Introduction
Lung cancer is the most common cause of
cancer-related mortality in humans and is characterised as either
non-small cell lung cancer (NSCLC) or small cell lung cancer (SCLC)
based on the histological type, size and appearance of the
malignant cells (1). NSCLC cases
comprise ~80% of all lung cancer cases. NSCLC is a heterogeneous
cancer that can be classified into three major subtypes: lung
squamous carcinoma, lung adenocarcinoma (LAC) and large cell
carcinoma (2). Common lung cancer
treatment includes chemotherapy, surgery and radiotherapy. Due to
the lack of an efficient diagnostic procedure for early-stage lung
cancer the majority of lung cancer patients are diagnosed with
end-stage disease, resulting in undesirable therapeutic outcomes. A
biomarker-driven approach in the pre-metastatic phase could aid
cancer diagnosis. However, current markers, such as Th19 fragment
antigen 21–1 and neuron-specific enolase, have only aided in the
diagnosis of 37.3% of lung cancer patients (3).
Recently, DNA methylation, a promising biomarker in
cancer diagnosis, was found to be significantly associated with
tumour formation. DNA methylation can regulate gene expression via
complex mechanisms (4). For
instance, hypermethylated cytosines inhibit the combination of DNA
and trans-acting factors in the context of cytosine-guanine
dinucleotides (CpGs), while demethylated cytosines promote this
combination. In cancer cells, CpG islands located in the promoter
of tumour-suppressor genes and 'housekeeping̓ genes become
hypermethylated, which decreases the expression of these genes and
in turn activates oncogenes (5–10).
Lokk et al observed the hypermethylation of 496 CpGs in 379
genes and hypomethylation of 373 CpGs in 335 genes in NSCLC
(7). Aberrant DNA methylation
reportedly serves as a predictive biomarker for the early-stage
diagnosis of cancer (9–13).
The eyes absent (EYA) protein family is a component
of a conserved regulatory network that is involved in cell-fate
determination and is associated with cell proliferation and
survival (14). They encode four
EYA proteins in humans, including EYA2, which is required in the
retinal determination network and is essential during development.
The single nucleotide polymorphisms (SNPs) of the eya2 gene
were found to be associated with the overall survival rate of NSCLC
patients (15). Studies suggest
that EYA2 is overexpressed in epithelial ovarian cancer, cervical
carcinogenesis and breast cancers and its overexpression is
correlated with poor prognosis, especially in breast cancer
(16–18). A recent study revealed that the
downregulation of EYA2 by tristetraprolin protein may reduce cell
viability (19). The eya2
gene is reportedly methylated in colorectal cancers but not in
normal tissues (20), and
epigenetic silencing of the eya2 gene has been shown to
promote pancreatic tumour growth (21). As a dual-functional transcription
factor/phosphatase, EYA2 has been found to regulate cell
development, differentiation and mortality and play an important
role in DNA damage and repair (22–24).
However, the fragment of the eya2 gene that was used to
conduct the methylation analysis in previous studies included only
~200 bases of the 5′-UTR and little information is available on the
methylation status of other CpGs in the eya2 gene. Thus, the
precise effect of EYA2 in NSCLC remains unclear, and additional
studies of the methylation levels of all CpG islands in the
eya2 gene and the precise role of EYA2 in NSCLC are
warranted.
In the present study, we evaluated the expression of
EYA2 in NSCLC cell lines and a normal lung cell line to identify
the association between EYA2 and oncogenesis in NSCLC. We then
examined the methylation levels of the eya2 gene in a LAC
cell line and human LAC tissues. The results suggest that the
aberrant expression and methylation of the eya2 gene are
associated with LAC oncogenesis. Transfecting LAC cells with small
interfering RNA (siRNA) inhibited the expression of EYA2, and
stable knockdown of this gene in cells was verified; these cells
were utilised in further research. Furthermore, the proliferation,
cell cycle, apoptosis, migration and invasion of cells were
assessed. Our data indicated that the inhibition of EYA2 suppressed
the growth of tumour cells by attenuating cell proliferation,
arresting the cell cycle, increasing apoptosis and repressing cell
migration and invasion. Our findings identified the eya2
gene as a cancer-related gene and a potential diagnostic biomarker
for LAC.
Materials and methods
Ethics statement
All cells used in the present study were purchased
from the American Type Culture Collection and all lung cancer
tissues used in the present study were obtained from the First
Affiliated Hospital of Kunming Medical University. All experimental
protocols were reviewed and approved by the Yunnan University
Ethics Committee and all subjects involved in the present study
provided informed consent.
Cell culture and sample
The human lung adenocarcinoma cell line A549
(CRM-CCL-185™; ATCC, Manassas, VA, USA), the human large cell
carcinoma cell line H661 (HTB-183D™; ATCC) and the human lung
squamous carcinoma cell line SKMES1 (HTB-58™; ATCC), were cultured
in Dulbecco's modified Eagle's medium (DMEM) (SH30022; Thermo
Fisher Scientific, Waltham, MA, USA) supplemented with 10% foetal
bovine serum (FBS, BI, AU, 04-12-1). The BEAS-2B cell line
(CRL-9609™; ATCC), a human normal pulmonary epithelial cell line,
was cultured in RPMI-1640 (SH30027; Thermo Fisher Scientific)
supplemented with 10% FBS, and 10 paraffin-embedded lung
adenocarcinoma tissues were obtained from the First Affiliated
Hospital of Kunming Medical University. Participants in the present
study group ranged from 42 to 61 years of age (mean age of males
n=4, 52.25 years; mean age of females n=6, 55.83 years). The
patients did not undergo any preoperative chemotherapy or
radiotherapy.
Western blot analysis
The total protein of the A549 and BEAS-2B cells was
isolated on ice with a Tissue or Cell Total Protein Extraction kit
(BSP003; Sangon, Shanghai, China). For each sample, 80 µg of
total protein was electrophoresed on a 10% SDS-polyacrylamide gel
followed by electrophoretic transfer to a polyvinylidene fluoride
membrane. Immunoblotting was performed using an EYA2
(N-16)-specific goat polyclonal antibody (sc-15097; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). The results were
visualised by chemiluminescence with Super Signal West Pico (34080;
Thermo Fisher Scientific). The expression level of the target
proteins was normalised to the expression of GAPDH (Affinity, USA).
The expression data were obtained with ImageJ.
Quantitative real-time PCR
Total RNA was extracted from the cultured cells and
reverse transcribed using the PrimeScript™ RT reagent kit with gDNA
Eraser (RR047; Takara, Otsu, Shiga, Japan). The transcription
levels of the eya2 gene were quantitatively analysed using
real-time PCR (25) on a 7300
Real-Time PCR system with the SYBR Premix Ex Taq™ (RR420, Takara).
The amount of target cDNA in each sample was measured by
determining a fractional PCR threshold cycle number (Ct) and
estimated by interpolating from a standard curve. The standard
curve was prepared from known amounts of the corresponding product
with the same primer sets and was run on each PCR plate. The
transcription levels of the target gene were normalised to the
transcription of the GAPDH gene. The primer sequences are shown in
Table II.
 | Table IIReverse transcription and real-time
quantitative PCR primers. |
Table II
Reverse transcription and real-time
quantitative PCR primers.
| Gene | Sense primer (5′ to
3′) | Antisense primer
(5′ to 3′) |
|---|
| EYA2 |
CAAGGAGGAAATGGACTGGG |
GGGTTGTAGGATGAGCCGTAA |
| GAPDH |
CACTCCTCCACCTTTGACGC |
TGCTGTAGCCAAATTCGTTGT |
CpG island analysis
The CpG islands of the eya2 gene were
analysed with the Methyl Primer Express v1.0 and MethPrimer
software available online (http://www.urogene.org/methprimer/) as follows: a
region of >200 bases with a G+C content of at least 50% and a
ratio of observed to statistically expected CpG frequencies of at
least 0.6 CpG dinucleotides (4).
DNA extraction and bisulphite
modification
The DNA was extracted from the cells using the
Wizard Genomic DNA purification kit (A1120; Promega, Madison, WI,
USA), while the QIAamp DNA FFPE Tissue Kit (50) (56404; Qiagen,
Hilden, Germany) was used to extract DNA from paraffin-embedded
tissues (2.4 cm3 of tumour and matching paracancerous
tissues). The DNA yield and purity were determined using a NanoDrop
2000 spectrophotometer. From each sample, 500 ng of genomic DNA was
bisulphite-modified using a Methyledge™ bisulphite conversion
system (1301; Promega) according to the manufacturer's
recommendations.
Bisulphite genomic-PCR
The CpG islands of the eya2 gene were
produced using nested PCR (26) and
detected with agarose gel electrophoresis. The modified DNA was
subjected to stage-1 PCR with Tm touchdown from 68 to 48°C,
followed by 20 cycles with Tm of 48°C. The product of the stage was
then used as a template for the stage 2 PCR. Six pairs of primers
(Table I; fragment 1, 2, 3, 4 and
5) were used for the CpG island of the eya2 gene in the cell
lines and four pairs of primers (Table
I; fragment 1, 2, 3 and 5) were used for the tissues. The
product DNA was visualised on 2% agarose gels and purified with the
Takara MiniBEST Agarose Gel DNA Extraction kit ver. 4.0 (9762;
Takara).
 | Table IThe CpG island of EYA2 PCR regions
and primers. |
Table I
The CpG island of EYA2 PCR regions
and primers.
| Fragment site | Primer (5′ to
3′) | Product size
(bp) |
|---|
1
(−163) to (−36) | Stage-1 (2) | Sense:
GAATGTTAGTGTTATTATTGAGGTTTTT | 128 |
| Antisense:
CCTCCCCACCCACCAAC |
2
(−36) to (200) | Stage-1 (2) | Sense:
GAGGTTGGGTTTTGGTTTTTA | 237 |
| Antisense:
ACCCCTTCTCCTCCCTAAAC |
3
(181) to (444) | Stage-1 (2) | Sense:
GTTTAGGGAGGAGAAGGGGT | 264 |
| Antisense:
AAAAAATCCCYATAAACAACTCC |
4
(422) to (543) | Stage-1 | Sense:
TTAGGGAGGAGAAGGGGT | 122 |
| Antisense:
CCCTTATACCTTCCTAACCC |
| Stage-2 | Sense:
GGAGTTGTTTATYGGGATTTTT | |
| Antisense:
CCCTTATACCTTCCTAACCCT |
5
(556) to (788) | Stage-1 (2) | Sense:
TAGTAGAGTTTTTTTTTGGAAAGGT | 233 |
| Antisense:
CCATAAACACTACCTAAAAACTTAAAT |
DNA cloning and sequencing
The purified CpG island DNA was cloned with the
pMDTM18-T vector cloning kit (Takara, 6011) and Dh5α competent
cells (Takara, 9057). The clone products were sequenced at the
Beijing Genomics Institute (BGI, Shenzhen, China). The gene
methylation status was analysed with the BIQ Analyser (27).
Liposome-mediated lung adenocarcinoma
cell transfection
The experiment was divided into three groups: the
EYA2-special siRNA-FAM A549 cell group, the negative control-FAM
A549 cell group (scrambled) and the untreated A549 cell group
(mock). EYA2-specific siRNA sequences were forward,
CAGCGAUUGUCUGGAUAAATT and reverse, UUUAUCCAGACAAUCGCUGTT. The
scrambled siRNA sequences were forward, UUCUCCGAACGUGUCACGUTT and
reverse, ACGUGACACGUUCGGAGAATT (A10005; GenePharma, Shanghai,
China). For liposome trans-fection, the cells were plated in 6-well
plates (1×105/well) 12 h prior to transfection. All
siRNAs (5 µg/well) were transfected with Lipofectamine 3000
(L3000001; Invitrogen, Carlsbad, CA USA).
Cell proliferation assay
The cell viability was evaluated with a
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
inner salt (MTS) colorimetric assay using the CellTiter 96 Aqueous
One Solution Cell Proliferation Assay (G3582; Promega) according to
the manufacturer's instructions. All groups were dispensed at a
density of 1,000 cells/well in 96-well plates with DMEM containing
10% FBS and the corresponding transfection solution after
transfection for 48 h. After various intervals of incubation, 40
µl of MTS reagent was added to the medium and incubated for
4 h at 37°C in 5% CO2 and the absorbance was then read
at 490 nm in a microplate absorbance reader.
Cell cycle and apoptosis analysis
To analyse the cell cycle, the cells were collected
48 h after transfection. The cells were then washed with PBS and
stained with 50 µg/ml propidium iodide (PI) in the dark at
room temperature for 5 min. The intracellular DNA content was
analysed using a flow cytometry (Accuri™ C6; BD).
To analyse apoptosis, the apoptotic cells were
quantified using flow cytometry 72 h after transfection using an
Annexin V-FITC/PI Apoptosis Detection kit (556547; BD) according to
the manufacturer's instructions. The cells were collected, washed
with PBS and re-suspended with 100 µl of 1X Annexin
V-binding buffer. The samples were then stained with 5 µl of
FITC Annexin V and 5 µl of PI at room temperature for 15 min
in the dark. In addition, 400 µl 1X Annexin V-binding buffer
was added prior to the flow cytometric analysis.
Migration assay
The migratory ability of the A549 cells was
evaluated using the wound healing method, as previously described
(28). Three groups of cells
(transfected with 0, 100 or 200 nM siRNA) were grown to confluency
in 6-well plates. A line of cells was scraped away in each well
using a sterile 200 µl pipette tip. The wells were then
immediately filled with 1 ml of DMEM containing 10% FBS and the
corresponding transfection reagent.
Immediately after the scratch and at 24 h, two
images of five locations of the same scraped area were captured
with a microscope set at a magnification ×100. The remaining
wounded area per image was measured. Three independent experiments
were performed.
Invasion assay
The invasive potential of the cells was assessed in
a Transwell, which allowed the cells to pass through a
polycarbonate membrane (8-µm pore size) coated with Matrigel
(356234; Haoran, Shanghai, China). Briefly, the Transwell was
coated with Matrigel (2 mg/ml) diluted with serum-free DMEM for 30
min at 37°C. Three groups of cells (transfected with 0, 100 or 200
nM siRNA) (5×104 cells) were then re-suspended in
serum-free DMEM (200 µl) and deposited into the upper
chamber of each well. The lower chambers were also filled with 750
µl of DMEM containing 10% FBS. The cells were incubated for
24 h at 37°C in 5% CO2. The cells on the upper chamber
were removed via gentle scraping. The cells on the lower surface
were then fixed with 4% paraformaldehyde for 20 min, followed by
Giemsa staining for 20 min in the dark. Four images of the invading
cells were captured at a magnification ×100 in each well. Three
independent experiments were performed.
Statistical analysis
Statistical analysis was carried out using the
GraphPad Prism software (version 5.0) (29), and all results are expressed as the
mean ± SD. A t-test was performed to identify the differences in
the DNA methylation levels of the eya2 gene between the A549
and BEAS-2B cells. A one-way ANOVA was performed to identify the
differences in expression intensities of the EYA2 protein among
four groups of examined cells, changes in proliferation, the cell
cycle, apoptosis, migration capacity and invasive capability after
the knockdown of EYA2 in A549 cells. P-values of <0.05 were
considered statistically significant.
Results
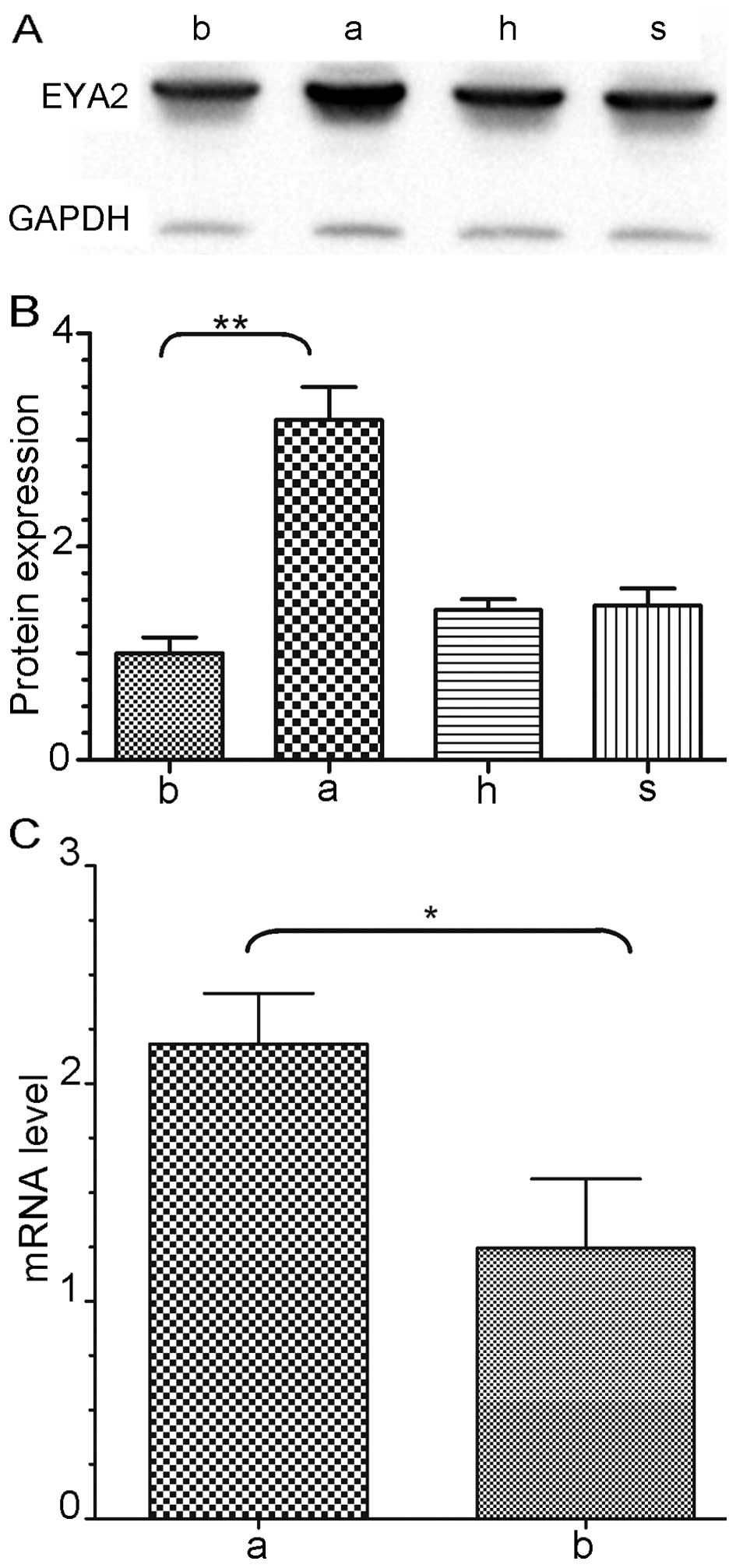
Upregulated expression of EYA2 in lung
adenocarcinoma cells
The EYA2 protein expression levels were quantified
(Fig. 1A). The results of the
western blot analysis revealed that the expression of EYA2 protein
was significantly upregulated 3.2-fold in the A549 cells compared
to that in the BEAS-2B cells (P<0.01, Fig. 1B), while obvious differences were
absent in the H661 and SKMES1 cells compared to that in the BEAS-2B
cells. The mRNA and protein levels were detected by quantitative
real-time RT-PCR and western blot analysis, respectively
(n=3/group). The EYA2 mRNA transcriptional level was upregulated
2.1-fold in the A549 cells compared to this level in the BEAS-2B
cells and this upregulation was significant (P<0.05, Fig. 1C).
Aberrant hypomethylation of the eya2 gene
in lung adenocarcinoma
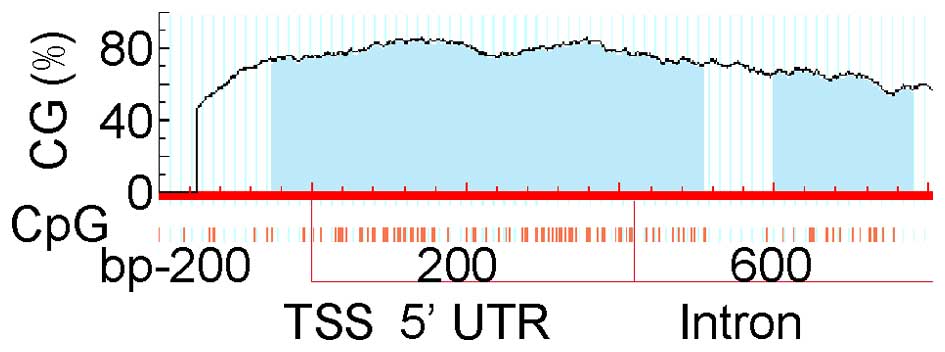
The CpG islands of the eya2 gene are shown in
Fig. 2. The entire eya2 gene
sequence, including the 2000-bp upstream region of the TSS, was
analysed. CpG islands were not found upstream of TSS and a long
fragment of 951 bp (from the 163 bp upstream to the 788 bp
downstream of the eya2 gene TSS, containing the upstream
region of TSS, 5′-UTR and the first intron) that included 105 CpGs
was selected. The selected area was divided into 5 DNA fragments to
perform PCR. Five fragment locations and the primers for PCR are
listed in Table I.
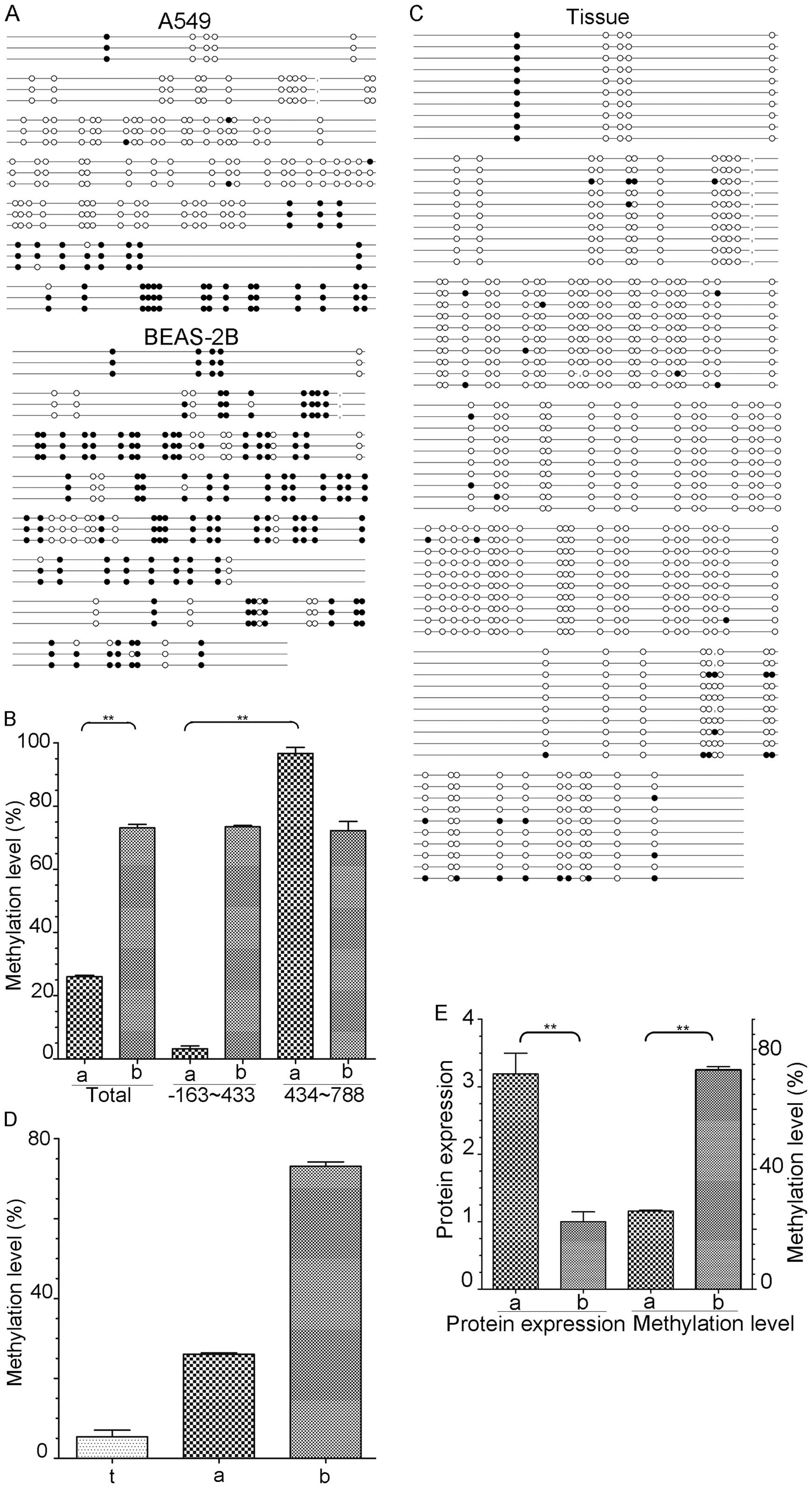
A total of three clones were detected in the two
cell lines. The methylation statuses of the eya2 gene in the
A549 and BEAS-2B cells are described (Fig. 3A). The data were corrected for
reversed sequences, erroneous bases and identical clones. On
average 27 CpG dinucleotides out of 105 CpGs were methylated in the
A549 cells and 76 CpGs out of 105 CpGs were methylated in the
BEAS-2B cells. The total methylation level of CpGs significantly
differed between the A549 and BEAS-2B cells at 25.71 and 72.38%,
respectively (P<0.01, Fig. 3B).
Moreover, the methylation level of the A549 cells significantly
varied by fragment. Two out of 75 CpGs were methylated in the TSS
163-bp upstream to 433-bp downstream fragment and 30 CpGs were
methylated in the TSS 434 to 788 bp downstream fragment; the
overall methylation levels were 2.67 and 96.67%, respectively
(P<0.01). These two fragments did not significantly differ and
the methylation levels were 73.43 and 72.23% in the BEAS-2B
cells.
In total, 10 tumour tissues were detected by BSP.
The methylation statuses of the tissues are displayed in Fig. 3C, and the data were corrected for
reversed sequences, erroneous bases and identical clones. On
average, 5 CpG dinucleotides out of 96 CpGs were methylated in the
tumour tissues (4 fragments, including 9 CpGs, were excluded
because most of the tumour tissues did not yield PCR products). The
total methylation levels of the eya2 gene were low in both
the A549 cells and tumour tissues compared with the level in the
BEAS-2B cells (Fig. 3D). However,
the methylation levels in the tumour tissues were lower than those
in the A549 cells at 5.31 and 25.71%, respectively. The obvious
difference between the TSS 163-bp upstream to 433-bp downstream
fragment and the TTS 434 to 788 bp downstream fragment was absent
in the tumour tissues.
To research the significance of EYA2 in LAC, the
relevance of EYA2 expression to the methylation status of the
eya2 gene was analysed in the A549 and BEAS-2B cells. The
distribution of the eya2 gene methylation levels was
inversely correlated with the EYA2 expression levels in these two
cell lines (Fig. 3E).
Knockdown of EYA2 effects lung
adenocarcinoma cell proliferation by arresting the cell cycle and
increasing apoptosis
To further investigate the biological role of EYA2
in LAC, an RNA interference approach was utilised to downregulate
the expression of EYA2 in the A549 cell line. Moreover, we
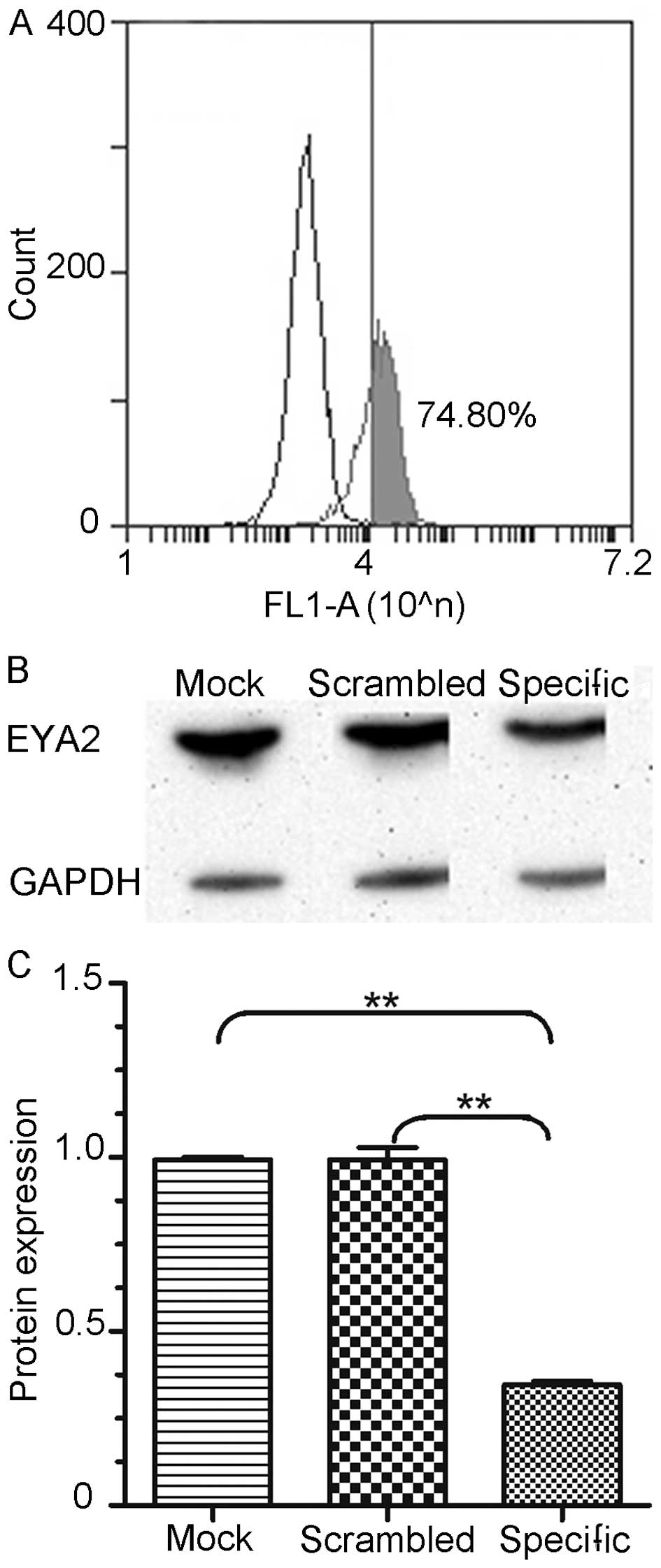
evaluated the knockdown efficiencies of EYA2-specific siRNA-FAM in
the A549 cells by flow cytometry and western blot analysis
(n=3/group) 48 h after transfection. We detected that 74.80% of the
processed cells were successfully trans-fected with siRNA-FAM
(Fig. 4A). The expression level of
EYA2 was significantly decreased in the cells transfected with the
EYA2-specific siRNA 48 h after transfection compared with the
negative control and the untreated group (Fig. 4B). The differences between the
specific and mock construct and between the specific and scrambled
construct were both significant (0.3472 vs. 0.9934, P<0.01 and
0.3472 vs. 0.9925, P<0.01; Fig.
4C). These results indicated that the EYA2-specific siRNA
effectively suppressed EYA2 expression.
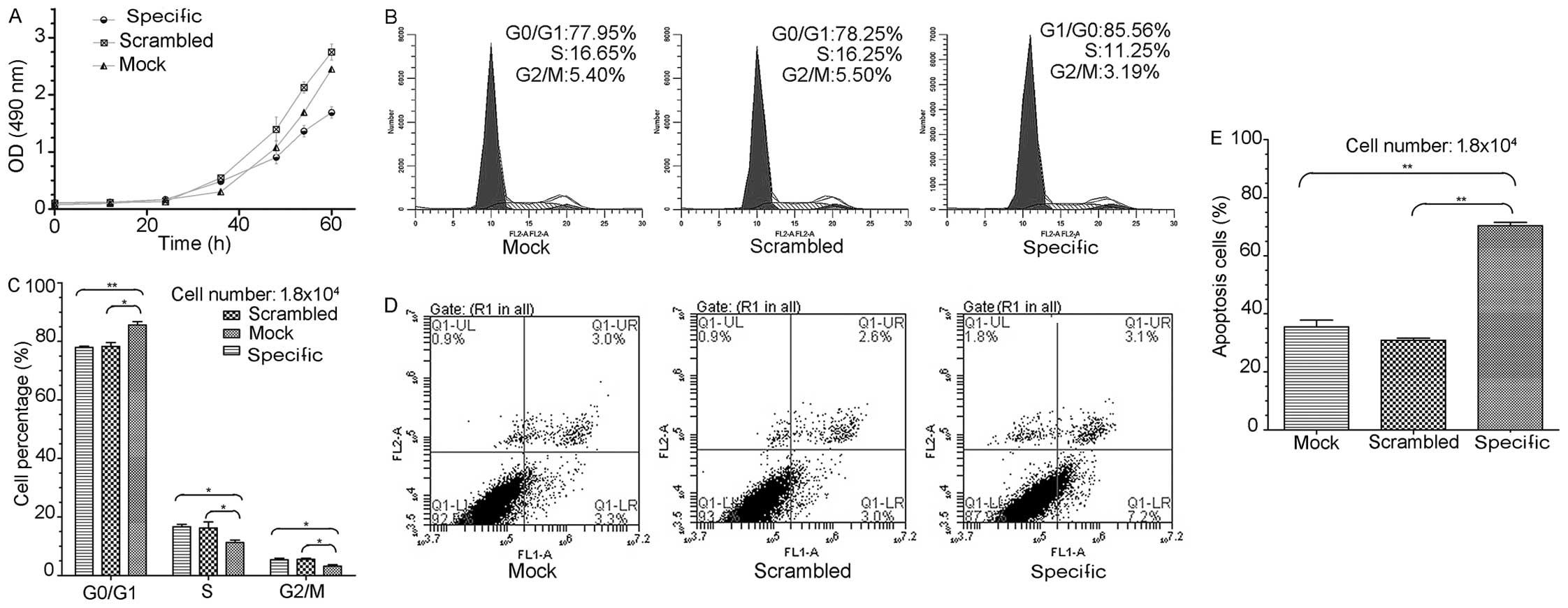
We assessed the effects of EYA2 knockdown on the
proliferation of A549 cells with an MTS colorimetric assay
(n=5/group). Our data revealed that the inhibition of EYA2
significantly inhibited the proliferation of the A549 cells
(Fig. 5A). Our findings demonstrate
that EYA2 contributed to the proliferation of A549 cells in
vitro.
Because the knockdown of EYA2 by siRNA suppressed
the proliferation of A549 cells, we examined whether this effect
was mediated by a change in the cell cycle. We detected changes in
the cell cycles in the three groups of siRNA-treated cells
(specific, scrambled and mock) 48 h after transfection using PI
staining and flow cytometry (n=3/group) (Fig. 5B). The number of cells in the G0/G1
phase was significantly increased and those in the S and G2/M
phases were significantly decreased in the cells transfected with
the specific siRNA compared to the mock group and the scrambled
group (G0/G1: 85.56 vs. 77.95%, P<0.01 and 85.56 vs. 78.25%,
P<0.05; S: 11.25 vs. 16.65%, P<0.05 and 11.25 vs. 16.25%,
P<0.05; G2/M: 3.19 vs. 5.40%, P<0.05 and 3.19 vs. 5.5%,
P<0.05; Fig. 5C). These results
suggest that the downregulation of EYA2 arrested most cells in the
G1 phase and prevented mitosis in the A549 cell line.
To explore the potential effect of the knockdown of
EYA2 on apoptosis in the A549 cells, we examined differences in
apoptosis between the three siRNA-transfected groups of cells
(specific, scrambled and mock) 60–72 h after transfection using
Annexin V-FITC and PI double-staining in conjunction with flow
cytometry (n=3/group) (Fig. 5D).
The percentage of apoptotic cells in the specific group was
markedly increased compared to the scrambled group and the mock
group (7.2 vs. 3.0%, P<0.01 and 7.2 vs. 3.3%, P<0.01;
Fig. 5E). These results imply that
the inhibition of EYA2 promoted the apoptosis of the A549
cells.
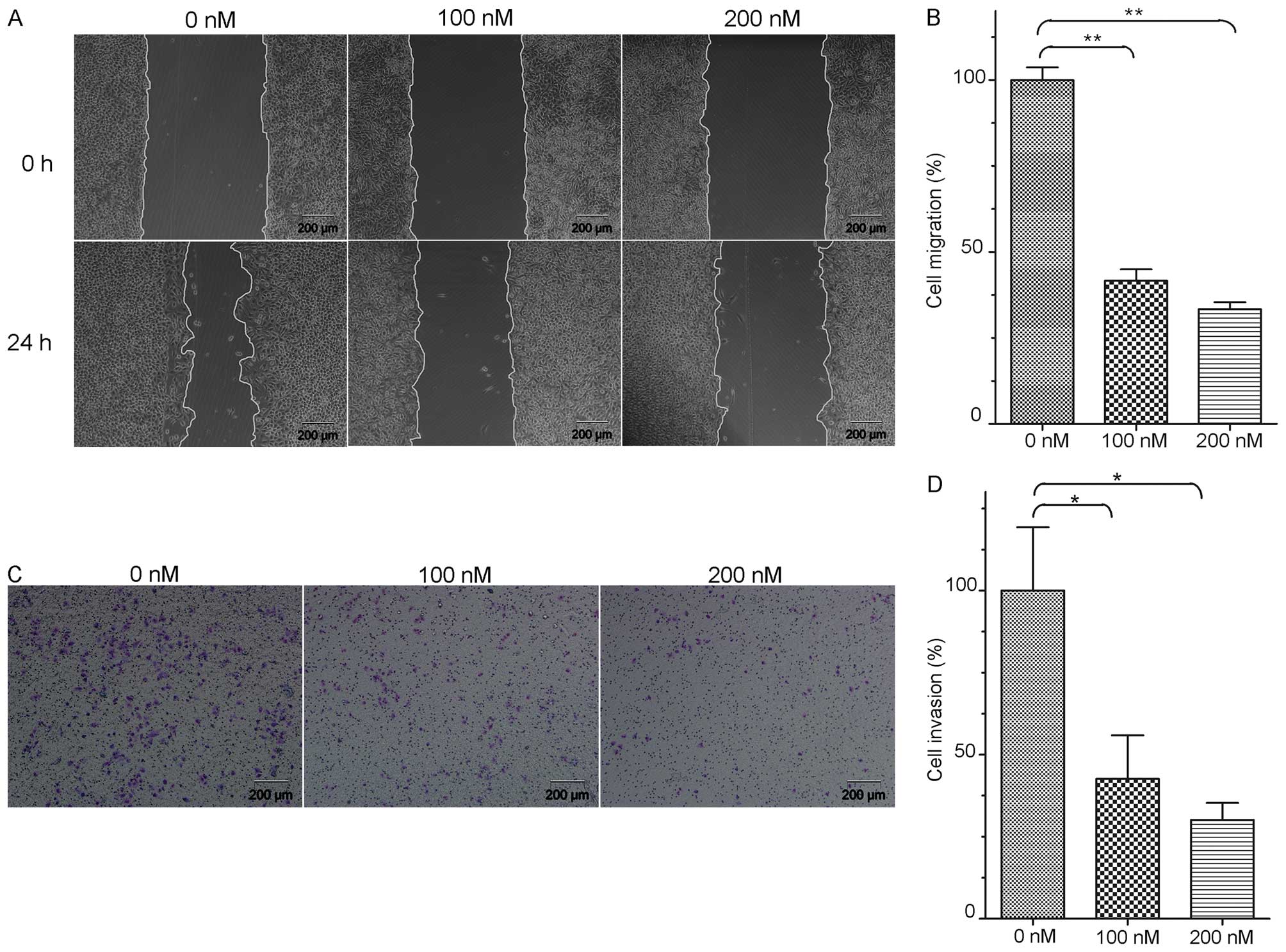
Downregulation of EYA2 inhibits the
migratory and invasive potential of lung adenocarcinoma cells
A wound healing assay (WH) was used to detect the
change in the migratory capability of the A549 cells (Fig. 6A). After transfection with 100 and
200 nM siRNA for 24 h, the percentages of A549 cells that
penetrated the membrane were significantly decreased by 41.70 and
33.41% compared with that in the control group, respectively
(P<0.01, Fig. 6B). These results
indicated that the downregulation of EYA2 suppressed the migratory
capacity of the A549 cells.
Additionally, we tested the invasive capacity of the
A549 cells after the knockdown of EYA2 (Fig. 6C). After transfection with 100 and
200 nM siRNA for 24 h, the abilities of the cells to enzymatically
degrade the Matrigel and migrate to a new site was significantly
decreased by 42.63 and 30.13% compared with the control group
(P<0.05, Fig. 6D). Therefore,
the results revealed that the knockdown of EYA2 decreased the
invasive capacity of the A549 cells.
Discussion
EYA2 is a key regulator that can prevent adverse
cardiac remodelling under pressure overload by altering metabolic
gene expression and preserving the PI3K/Akt/mTOR signalling pathway
(30,31). Furthermore, the physical complex of
EYA2 and SIX1 plays a critical role in the preservation of
mitochondrial integrity in response to pressure overload due to
physiological hypertrophy and is essential for mammalian
development (32); a loss of this
function may cause branchiootorenal (BOR) syndrome (33,34).
Moreover, EYA2 is associated with G protein-mediated signalling,
retinoid-induced limb malformations and hypaxial somitic myogenesis
(35–37). Thus, EYA2 may exert a key effect on
disease development and tumourigenesis. In the present study, we
analysed the expression of EYA2 in NSCLC cells and studied the
methylation of the eya2 gene in the A549 and BEAS-2B cell
lines as well as in LAC tissues. In addition, we knocked down the
expression of EYA2 and conducted various functional studies
associated with tumour cell growth and metastasis in these
cells.
Farabaugh et al found that EYA2 functions as
a prometastatic factor in breast cancer (17). Vincent and his collaborators
hypothesised that EYA2 expression is reduced due to epigenetic
silencing, which promotes tumour growth in pancreatic cancer
(21). Zou et al reported
that EYA2 is hypermethylated in colorectal neoplasms (20). In the present study, the eya2
gene was hypomethylated in the A549 cell line and cancer tissues
and hypermethylated in the BEAS-2B cell line. The results showed
that the DNA methylation levels significantly differed between the
LAC and the normal control samples. Moreover, the selected intron
region significantly differed between the paraffin-embedded tissues
and the A549 cell line. The growth environment of the A549 cell
line in vitro differs from the in vivo environment
and further research is warranted to determine whether the
developmental conditions of the cells influence their DNA
methylation levels. Furthermore, the EYA2 expression was markedly
increased in the lung adenocarcinoma cell line compared with the
normal pulmonary epithelial cell line and the relevance of the
methylation levels of the eya2 gene and EYA2 expression
levels were markedly inversely correlated, which is consistent with
a study by Zhang et al (18). In contrast to previous studies in
which the methylation levels of short sequences of the eya2
gene 5′-UTR region were analysed, we analysed the methylation
status of the entire CpG islands of the eya2 gene, which
containing almost all selected fragments in previously methylated
studies of the eya2 gene. Thus, our data constitute
conclusive proof of the association between the aberrant
methylation of EYA2 and oncogenesis in NSLCL. The function of EYA2
may be organ- or cell-specific. Further studies are warranted to
elucidate the complex function of EYA2 in tumours.
Additionally, deregulated cell cycle progression is
one of the primary characteristics of human cancer cells (38). EYA2 was found to increase the
expression of c-Myc, cyclin A, D1 and E, which are all involved in
G1/S cell cycle progression in breast cancer cells (39). Clark et al discovered that
inappropriate changes in the steady state levels of EYA proteins
can trigger programmed cell death during development (40). To identify the role that EYA2 plays
in the function of A549 cells, we knocked down the expression of
EYA2 in A549 cells via RNA interference technology in vitro.
First, we confirmed that the specific siRNA could efficiently
reduce the EYA2 expression in the A549 cells. The proliferation
rate of the A549 cells decreased when the endogenous EYA2 was
downregulated compared with the control groups. Moreover, we
examined the cycle distribution of the A549 cells after the
inhibition of EYA2. Our data revealed that the knockdown of EYA2
arrested cells in the G0/G1 phase. Furthermore, the percentage of
apoptotic A549 cells was increased when EYA2 was knocked down
compared with the control group. Thus, we concluded that reduction
of EYA2 may suppress A549 cell proliferation by inhibiting the G1/S
transition of the cell cycle and increasing apoptosis. In addition,
previous studies also showed that EYA2 is associated with cell
proliferation, the cell cycle and apoptosis (41–44).
Pandey et al discovered that EYA3, which
belongs to the EYA family of proteins, promoted not only tumour
cell proliferation, but also their migration and invasion (45). EYA2 is a homologous protein of EYA3
and may play the same important role in tumour cell metastasis.
Therefore, we investigated the effect of EYA2 knockdown on cell
migration and invasion in the A549 cells. As expected,
downregulation of EYA2 reduced the migratory and invasive
capacities of the A549 cells, consistent with prior research by
Krueger et al, who implicated EYA2 in tumour development
(46).
In conclusion, we found that the methylation status
and expression of the eya2 gene were altered in lung
adenocarcinoma and that the knockdown of EYA2 could change various
functions in lung adenocarcinoma cells. Previous studies have shown
that the eya2 gene may serve as a promising early-stage
diagnostic biomarker and key treatment target gene in lung
adenocarcinoma. These findings imply the importance of EYA2 in
NSCLC development and progression. We plan to explore whether the
growth status of tumour cells can deteriorate by specifically
modulating the methylation of the eya2 gene. To this end, a
large number of specific inhibitors should be screened to
ameliorate the aberrant expression of EYA2 in tumour cells and
identify an inhibitor that can suppress the growth of tumour cells.
Thus, clearly further investigation are required.
Acknowledgments
We express our sincere appreciation to all patients
that participated in the present study. The present study was
financially supported by grants from the National Natural Science
Foundation of China (no. 81372360) to C.X. and from the National
Natural Science Foundation of China (no. 81460435) and the Key
Projects of Applied Basic Reasearch of Yunnan Province (no.
2014FA022) to S.Z.
Abbreviations:
|
EYA2
|
eyes absent homologue 2 protein
(Accession no. CAA71310)
|
|
eya2
|
eyes absent homologue 2 gene (gene ID:
2139)
|
|
NSCLC
|
non-small lung cancer
|
|
LAC
|
lung adenocarcinoma
|
|
SCLC
|
small cell lung cancer
|
|
CpGs
|
cytosine-guanine dinucleotides
|
|
TSS
|
transcriptional start site
|
References
|
1
|
Kumar V; Abass KA, Fausto N and Mitchell
R: Robbins Basic Pathology. 8th edition. Saunders Elsevier;
Philadelphia, PA: 2007
|
|
2
|
Brambilla E, Travis WD, Colby TV, Corrin B
and Shimosato Y: The new World Health Organization classification
of lung tumours. Eur Respir J. 18:1059–1068. 2001. View Article : Google Scholar
|
|
3
|
Chu XY, Hou XB, Song WA, Xue ZQ, Wang B
and Zhang LB: Diagnostic values of SCC, CEA, Cyfra21-1 and NSE for
lung cancer in patients with suspicious pulmonary masses: A single
center analysis. Cancer Biol Ther. 11:995–1000. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takai D and Jones PA: Comprehensive
analysis of CpG islands in human chromosomes 21 and 22. Proc Natl
Acad Sci USA. 99:3740–3745. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grønbaek K, Hother C and Jones PA:
Epigenetic changes in cancer. APMIS. 115:1039–1059. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lokk K, Vooder T, Kolde R, Välk K, Võsa U,
Roosipuu R, Milani L, Fischer K, Koltsina M, Urgard E, et al:
Methylation markers of early-stage non-small cell lung cancer. PLoS
One. 7:e398132012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tekpli X, Landvik NE, Anmarkud KH, Skaug
V, Haugen A and Zienolddiny S: DNA methylation at promoter regions
of interleukin 1B, interleukin 6, and interleukin 8 in non-small
cell lung cancer. Cancer Immunol Immunother. 62:337–345. 2013.
View Article : Google Scholar
|
|
9
|
Sato T, Arai E, Kohno T, Tsuta K, Watanabe
S, Soejima K, Betsuyaku T and Kanai Y: DNA methylation profiles at
precancerous stages associated with recurrence of lung
adenocarcinoma. PLoS One. 8:e594442013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pesta M, Kulda V, Topolcan O, Safranek J,
Vrzalova J, Cerny R and Holubec L: Significance of methylation
status and the expression of RECK mRNA in lung tissue of patients
with NSCLC. Anticancer Res. 29:4535–4539. 2009.PubMed/NCBI
|
|
11
|
Suzuki M, Shiraishi K, Eguchi A, Ikeda K,
Mori T, Yoshimoto K, Ohba Y, Yamada T, Ito T, Baba Y, et al:
Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small
cell lung cancer. Oncol Rep. 29:1308–1314. 2013.PubMed/NCBI
|
|
12
|
Grimminger PP, Maus MK, Schneider PM,
Metzger R, Hölscher AH, Sugita H, Danenberg PV, Alakus H and
Brabender J: Glutathione S-transferase PI (GST-PI) mRNA expression
and DNA methylation is involved in the pathogenesis and prognosis
of NSCLC. Lung Cancer. 78:87–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan H, He Z, Ma J, Zhang B, Sheng Z, Bin
P, Cheng J, Niu Y, Dong H, Lin H, et al: Global and MGMT promoter
hypomethylation independently associated with genomic instability
of lymphocytes in subjects exposed to high-dose polycyclic aromatic
hydrocarbon. Arch Toxicol. 87:2013–2022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tadjuidje E and Hegde RS: The eyes absent
proteins in development and disease. Cell Mol Life Sci.
70:1897–1913. 2013. View Article : Google Scholar :
|
|
15
|
Huang YT, Heist RS, Chirieac LR, Lin X,
Skaug V, Zienolddiny S, Haugen A, Wu MC, Wang Z, Su L, et al:
Genome-wide analysis of survival in early-stage non-small-cell lung
cancer. J Clin Oncol. 27:2660–2667. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bierkens M, Krijgsman O, Wilting SM, Bosch
L, Jaspers A, Meijer GA, Meijer CJ, Snijders PJ, Ylstra B and
Steenbergen RD: Focal aberrations indicate EYA2 and hsa-miR-375 as
oncogene and tumor suppressor in cervical carcinogenesis. Genes
Chromosomes Cancer. 52:56–68. 2013. View Article : Google Scholar
|
|
17
|
Farabaugh SM, Micalizzi DS, Jedlicka P,
Zhao R and Ford HL: Eya2 is required to mediate the pro-metastatic
functions of Six1 via the induction of TGF-β signaling,
epithelial-mesenchymal transition, and cancer stem cell properties.
Oncogene. 31:552–562. 2012.
|
|
18
|
Zhang L, Yang N, Huang J, Buckanovich RJ,
Liang S, Barchetti A, Vezzani C, O'Brien-Jenkins A, Wang J, Ward
MR, et al: Transcriptional coactivator Drosophila eyes absent
homologue 2 is up-regulated in epithelial ovarian cancer and
promotes tumor growth. Cancer Res. 65:925–932. 2005.PubMed/NCBI
|
|
19
|
Yeh PA, Yang WH, Chiang PY, Wang SC, Chang
MS and Chang CJ: Drosophila eyes absent is a novel mRNA target of
the tristetraprolin (TTP) protein DTIS11. Int J Biol Sci.
8:606–619. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zou H, Harrington JJ, Shire AM, Rego RL,
Wang L, Campbell ME, Oberg AL and Ahlquist DA: Highly methylated
genes in colorectal neoplasia: implications for screening. Cancer
Epidemiol, Biomarkers Prev. 16:2686–2696. 2007. View Article : Google Scholar
|
|
21
|
Vincent A, Hong SM, Hu C, Omura N, Young
A, Kim H, Yu J, Knight S, Ayars M, Griffith M, et al: Epigenetic
silencing of EYA2 in pancreatic adenocarcinomas promotes tumor
growth. Oncotarget. 5:2575–2587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jemc J and Rebay I: Identification of
transcriptional targets of the dual-function transcription
factor/phosphatase eyes absent. Dev Biol. 310:416–429. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jemc J and Rebay I: The eyes absent family
of phosphotyrosine phosphatases: Properties and roles in
developmental regulation of transcription. Annu Rev Biochem.
76:513–538. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krishnan N, Jeong DG, Jung SK, Ryu SE,
Xiao A, Allis CD, Kim SJ and Tonks NK: Dephosphorylation of the
C-terminal tyrosyl residue of the DNA damage-related histone H2A.X
is mediated by the protein phosphatase eyes absent. J Biol Chem.
284:16066–16070. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Spackman E and Suarez DL: Type A influenza
virus detection and quantitation by real-time RT-PCR. Methods Mol
Biol. 436:19–26. 2008.PubMed/NCBI
|
|
26
|
Kobayashi H and Kono T: DNA methylation
analysis of germ cells by using bisulfite-based sequencing methods.
Methods Mol Biol. 825:223–235. 2012. View Article : Google Scholar
|
|
27
|
Becker D, Lutsik P, Ebert P, Bock C,
Lengauer T and Walter J: BiQ Analyzer HiMod: An interactive
software tool for high-throughput locus-specific analysis of
5-methylcytosine and its oxidized derivatives. Nucleic Acids Res.
42(W1): W501–W507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arranz-Valsero I, Soriano-Romaní L,
García-Posadas L, López-García A and Diebold Y: IL-6 as a corneal
wound healing mediator in an in vitro scratch assay. Exp Eye Res.
125:183–192. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo J, Lin P, Zhao X, Zhang J, Wei X, Wang
Q and Wang C: Etazolate abrogates the lipopolysaccharide
(LPS)-induced downregulation of the cAMP/pCREB/BDNF signaling,
neuro-inflammatory response and depressive-like behavior in mice.
Neuroscience. 263:1–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang DK, Choi BY, Lee YH, Kim YG, Cho MC,
Hong SE, Kim H, Hajjar RJ and Park WJ: Gene profiling during
regression of pressure overload-induced cardiac hypertrophy.
Physiol Genomics. 30:1–7. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee SH, Yang DK, Choi BY, Lee YH, Kim SY,
Jeong D, Hajjar RJ and Park WJ: The transcription factor Eya2
prevents pressure overload-induced adverse cardiac remodeling. J
Mol Cell Cardiol. 46:596–605. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee SH, Kim J, Ryu JY, Lee S, Yang DK,
Jeong D, Kim J, Lee SH, Kim JM, Hajjar RJ, et al: Transcription
coactivator Eya2 is a critical regulator of physiological
hypertrophy. J Mol Cell Cardiol. 52:718–726. 2012. View Article : Google Scholar
|
|
33
|
Patrick AN, Cabrera JH, Smith AL, Chen XS,
Ford HL and Zhao R: Structure-function analyses of the human
SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome.
Nat Struct Mol Biol. 20:447–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ohto H, Kamada S, Tago K, Tominaga SI,
Ozaki H, Sato S and Kawakami K: Cooperation of six and eya in
activation of their target genes through nuclear translocation of
Eya. Mol Cell Biol. 19:6815–6824. 1999.PubMed/NCBI
|
|
35
|
Grifone R, Demignon J, Giordani J, Niro C,
Souil E, Bertin F, Laclef C, Xu PX and Maire P: Eya1 and Eya2
proteins are required for hypaxial somitic myogenesis in the mouse
embryo. Dev Biol. 302:602–616. 2007. View Article : Google Scholar
|
|
36
|
Embry AC, Glick JL, Linder ME and Casey
PJ: Reciprocal signaling between the transcriptional co-factor Eya2
and specific members of the Galphai family. Mol Pharmacol.
66:1325–1331. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ali-Khan SE and Hales BF: Novel retinoid
targets in the mouse limb during organogenesis. Toxicol Sci.
94:139–152. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Molinari M: Cell cycle checkpoints and
their inactivation in human cancer. Cell Prolif. 33:261–274. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J,
Cheng L, Fan Z, Yuan B, Tian P, et al: miR-30a suppresses breast
cancer cell proliferation and migration by targeting Eya2. Biochem
Biophys Res Commun. 445:314–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Clark SW, Fee BE and Cleveland JL:
Misexpression of the eyes absent family triggers the apoptotic
program. J Biol Chem. 277:3560–3567. 2002. View Article : Google Scholar
|
|
41
|
Matt N, Dupé V, Garnier JM, Dennefeld C,
Chambon P, Mark M and Ghyselinck NB: Retinoic acid-dependent eye
morphogenesis is orchestrated by neural crest cells. Development.
132:4789–4800. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li X, Oghi KA, Zhang J, Krones A, Bush KT,
Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al: Eya
protein phosphatase activity regulates Six1-Dach-Eya
transcriptional effects in mammalian organogenesis. Nature.
426:247–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kohrt D, Crary J, Zimmer M, Patrick AN,
Ford HL, Hinds PW and Grossel MJ: CDK6 binds and promotes the
degradation of the EYA2 protein. Cell Cycle. 13:62–71. 2014.
View Article : Google Scholar :
|
|
44
|
Bonini NM, Leiserson WM and Benzer S: The
eyes absent gene: Genetic control of cell survival and
differentiation in the developing Drosophila eye. Cell. 72:379–395.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pandey RN, Rani R, Yeo EJ, Spencer M, Hu
S, Lang RA and Hegde RS: The Eyes Absent phosphatase-transactivator
proteins promote proliferation, transformation, migration, and
invasion of tumor cells. Oncogene. 29:3715–3722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Krueger AB, Drasin DJ, Lea WA, Patrick AN,
Patnaik S, Backos DS, Matheson CJ, Hu X, Barnaeva E, Holliday MJ,
et al: Allosteric inhibitors of the Eya2 phosphatase are selective
and inhibit Eya2-mediated cell migration. J Biol Chem.
289:16349–16361. 2014. View Article : Google Scholar : PubMed/NCBI
|