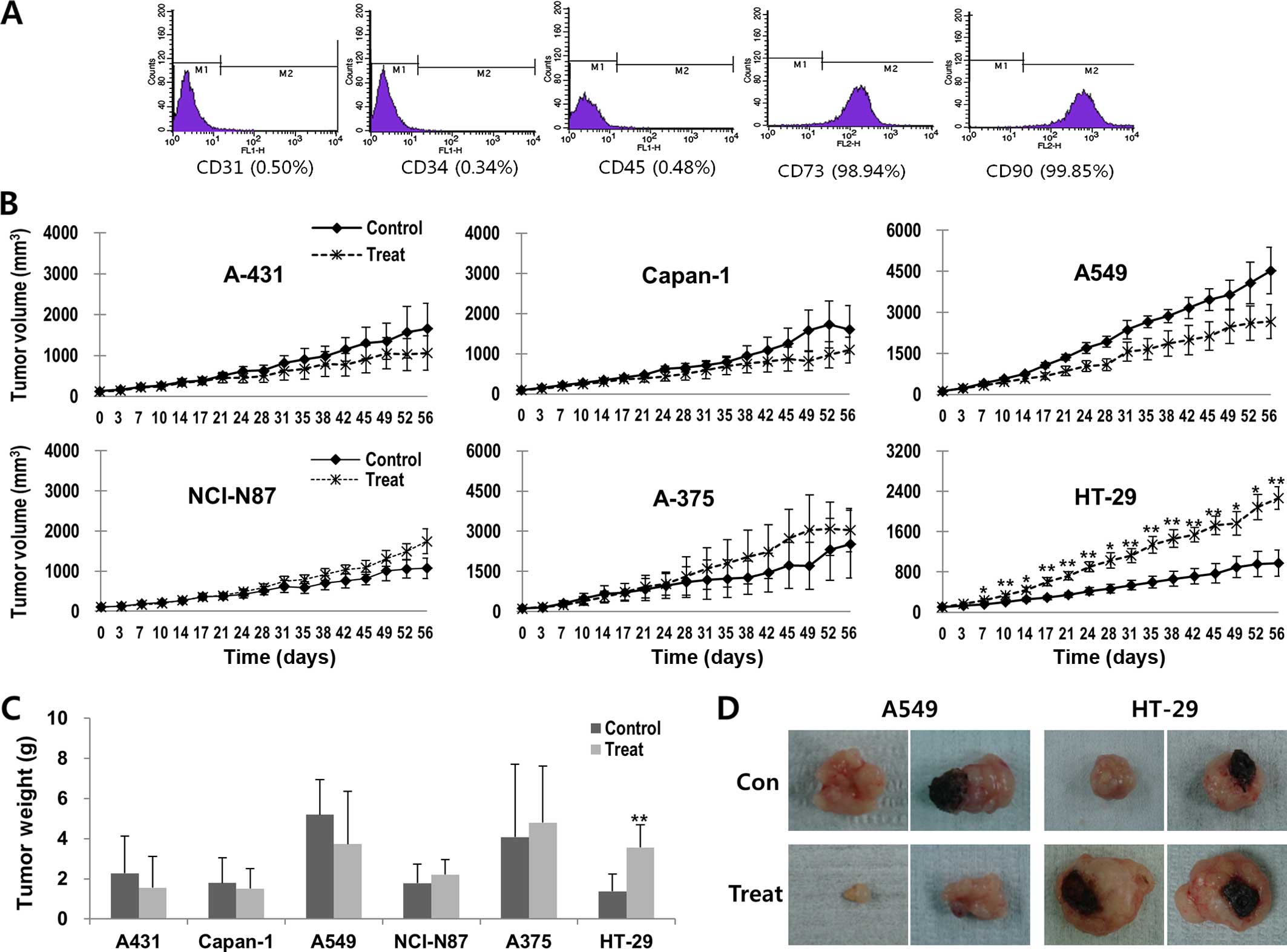
hATMSCs, surface marker expression was examined by flow cytometry.
The hATMSCs expressed high levels of MSC markers CD73 and CD90,
whereas the hATMSCs were negative for cell adhesion molecule marker
CD31 and hematopoietic stem cell markers CD34 and CD45 (Fig. 1A).
In Korea, the stomach, colon and lung have been
reported to be the five leading primary sites of cancer both in
males and females in 2013 (28). In
addition to xenograft models for these cancers, three well-known
xenografts, A-375 melanoma, skin squamous cell carcinoma A-431 and
Capan-1 pancreatic xenografts (29–31),
were used in order to investigate the dual effects of the hATMSCs
on tumor growth. The tumor size was measured for 8 weeks after the
first subcutaneous hATMSC injection in the BALB/c-nude mice
subcutaneously inoculated with the cancer cells. The rates of
growth of the A431, Capan-1 and A549 tumors were reduced in the
hATMSC-injected groups compared with the respective control groups,
although the differences did not reach statistical significance
(Fig. 1B). In contrast, NCI-N87 and
A375 tumors showed the tendency toward increased volume in the
hATMSC-injected groups in comparison to the respective control
groups. Particularly, the tumor weight of the HT-29 tumors in the
hATMSC group was significantly greater than that in the control
group. The differences in tumor weights between the hATMSC and the
respective control groups showed consistent patterns with the
changes in tumor volume (Fig. 1C and
D).
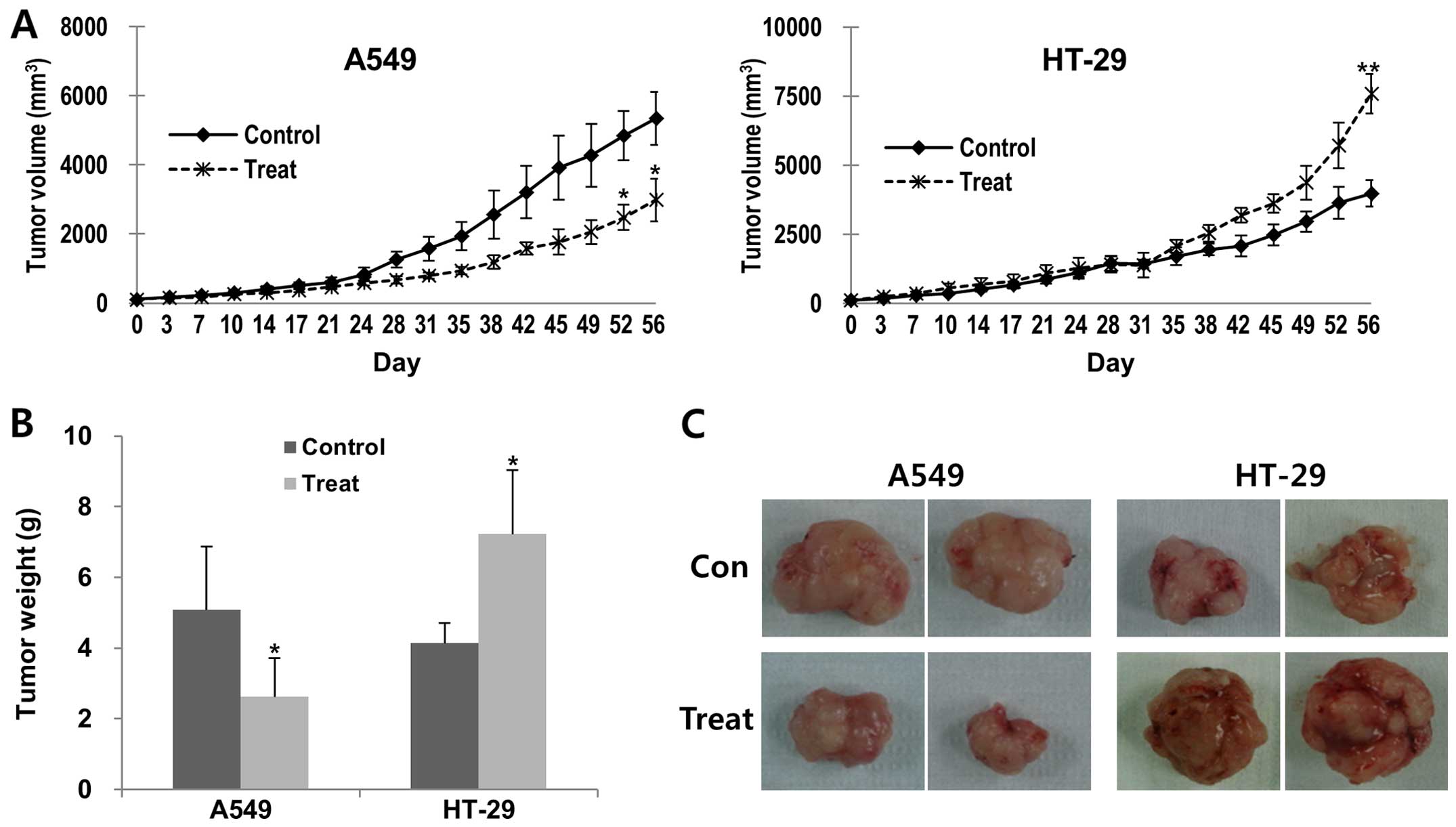
Based on the changes in mean volumes and weights by
the hATMSC injection in the BALB/c-nude mice, we selected the A549
and the HT-29 tumors for additional in vivo experiments
using hairless SCID mice, and confirmed similar dual effects of the
hATMSCs in these two types of tumors. The volume and weight of the
A549 tumors were significantly reduced in the hATMSC group compared
with the parameters in the control group, whereas the tumor volume
and weight of the hATMSC-treated HT-29 tumors were significantly
greater than the control group (Fig.
2).
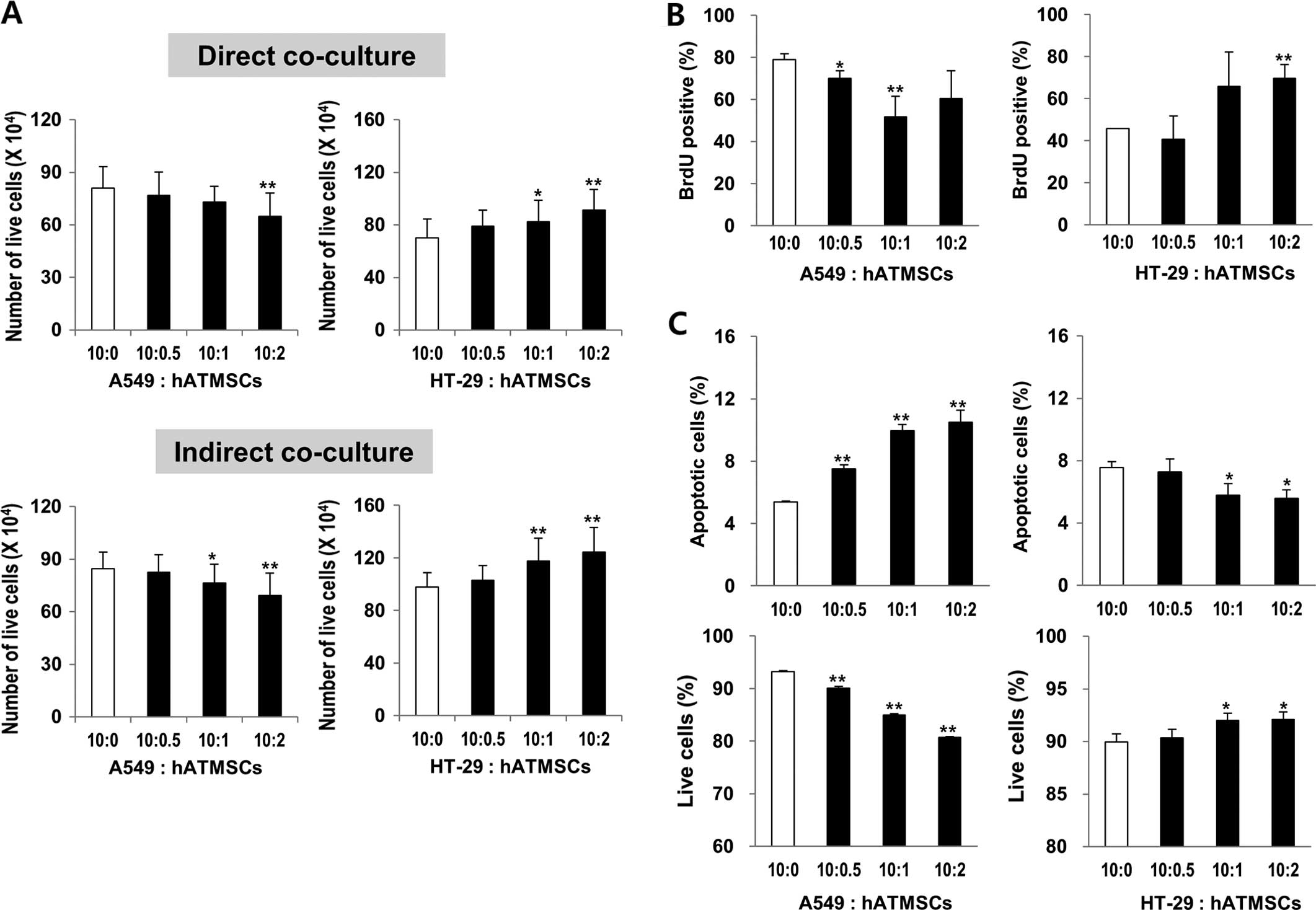
Co-culture effect of the hATMSCs on the
viability of cancer cell lines A549 and HT-29 in vitro
Tian et al (8) reported opposite effects of MSCs in
vivo and in vitro; human BM-MSCs exerted a tumor
inhibitory effect in vitro, yet surprisingly had a promoting
effect on tumor growth in vivo. To confirm the effects of
the hATMSCs on tumor growth in vivo, the A549 and HT-29
cells were co-cultured with the hATMSCs at different ratios of
10:0.5, 10:1 and 10:2 (Fig. 3A).
The direct co-culture of the A549 cells with the hATMSCs at the
ratios of 10:1 and 10:2 attenuated the proliferation rates of the
cells by 9.78 and 19.82%, respectively, compared to the control
group (10:0), whereas the direct co-culture of the HT-29 cells with
the hATMSCs at the ratios of 10:1 and 10:2 increased the
proliferation rates of the cells by 17.22 and 29.78%, respectively,
compared to the control group (10:0). This probably resulted from
the proliferation rates of the cancer cells changed by the hATMSCs.
However, further evidence that excluded the confounding effects
associated with changes in the proliferation rates of the hATMSCs
by the cancer cells was needed. Thus, indirect co-culture of the
cancer cells with the hATMSCs using cell inserts was conducted in
order to prevent cell-to-cell contact. The same trends were
observed in the indirect co-culture. The proliferation rates of the
A549 cells were reduced by 9.72 and 18.57%, respectively, whereas
those of the HT-29 cells were increased by 19.90 and 25.79%,
respectively, compared to the control group (10:0).
Co-culture effect of the hATMSCs on the
proliferation and apoptosis of cancer cells
The proliferation of the cancer cells indirectly
co-cultured with the hATMSCs was measured by BrdU staining and flow
cytometry. The percentage of BrdU-positive A549 cells was reduced
by the hATMSC co-culture by 34 and 23% at the ratios of 10:1 and
10:2, respectively, whereas there was a significant induction of
the percentage of BrdU-positive HT-29 cells co-cultured with the
hATMSCs at the ratios of 10:2 (Fig.
3B). The apoptotic rates of the cancer cells co-cultured with
the hATMSCs were analyzed by Annexin V/PI double staining and flow
cytometry. While the percentage of live A549 cells was
significantly diminished by co-culture with the hATMSCs, the
percentage of apoptotic A549 cells was elevated by 40, 85 and 95%
at the ratios of 10:0.5, 10:1 and 10:2, respectively (Fig. 3C). However, the hATMSCs had opposite
effects on the HT-29 cells; there was an increase in live HT-29
cells and a decrease in apoptotic HT-29 cells of 23 and 26% at the
ratios of 10:1 and 10:2, respectively. Collectively, the hATMSCs
inhibited the proliferation, yet elevated the apoptosis in the A549
cancer cells, whereas it promoted the proliferation and reduced
apoptosis in the HT-29 cancer cells.
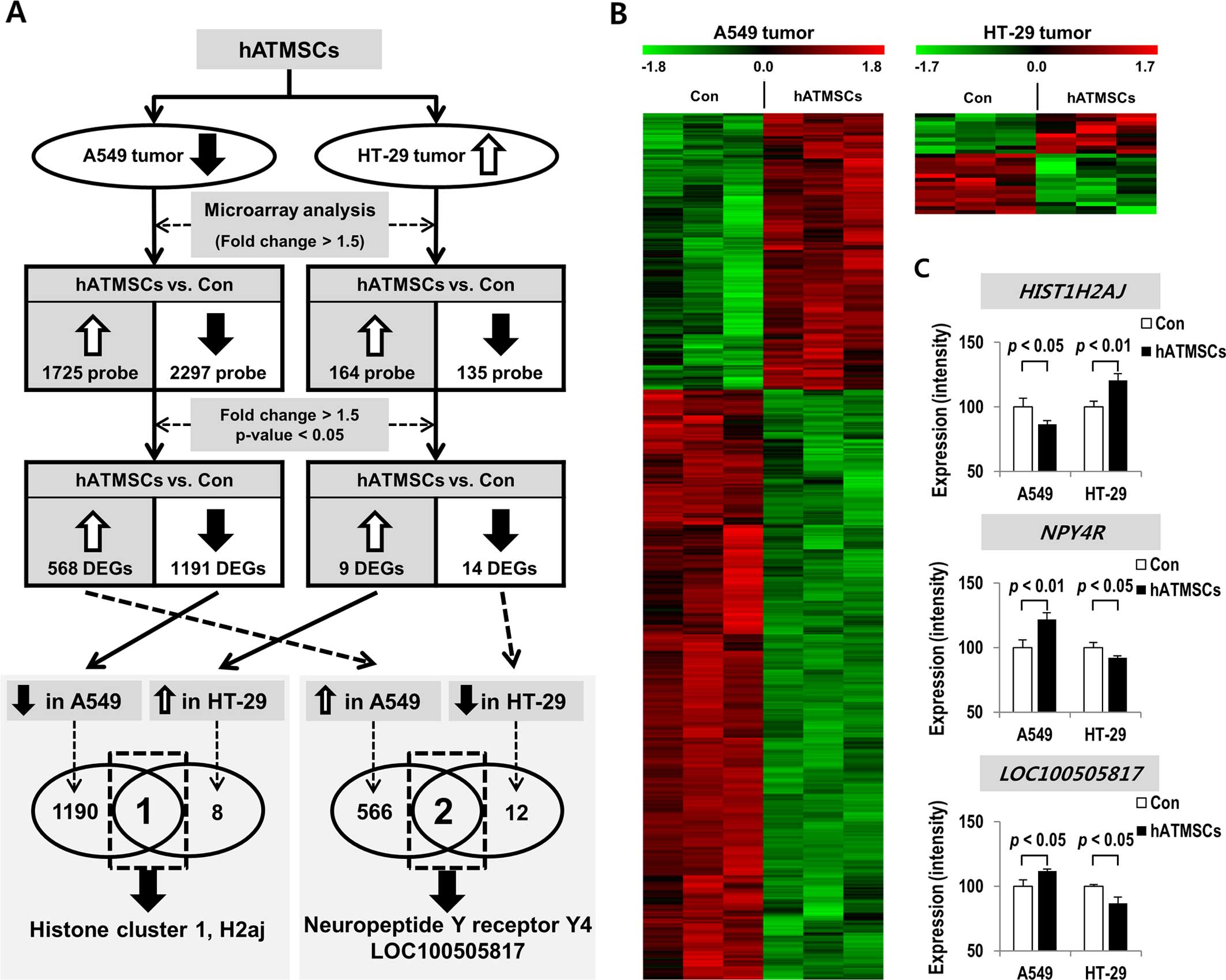
Global differences in gene expression in
the xenograft tumors in the presence or absence of the hATMSCs
In the microarray experiment with the A549 tumors of
the hairless SCID mice, 4,022 probe sets were used to identify
significant differences between the hATMSC and the control group in
regards to expression levels of genes (fold-change >1.5)
(Fig. 4A and B). Particularly, we
found that the expression levels of 1,725 probes were increased and
those of 2,297 probes were decreased in the hATMSC group. In
contrast, in the microarray experiment with the HT-29 tumors of the
hairless SCID mice, 164 probes were found expressed more highly in
the hATMSC group, whereas 135 probes were found to be more lowly
expressed in the hATMSC group. Based on more stringent statistical
criteria (fold-change >1.5 and p<0.05), 568 genes were
expressed at a higher level and 1,191 genes were expressed at a
lower level in the hATMSC group compared with the control group in
the A549 tumors (Table I). In
contrast, 9 genes were expressed at a higher level and 14 genes
were expressed at a lower level in the hATMSC group when compared
with the control group in the HT-29 tumors (Table II).
 | Table ITop 30 genes showing differential
expression in the hATMSC-treated A549 tumors ranked by
fold-change. |
Table I
Top 30 genes showing differential
expression in the hATMSC-treated A549 tumors ranked by
fold-change.
| Probe set | Gene | Gene
description | Fold-changea |
|---|
| Genes expressed at
a higher level in A549 tumors after hATMSC injection (p<0.05,
fold-change >1.5) |
| 16863393 | IGFL2 | IGF-like family
member 2 | 3.68 |
| 16976821 | PPBP | Pro-platelet basic
protein [chemokine (C-X-C motif) ligand 7] | 3.18 |
| 17061759 | NRCAM | Neuronal cell
adhesion molecule | 2.77 |
| 16852322 | MAPK4 | Mitogen-activated
protein kinase 4 | 2.47 |
| 16873513 |
LOC400706 | Uncharacterized
LOC400706 | 2.46 |
| 17083433 | IL33 | Interleukin 33 | 2.45 |
| 16745990 | miR3167 | MicroRNA 3167 | 2.17 |
| 16909319 | DNER | Δ/notch-like EGF
repeat containing | 2.07 |
| 17022996 | ROS1 | c-ros oncogene 1,
receptor tyrosine kinase | 1.97 |
| 16713760 |
HNRNPA1P33 | Heterogeneous
nuclear ribonucleoprotein A1 pseudogene 33 | 1.97 |
| 16733325 |
KIRREL3-AS2 | KIRREL3 antisense
RNA 2 | 1.87 |
| 16704464 | NPY4R | Neuropeptide Y
receptor Y4 | 1.86 |
| 16742086 | PGM2L1 | Phosphoglucomutase
2-like 1 | 1.85 |
| 16710126 | HTRA1 | HtrA serine
peptidase 1 | 1.79 |
| 17012392 | RSPO3 | R-spondin 3 | 1.69 |
| 16737590 | SYT13 | Synaptotagmin
XIII | 1.66 |
| 16997795 |
VCAN-AS1 | VCAN antisense RNA
1 | 1.66 |
| 16852849 |
SERPINB11 | Serpin peptidase
inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene) | 1.63 |
| 16958382 | MUC13 | Mucin 13, cell
surface associated | 1.62 |
| 16713762 | ANXA8L1 | Annexin A8-like
1 | 1.61 |
| 16851950 | miR3975 | MicroRNA 3975 | 1.59 |
| 17060820 | MUC12 | Mucin 12, cell
surface associated | 1.58 |
| 16705934 | CHST3 | Carbohydrate
(chondroitin 6) sulfotransferase 3 | 1.58 |
| 16677556 | TGFB2 | Transforming growth
factor, β2 | 1.57 |
| 16704614 | ANXA8 | Annexin A8 | 1.55 |
| 16704516 | ANXA8L2 | Annexin A8-like
2 | 1.55 |
| 16957304 | ZBED2 | Zinc finger,
BED-type containing 2 | 1.54 |
| 16960922 | RARRES1 | Retinoic acid
receptor responder (tazarotene induced) 1 | 1.51 |
| 17067696 | NRG1 | Neuregulin 1 | 1.50 |
| 16697674 |
LINC00862 | Long intergenic
non-protein coding RNA 862 | 1.50 |
| Genes expressed at
a lower level in A549 tumors after hATMSC injection (p<0.05,
fold-change >1.5) |
| 16971272 | EDNRA | Endothelin receptor
type A | −4.75 |
| 17113448 | IL13RA2 | Interleukin 13
receptor, α2 | −4.58 |
| 17112149 | XIST | X inactive specific
transcript (non-protein coding) | −4.44 |
| 16965377 | SLIT2 | Slit homolog 2
(Drosophila) | −4.32 |
| 16977241 | NAA11 |
N(α)-acetyltransferase 11, NatA catalytic
subunit | −4.22 |
| 17055447 | MEOX2 | Mesenchyme homeobox
2 | −4.11 |
| 17101570 |
LOC100133123 | Uncharacterized
LOC100133123 | −4.05 |
| 16870838 | ZNF208 | Zinc finger protein
208 | −4.01 |
| 17001578 | PDGFRB | Platelet-derived
growth factor receptor, β polypeptide | −3.93 |
| 16732840 | OR8G2 | Olfactory receptor,
family 8, subfamily G, member 2 | −3.91 |
| 16736764 | MUC15 | Mucin 15, cell
surface associated | −3.86 |
| 16870821 | ZNF100 | Zinc finger protein
100 | −3.83 |
| 16909828 | COL6A3 | Collagen, type VI,
α3 | −3.72 |
| 16997383 | F2RL2 | Coagulation factor
II (thrombin) receptor-like 2 | −3.68 |
| 16979875 | PCDH18 | Protocadherin
18 | −3.62 |
| 17058002 |
LOC650226 | Ankyrin repeat
domain 26 pseudogene | −3.56 |
| 16942958 | EPHA3 | EPH receptor
A3 | −3.52 |
| 17108003 | BGN | Biglycan | −3.51 |
| 16852966 |
CCDC102B | Coiled-coil domain
containing 102B | −3.51 |
| 17097661 | TNC | Tenascin C | −3.45 |
| 17078870 | MMP16 | Matrix
metallopeptidase 16 (membrane-inserted) | −3.32 |
| 16772888 |
RNU6-52P | RNA, U6 small
nuclear 52, pseudogene | −3.29 |
| 16870854 | ZNF676 | Zinc finger protein
676 | −3.21 |
| 16888610 | COL3A1 | Collagen, type III,
α1 | −3.13 |
| 16923842 | COL6A1 | Collagen, type VI,
α1 | −3.13 |
| 16666509 | IFI44 | Interferon-induced
protein 44 | −3.08 |
| 17107835 | MAGEA4 | Melanoma antigen
family A, 4 | −3.04 |
| 17107867 | MAGEA6 | Melanoma antigen
family A, 6 | −3.02 |
| 16673075 | DDR2 | Discoidin domain
receptor tyrosine kinase 2 | −3.00 |
| 16885596 | POTEJ | POTE ankyrin domain
family, member J | −2.97 |
 | Table IIGenes showing differential expression
in the hATMSC-treated HT-29 tumors ranked by fold-change. |
Table II
Genes showing differential expression
in the hATMSC-treated HT-29 tumors ranked by fold-change.
| Probe set | Gene | Gene
description | Fold-changea |
|---|
| Genes expressed at
a higher level in the HT-29 tumors after hATMSC injection
(p<0.05, fold-change >1.5) |
| 16703429 |
RNA5SP307 | RNA, 5S ribosomal
pseudogene 307 | 0.97 |
| 17016490 |
HIST1H2AJ | Histone cluster 1,
H2aj | 0.91 |
| 16761541 | PRB4 | Proline-rich
protein BstNI subfamily 4 | 0.85 |
| 17063776 | TRBV6-8 | T cell receptor β
variable 6-8 | 0.77 |
| 16854216 |
miR133A1 | MicroRNA
133a-1 | 0.74 |
| 16943350 | GPR128 | G protein-coupled
receptor 128 | 0.67 |
| 17117463 |
LOC100509635 | Uncharacterized
LOC100509635 | 0.64 |
| 17063800 | TRBV7-4 | T cell receptor β
variable 7-4 (gene/pseudogene) | 0.63 |
| 16904484 |
SNORA70F | Small nucleolar
RNA, H/ACA box 70F | 0.61 |
| Genes expressed at
a lower level in the HT-29 tumors after hATMSC injection
(p<0.05, fold-change >1.5) |
| 16691574 |
RNA5SP56 | RNA, 5S ribosomal
pseudogene 56 | −1.05 |
| 16798050 | PAR-SN | Paternally
expressed transcript PAR-SN | −0.74 |
| 17114740 | SPANXN3 | SPANX family,
member N3 | −0.74 |
| 16798146 |
SNORD116-5 | Small nucleolar
RNA, C/D box 116-5 | −0.73 |
| 16798150 |
SNORD116-5 | Small nucleolar
RNA, C/D box 116-5 | −0.73 |
| 16734339 | miR4298 | MicroRNA 4298 | −0.72 |
| 16716369 |
miR4679-2 | MicroRNA
4679-2 | −0.70 |
| 16743207 |
TRIM51EP | Tripartite
motif-containing 51E, pseudogene | −0.68 |
| 16853028 |
LOC100505817 | Uncharacterized
LOC100505817 | −0.64 |
| 16767367 |
miR3913-1 | MicroRNA
3913-1 | −0.63 |
| 17114772 | miR891B | MicroRNA 891b | −0.60 |
| 16696120 | DPT | Dermatopontin | −0.59 |
| 17096623 | BAAT | Bile acid CoA:
amino acid N-acyltransferase (glycine N-choloyltransferase) | −0.59 |
| 16704464 | NPY4R | Neuropeptide Y
receptor Y4 | −0.59 |
Among the genes differentially expressed in the
xenograft tumors after the hATMSC injection, histone cluster 1,
H2aj (HIST1H2AJ) was expressed in direct proportion to the
xenograft tumor size. In other words, it was expressed at lower
level in the A549 tumors and was expressed at a higher level in the
HT-29 tumors after the hATMSC injection in comparison to the
respective control groups. In addition, neuropeptide Y receptor Y4
(NPY4R) and LOC100505817 were expressed in inverse
proportion to the xenograft tumor size. They were expressed at a
higher level in the A549 tumors and were expressed at a lower level
in the HT-29 tumors after the hATMSC injection in comparison to the
respective control groups (Fig. 4A and
C).
Functional classification of genes
showing differential expression in the hATMSC-treated xenograft
tumors
In order to identify the biological function of the
genes differentially expressed following the hATMSC injection in
the A549 and HT-29 tumor models using hairless SCID mice, GO
analysis was performed. The statistically significant categories of
GO terms [p<0.01; false discovery rate (FDR) <0.05] are shown
in Table III. No enriched GO term
was altered following treatment with the hATMSCs in the HT-29
tumors, whereas the genes involved in biological processes, such as
'Regulation of growth' and 'Regulation of cell growth' were
upregulated in the hATMSC-treated A549 tumors. The genes involved
in biological processes, such as 'Nucleosome assembly', 'Nucleosome
organization', 'Protein-DNA complex assembly', 'Chromatin assembly
or disassembly', 'Cell-cell adhesion' and 'Regulation of
transcription (DNA-dependent)' were downregulated in the
hATMSC-treated A549 tumors.
 | Table IIIGO terms associated with upregulated
and downregulated trends in the hATMSC-treated A549 tumors
(p<0.01, FDR<0.05). |
Table III
GO terms associated with upregulated
and downregulated trends in the hATMSC-treated A549 tumors
(p<0.01, FDR<0.05).
| GO ID | Biological process
term | Gene count | P-value | Benjamini | FDR |
|---|
| Upregulated
trend |
| GO:0040008 | Regulation of
growth | 24 | 1.19E-05 | 0.0258 | 0.021 |
| GO:0001558 | Regulation of cell
growth | 17 | 2.37E-05 | 0.0256 | 0.041 |
| Downregulated
trend |
| GO:0006334 | Nucleosome
assembly | 24 | 9.31E-11 | 2.44E-07 | 1.65E-07 |
| GO:0034728 | Nucleosome
organization | 25 | 1.41E-10 | 1.85E-07 | 2.51E-07 |
| GO:0031497 | Chromatin
assembly | 24 | 2.02E-10 | 1.77E-07 | 3.58E-07 |
| GO:0065004 | Protein-DNA complex
assembly | 24 | 5.37E-10 | 3.53E-07 | 9.52E-07 |
| GO:0006323 | DNA packaging | 27 | 8.47E-10 | 4.45E-07 | 1.50E-06 |
| GO:0006333 | Chromatin assembly
or disassembly | 28 | 1.15E-09 | 5.02E-07 | 2.03E-06 |
| GO:0006350 | Transcription | 175 | 9.61E-09 | 3.60E-06 | 1.70E-05 |
| GO:0006355 | Regulation of
transcription, DNA-dependent | 151 | 3.65E-08 | 1.20E-05 | 6.47E-05 |
| GO:0051252 | Regulation of RNA
metabolic process | 153 | 5.05E-08 | 1.47E-05 | 8.96E-05 |
| GO:0034621 | Cellular
macromolecular complex subunit organization | 45 | 5.14E-07 | 1.35E-04 | 9.12E-04 |
| GO:0034622 | Cellular
macromolecular complex assembly | 41 | 1.01E-06 | 2.41E-04 | 0.002 |
| GO:0006325 | Chromatin
organization | 46 | 1.01E-06 | 2.22E-04 | 0.002 |
| GO:0016339 | Calcium-dependent
cell-cell adhesion | 10 | 1.91E-06 | 3.85E-04 | 0.003 |
| GO:0045449 | Regulation of
transcription | 193 | 6.37E-06 | 0.0012 | 0.011 |
| GO:0051276 | Chromosome
organization | 52 | 7.69E-06 | 0.0013 | 0.014 |
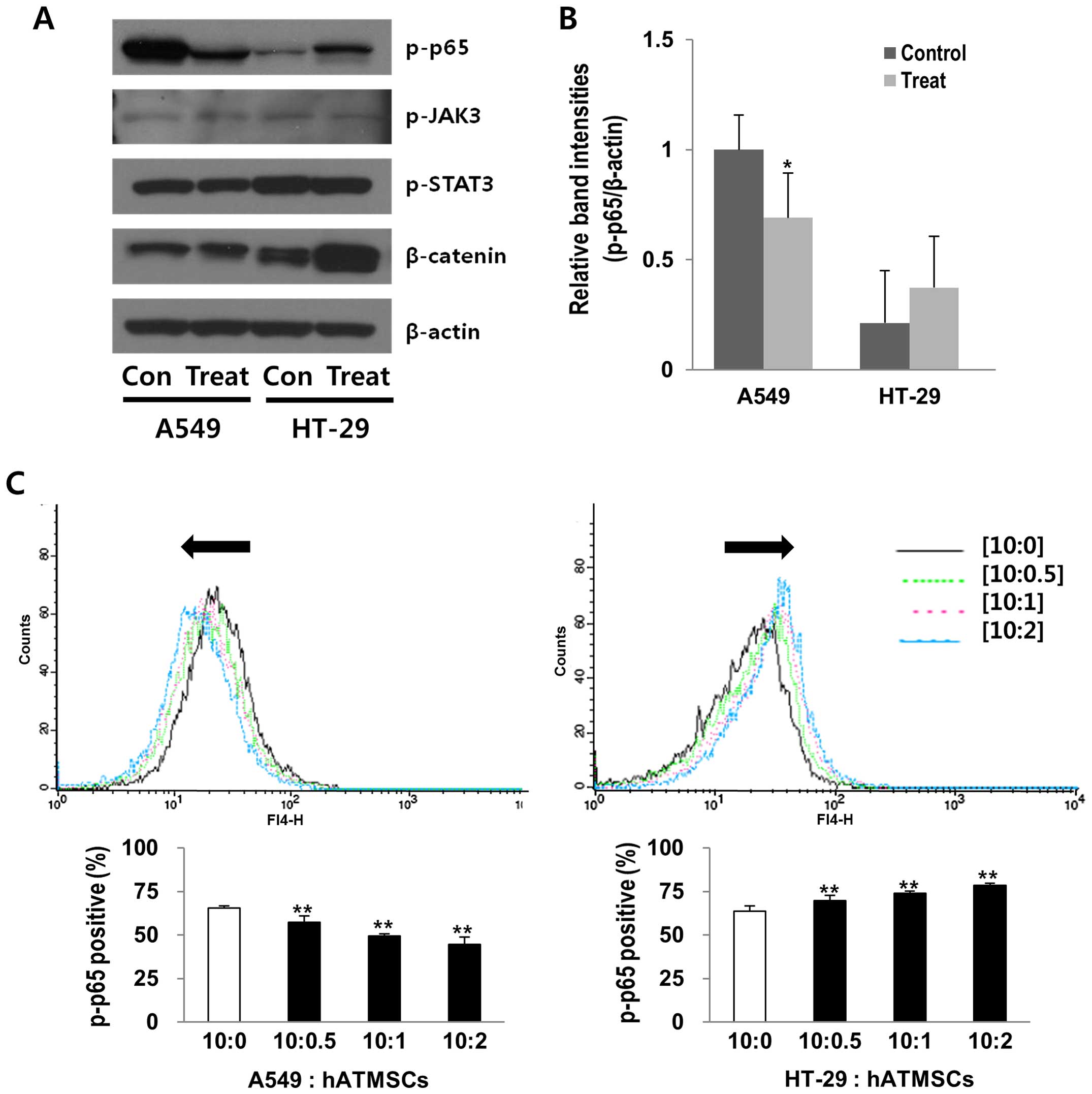
Effects of the hATMSCs on the
phosphorylation of NF-κB p65 in the A549 and HT-29 tumors in vivo
and in vitro
The proteins extracted from the xenograft tumor
masses of the hairless SCID mice were analyzed by western blot
analysis to investigate whether there is a relationship between
certain cell signaling pathways and the hATMSC-associated
modulation of tumor growth. Among several proteins related to cell
signaling pathways, the expression level of phosphorylated NF-κB
p65 was reduced by 31% in the hATMSC-injected A549 tumor, whereas
it was increased by 77% in the hATMSC-injected HT-29 tumors
(Fig. 5A and B). The levels of
other cell signaling-related proteins, such as p-JAK3 and p-STAT3,
were not affected by the hATMSCs in the A549 and HT-29 tumors, and
the β-catenin protein level was elevated by the hATMSCs in the
HT-29 tumors, yet not affected in the A549 tumors, providing a
possible correlation between the NF-κB pathway and hATMSC-related
tumor modulation, at least in the A549 and the HT-29 tumors.
Changes in phosphorylated NF-κB p65 in the tumor
masses in vivo were confirmed using flow cytometric analysis
in the cancer cells indirectly co-cultured with the hATMSCs, which
revealed significant reductions in phosphorylated NF-κB p65 by 12,
25 and 32% in the co-culture of the A549 cancer cells with the
hATMSCs at the ratios of 10:0.5, 10:1 and 10:2, respectively,
compared with the control (10:0) (Fig.
5C). However, there was an inverse effect in the HT-29 cancer
cells, showing a profound increase in phosphorylated NF-κB p65 by
15% in the co-culture of HT-29 cancer cells with the hATMSCs at the
ratio of 10:2.
Discussion
In the present study, we found conflicting results
concerning the effects of the hATMSCs on tumor growth, particularly
in the A549 and HT-29 tumors which were suppressed and supported,
respectively, by the hATMSCs. It is possible that the dual effect
of MSCs can be dependent on tumor type, at least partially,
although these two cell lines do not represent the types of cancer
that they are derived from. Thus, further analysis is needed to
verify the actual factors in different microenvironment induced by
different cancer cells rather than attempting to propose the type
of tumor as the reason.
A recent study showed that direct cell-to-cell
interaction between cancer cells and MSCs is responsible for the
effects of MSCs on tumor growth (32). Although we also observed this result
when hATMSCs were directly injected at the tumor site in the
xenograft tumor models in vivo, our hypothesis is that this
effect of hATMSCs on tumor growth could be indirect according to
the results of previous studies (14,33).
The hATMSCs showed dual effects such as a strong inhibitory effect
on the A549 cancer cell viability and a dramatic promoting effect
on the HT-29 cancer cell viability in vitro. In particular,
in consistent with a study by Fierro et al (34), the hATMSCs showed a similar impact
on cell viability between direct and indirect co-culture in our
experiments using in vitro co-culture systems, indicating
that the hATMSCs affected tumor cell growth even without direct
cell-to-cell contact possibly due to certain soluble factors in the
conditioned medium released from the hATMSCs. Among the many
MSC-related soluble factors, proangiogenic factors, such as
vascular endothelial growth factor (34), fibroblast growth factor (35), and angiopoietin-1 (36), promote endothelial and smooth muscle
migration and proliferation at the tumor site, facilitating
angiogenesis. In addition, there are factors such as interleukin 6
and tumor necrosis factor α (37)
that are produced by MSCs and have immunosuppressive effects and
eventually tumor supporting effects. Karnoub et al (21) reported that MSC-secreted chemokine
ligand 5 induced a transient pro-metastatic effect on breast cancer
cells. Meanwhile, ATMSCs were found to inhibit the proliferation in
primary leukemia cells by secreting DKK-1, which exhibits an
inhibitory effect on β-catenin signaling (15,24).
Further studies are necessary to analyze which hATMSC-releasing
factors contribute to the effect of hATMSCs on tumor growth.
The tumor growth was differentially mediated by
hATMSCs between the A549 and the HT-29 tumors with in vivo
xenograft models. For functional classification of the genes that
were expressed differentially following treatment with the hATMSCs
in the A549 and HT-29 tumors, GO and KEGG pathway analyses were
performed. Noteworthy, 17 enriched GO categories were identified in
the hATMSC-treated A549 tumors (p<0.01, FDR<0.05), whereas no
GO terms were identified in the HT-29 tumors. The GO analysis
clearly showed that the treatment of the A549 tumors with the
hATMSCs upregulated the expression of many genes associated with
'Regulation of cell growth', which has been known to be induced by
a potential anticancer drug (38).
In contrast, 'Regulation of transcription (DNA-dependent)' and
'Cell-cell adhesion', which have been previously reported to be
upregulated in xenograft tumor models and in cancer cell lines
(39,40), were suppressed in the A549 tumors
following treatment with the hATMSCs. Lo et al (41) showed that GO biological processes,
including 'Nucleosome assembly', 'Nucleosome organization',
'Protein-DNA complex assembly' and 'Chromatin assembly or
disassembly' were found to be altered in Asian and Caucasian lung
cancer patients. 'Nucleosome assembly' has also been known as a
process which alters the regulatory mechanisms of cancer cells
(42). In the present study, the
hATMSCs downregulated the above-mentioned biological processes in
the A549 tumors. In particular, the KEGG pathway analysis indicated
that 'Small cell lung cancer pathway' in the downregulated trend
gene set was significant in the hATMSC-treated A549 tumors
(p<0.05) (data not shown). These findings presented here,
suggesting that biological processes possibly associated with the
tumor modulation, are active in only A549 tumors, can provide
important implications to verify the dual effects of the
hATMSCs.
To identify possible genetic factors for
hATMSC-induced tumor modulation, the differentially expressed genes
in proportion to xenograft tumor size following hATMSC
administration were identified. Histone proteins, which are basic
nuclear proteins responsible for the nucleosome structure of the
chromosomal fiber, were expressed from four different clusters in
the human genome. In histone cluster 1, mainly histone 1 (e.g.
HIST1H1T) and histone 2 (e.g. HIST1H2BC,
HIST1H2AC and HIST1H2BD) genes were found to be
overexpressed in the cancer tissues (43). Although there are few reports of
HIST1H2AJ which is related to cancer directly,
HIST1H2AJ (histone cluster 1, H2aj) was found to be
expressed in direct proportion to the xenograft tumor size and is
likely to be functionally relevant in tumorigenesis. We also found
that NPY4R was differentially expressed in inverse
proportion to the xenograft A549 and HT-29 tumor size after hATMSC
administration. NPY family peptides, one of the most widely
distributed peptides in the central and peripheral nervous system,
regulate physiological processes associated with energy homeostasis
such as energy expenditure, lipid metabolism and insulin secretion
through their actions on Y receptors (44–47).
Moreover, the NPY system is known to be involved in cancer
development via effects on energy homeostasis, immune function, as
well as direct actions on tumor biology (47), raising the possibility of the
involvement of NPY4R expression in the regulation of tumor
growth following treatment with hATMSCs.
Cell signaling pathways associated with
translational changes have been referenced to explain the
mechanisms of the conflicting effects of MSCs on tumors. PI3K/AKT
was referred as a possible means related to the tumor-inhibitory
effect of umbilical cord MSCs (48). WNT (24) and JAK/STAT pathways (17) were also suggested as cell signaling
pathways involved in the effects of MSCs. We additionally
investigated the expression levels of certain proteins to determine
which cell signaling pathways are related to the effects of hATMSCs
on tumor growth. First of all, the hATMSCs affected the levels of
phosphorylated NF-κB p65, yet not p-JAK3, p-STAT3 or β-catenin that
are also key players in the effects of MSCs on tumors. Second, the
hATMSC-induced changes in phosphorylated NF-κB p65 levels were
correlated with the hATMSC-associated modulation on A549 and HT-29
tumor growth, suggesting the NF-κB pathway as a potential mediator
in the effects of hATMSCs on tumors. The NF-κB pathway is triggered
in response to infection or exposure to pro-inflammatory cytokines,
leading to inhibitor of κB (IκB) degradation and finally
translocation of p50/p65 heterodimers into the nucleus. In general,
NF-κB regulates a variety of downstream genes that govern immunity,
cell growth and apoptosis (49). In
the tumor environment, the NF-κB pathway is widely known to
regulate the proliferation of tumor cells through the transcription
of anti-apoptotic proteins, leading to the increased growth and
metastasis of various types of tumors (50). Actually, in the A549 and HT-29
cancer cells, correlations were reported in which cell
proliferation was promoted by activation of the NF-κB pathway and
suppressed by blocking the NF-κB pathway (51,52),
supporting our hypothesis on the possible link of the NF-κB pathway
to the tumor growth modulated by hATMSCs.
Given the possibility of MSCs as a novel therapeutic
tool for various diseases, it is critical to explain the
conflicting findings in tumor-MSC studies. Despite previous reports
on the possible factors of MSC-induced tumor modulation, the exact
mechanisms remain unclear. The most direct outcome of the present
study is that we confirmed the dual effects of hATMSCs in the two
types of cancer cells in different micro-environments in
vivo and in vitro. We also suggest the possible roles of
the identified gene as profiled and the NF-κB pathway in the dual
modulatory effects of hATMSCs on tumor growth. Our findings have
profound implication in understanding the interaction between MSCs
and tumors although further research is needed to confirm these
findings using various types of cancer.
Abbreviations:
|
hATMSCs
|
human adipose tissue-derived
mesenchymal stem cells
|
|
NF-κB
|
nuclear factor κB
|
|
MSCs
|
mesenchymal stem cells
|
|
BM-MSCs
|
bone marrow-derived mesenchymal stem
cells
|
|
SCID
|
severe combined immunodeficiency
|
References
|
1
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop Dj and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The International Society for Cellular
Therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong J, Uemura T, Shirasaki Y and Tateishi
T: Promotion of bone formation using highly pure porous beta-TCP
combined with bone marrow-derived osteoprogenitor cells.
Biomaterials. 23:4493–4502. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi HJ, Kim JM, Kwon E, Che JH, Lee JI,
Cho SR, Kang SK, Ra JC and Kang BC: Establishment of efficacy and
safety assessment of human adipose tissue-derived mesenchymal stem
cells (hATMSCs) in a nude rat femoral segmental defect model. J
Korean Med Sci. 26:482–491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Toyoshima KE, Asakawa K, Ishibashi N, Toki
H, Ogawa M, Hasegawa T, Irié T, Tachikawa T, Sato A, Takeda A, et
al: Fully functional hair follicle regeneration through the
rearrangement of stem cells and their niches. Nat Commun.
3:7842012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seo KW, Sohn SY, Bhang DH, Nam MJ, Lee HW
and Youn HY: Therapeutic effects of hepatocyte growth
factor-overexpressing human umbilical cord blood-derived
mesenchymal stem cells on liver fibrosis in rats. Cell Biol Int.
38:106–116. 2014. View Article : Google Scholar
|
|
6
|
Perez-Basterrechea M, Obaya AJ, Meana A,
Otero J and Esteban MM: Cooperation by fibroblasts and bone
marrow-mesenchymal stem cells to improve pancreatic rat-to-mouse
islet xenotransplantation. PLoS One. 8:e735262013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bas E, Van De Water TR, Lumbreras V,
Rajguru S, Goss G, Hare JM and Goldstein BJ: Adult human nasal
mesenchymal-like stem cells restore cochlear spiral ganglion
neurons after experimental lesion. Stem Cells Dev. 23:502–514.
2014. View Article : Google Scholar :
|
|
8
|
Tian LL, Yue W, Zhu F, Li S and Li W:
Human mesenchymal stem cells play a dual role on tumor cell growth
in vitro and in vivo. J Cell Physiol. 226:1860–1867. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ra JC, Shin IS, Kim SH, Kang SK, Kang BC,
Lee HY, Kim YJ, Jo JY, Yoon EJ, Choi HJ, et al: Safety of
intravenous infusion of human adipose tissue-derived mesenchymal
stem cells in animals and humans. Stem Cells Dev. 20:1297–1308.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
MacIsaac ZM, Shang H, Agrawal H, Yang N,
Parker A and Katz AJ: Long-term in-vivo tumorigenic assessment of
human culture-expanded adipose stromal/stem cells. Exp Cell Res.
318:416–423. 2012. View Article : Google Scholar :
|
|
11
|
Klopp AH, Gupta A, Spaeth E, Andreeff M
and Marini F III: Concise review: Dissecting a discrepancy in the
literature: do mesenchymal stem cells support or suppress tumor
growth? Stem Cells. 29:11–19. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maestroni GJ, Hertens E and Galli P:
Factor(s) from nonmacrophage bone marrow stromal cells inhibit
Lewis lung carcinoma and B16 melanoma growth in mice. Cell Mol Life
Sci. 55:663–667. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khakoo AY, Pati S, Anderson SA, Reid W,
Elshal MF, Rovira II, Nguyen AT, Malide D, Combs CA, Hall G, et al:
Human mesenchymal stem cells exert potent antitumorigenic effects
in a model of Kaposi's sarcoma. J Exp Med. 203:1235–1247. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li L, Tian H, Chen Z, Yue W, Li S and Li
W: Inhibition of lung cancer cell proliferation mediated by human
mesenchymal stem cells. Acta Biochim Biophys Sin. 43:143–148. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu W, Xu W, Jiang R, Qian H, Chen M, Hu
J, Cao W, Han C and Chen Y: Mesenchymal stem cells derived from
bone marrow favor tumor cell growth in vivo. Exp Mol Pathol.
80:267–274. 2006. View Article : Google Scholar
|
|
16
|
Xu WT, Bian ZY, Fan QM, Li G and Tang TT:
Human mesenchymal stem cells (hMSCs) target osteosarcoma and
promote its growth and pulmonary metastasis. Cancer Lett.
281:32–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsai KS, Yang SH, Lei YP, Tsai CC, Chen
HW, Hsu CY, Chen LL, Wang HW, Miller SA, Chiou SH, et al:
Mesenchymal stem cells promote formation of colorectal tumors in
mice. Gastroenterology. 141:1046–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Roorda BD, ter Elst A, Kamps WA and de
Bont ES: Bone marrow-derived cells and tumor growth: Contribution
of bone marrow-derived cells to tumor microenvironments with
special focus on mesenchymal stem cells. Crit Rev Oncol Hematol.
69:187–198. 2009. View Article : Google Scholar
|
|
19
|
Mishra PJ, Mishra PJ, Humeniuk R, Medina
DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW and Banerjee D:
Carcinoma-associated fibroblast-like differentiation of human
mesenchymal stem cells. Cancer Res. 68:4331–4339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Plumas J, Chaperot L, Richard MJ, Molens
JP, Bensa JC and Favrot MC: Mesenchymal stem cells induce apoptosis
of activated T cells. Leukemia. 19:1597–1604. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karnoub AE, Dash AB, Vo AP, Sullivan A,
Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R and Weinberg
RA: Mesenchymal stem cells within tumour stroma promote breast
cancer metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu YR, Yuan Y, Wang XJ, Wei LL, Chen YN,
Cong C, Li SF, Long D, Tan WD, Mao YQ, et al: The growth inhibitory
effect of mesenchymal stem cells on tumor cells in vitro and in
vivo. Cancer Biol Ther. 7:245–251. 2008. View Article : Google Scholar
|
|
23
|
Otsu K, Das S, Houser SD, Quadri SK,
Bhattacharya S and Bhattacharya J: Concentration-dependent
inhibition of angiogenesis by mesenchymal stem cells. Blood.
113:4197–4205. 2009. View Article : Google Scholar :
|
|
24
|
Qiao L, Xu ZL, Zhao TJ, Ye LH and Zhang
XD: Dkk-1 secreted by mesenchymal stem cells inhibits growth of
breast cancer cells via depression of Wnt signalling. Cancer Lett.
269:67–77. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kubota T, Yamaguchi H, Watanabe M,
Yamamoto T, Takahara T, Takeuchi T, Furukawa T, Kase S, Kodaira S,
Ishibiki K, et al: Growth of human tumor xenografts in nude mice
and mice with severe combined immunodeficiency (SCID). Surg Today.
23:375–377. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Denicolaï E, Baeza-Kallee N, Tchoghandjian
A, Carré M, Colin C, Jiglaire CJ, Mercurio S, Beclin C and
Figarella-Branger D: Proscillaridin A is cytotoxic for glioblastoma
cell lines and controls tumor xenograft growth in vivo. Oncotarget.
5:10934–10948. 2014.PubMed/NCBI
|
|
27
|
Yun JW, Lee TR, Kim CW, Park YH, Chung JH,
Lee YS, Kang KS and Lim KM: Predose blood gene expression profiles
might identify the individuals susceptible to carbon
tetrachloride-induced hepatotoxicity. Toxicol Sci. 115:12–21. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG
and Lee JS: Prediction of cancer incidence and mortality in Korea,
2013. Cancer Res Treat. 45:15–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Paine-Murrieta GD, Taylor CW, Curtis RA,
Lopez MH, Dorr RT, Johnson CS, Funk CY, Thompson F and Hersh EM:
Human tumor models in the severe combined immune deficient (scid)
mouse. Cancer Chemother Pharmacol. 40:209–214. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hao Y, Huang W, Liao M, Zhu Y, Liu H, Hao
C, Liu G, Zhang G, Feng H, Ning X, et al: The inhibition of
resveratrol to human skin squamous cell carcinoma A431 xenografts
in nude mice. Fitoterapia. 86:84–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ansari D, Bauden MP, Sasor A, Gundewar C
and Andersson R: Analysis of MUC4 expression in human pancreatic
cancer xenografts in immunodeficient mice. Anticancer Res.
34:3905–3910. 2014.PubMed/NCBI
|
|
32
|
Uchibori R, Tsukahara T, Mizuguchi H, Saga
Y, Urabe M, Mizukami H, Kume A and Ozawa K: NF-κB activity
regulates mesenchymal stem cell accumulation at tumor sites. Cancer
Res. 73:364–372. 2013. View Article : Google Scholar
|
|
33
|
Kéramidas M, de Fraipont F, Karageorgis A,
Moisan A, Persoons V, Richard MJ, Coll JL and Rome C: The dual
effect of mesenchymal stem cells on tumour growth and tumour
angiogenesis. Stem Cell Res Ther. 4:412013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fierro FA, Sierralta WD, Epuñan MJ and
Minguell JJ: Marrow-derived mesenchymal stem cells: Role in
epithelial tumor cell determination. Clin Exp Metastasis.
21:313–319. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Potapova IA, Gaudette GR, Brink PR,
Robinson RB, Rosen MR, Cohen IS and Doronin SV: Mesenchymal stem
cells support migration, extracellular matrix invasion,
proliferation, and survival of endothelial cells in vitro. Stem
Cells. 25:1761–1768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qiao L, Xu Z, Zhao T, Zhao Z, Shi M, Zhao
RC, Ye L and Zhang X: Suppression of tumorigenesis by human
mesenchymal stem cells in a hepatoma model. Cell Res. 18:500–507.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Abdel aziz MT, El Asmar MF, Atta HM,
Mahfouz S, Fouad HH, Roshdy NK, Rashed LA, Sabry D, Hassouna AA and
Taha FM: Efficacy of mesenchymal stem cells in suppression of
hepatocarcinorigenesis in rats: Possible role of Wnt signaling. J
Exp Clin Cancer Res. 30:492011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim BY, Lee J, Park SJ, Bang OS and Kim
NS: Gene expression profile of the A549 human non-small cell lung
carcinoma cell line following treatment with the seeds of
Descurainia sophia, a potential anticancer drug. Evid Based
Complement Alternat Med. 2013:5846042013.PubMed/NCBI
|
|
39
|
Creighton C, Kuick R, Misek DE, Rickman
DS, Brichory FM, Rouillard JM, Omenn GS and Hanash S: Profiling of
pathway-specific changes in gene expression following growth of
human cancer cell lines transplanted into mice. Genome Biol.
4:R462003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lu C, Xiong M, Luo Y, Li J, Zhang Y, Dong
Y, Zhu Y, Niu T, Wang Z and Duan L: Genome-wide transcriptional
analysis of apoptosis-related genes and pathways regulated by H2AX
in lung cancer A549 cells. Apoptosis. 18:1039–1047. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lo FY, Chang JW, Chang IS, Chen YJ, Hsu
HS, Huang SF, Tsai FY, Jiang SS, Kanteti R, Nandi S, et al: The
database of chromosome imbalance regions and genes resided in lung
cancer from Asian and Caucasian identified by array-comparative
genomic hybridization. BMC Cancer. 12:2352012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang B, Zhang J, Yin Y and Zhang Y:
Network-based inference framework for identifying cancer genes from
gene expression data. BioMed Res Int. 2013:4016492013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kavak E, Unlü M, Nistér M and Koman A:
Meta-analysis of cancer gene expression signatures reveals new
cancer genes, SAGE tags and tumor associated regions of
co-regulation. Nucleic Acids Res. 38:7008–7021. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Herzog H: Neuropeptide Y and energy
homeostasis: Insights from Y receptor knockout models. Eur J
Pharmacol. 480:21–29. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Renshaw D and Batterham RL: Peptide YY: A
potential therapy for obesity. Curr Drug Targets. 6:171–179. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huda MS, Wilding JP and Pinkney JH: Gut
peptides and the regulation of appetite. Obes Rev. 7:163–182. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang L, Bijker MS and Herzog H: The
neuropeptide Y system: Pathophysiological and therapeutic
implications in obesity and cancer. Pharmacol Ther. 131:91–113.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma Y, Hao X, Zhang S and Zhang J: The in
vitro and in vivo effects of human umbilical cord mesenchymal stem
cells on the growth of breast cancer cells. Breast Cancer Res
Treat. 133:473–485. 2012. View Article : Google Scholar
|
|
49
|
Escárcega RO, Fuentes-Alexandro S,
García-Carrasco M, Gatica A and Zamora A: The transcription factor
nuclear factor-kappa B and cancer. Clin Oncol. 19:154–161. 2007.
View Article : Google Scholar
|
|
50
|
Pikarsky E and Ben-Neriah Y: NF-kappaB
inhibition: A double-edged sword in cancer? Eur J Cancer.
42:779–784. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang J, Xu YJ, Xiong WN, Zhang ZX, Du CL,
Qiao LF, Ni W and Chen SX: Inhibition of NF-kappaB through
IkappaBalpha transfection affects invasion of human lung cancer
cell line A549. Ai Zheng. 27:710–715. 2008.In Chinese. PubMed/NCBI
|
|
52
|
Kuliková L, Mikeš J, Hýžďalová M, Palumbo
G and Fedoročko P: NF-κB is not directly responsible for
photoresistance induced by fractionated light delivery in HT-29
colon adenocarcinoma cells. Photochem Photobiol. 86:1285–1293.
2010. View Article : Google Scholar
|