Introduction
Acute leukemia treatment mainly comprises full
myeloablation, including leukemic cells, via chemotherapy and/or
total body irradiation; subsequent hematopoietic stem cell
transplantation is performed to reconstruct the hematopoietic
system (1–3). However, although rare, repeated
exposure to ionizing radiation (IR) can produce other leukemic
cells and/or radio-resistant leukemic cells and thus presents an
obstacle to treatment against treatment (4,5). Acute
promyelocytic leukemia (APL) is a unique subtype of acute myeloid
leukemia (AML), characterized by a block at the promyelocytic stage
of hematopoiesis (6,7).
Our previous study demonstrated that a model of
radiation-resistant APL (Res-HL60 cells) exhibited a high repair
capacity with normally functioning ATM/ATR and DNA-dependent
protein kinase (5); furthermore,
these cells exhibit resistance to phorbol 12-myristate
13-acetate-induced monocyte differentiation (8). However, little information is
available regarding the behavior of radio-resistant APL in terms of
gene expression profiles and intercellular communication. Our
present study investigated the characteristic mRNA patterns in
Res-HL60 and the transfer of related molecules between cells.
Recently, attention has focused on extracellular vesicles (EVs;
<200 nmφ), which transfer intracellular components and maintain
intercellular communication (9). It
is important to demonstrate clearly whether radio-resistant
behavior is maintained independently or via intercellular
communication when considering leukemic treatment strategies.
In this study, an mRNA expression analysis of both
intracellular and EV material was performed to clarify the genetic
network and target gene(s) in Res-HL60.
Materials and methods
Cell preparation and culture
The human APL cell line HL60 (Wt-HL60) was purchased
from RIKEN BioResource Center (Tsukuba, Japan). The Res-HL60 cell
line was established by subjecting Wt-HL60 to 4 Gy of
X-irradiation/week for 4 weeks. Approximately 2% surviving fraction
of wt-HL60 cells was shown following initial exposure of 4 Gy.
Wt-HL60 and Res-HL60 were maintained in RPMI-1640 medium (Life
Technologies, Carlsbad, CA, USA) supplemented with 10%
heat-inactivated fetal bovine serum (FBS; Japan Bioserum,
Hiroshima, Japan) and 1% penicillin/streptomycin (Life
Technologies) in a humidified atmosphere at 37°C and 5%
CO2.
Irradiation
X-ray irradiation (150 kVp, 20 mA with 0.5-mm
aluminum and 0.3-mm copper filters) was performed using an X-ray
generator (MBR-1520R-3; Hitachi Medical Co., Ltd., Tokyo, Japan),
with a distance of 45 cm between the focus and target. The dose was
monitored with a thimble ionization chamber placed next to the
sample during irradiation. The dose rate was 1 Gy/min.
cDNA microarray analysis
To compare the mRNA expression profiles of Res-HL60
and Wt-HL60 cells, a two-color mRNA microarray method was
performed. Total RNAs were extracted using an RNeasy isolation kit
(Qiagen, Hilden, Germany). Total RNA quality was confirmed using a
2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
mRNAs were labeled using Cy3 and Cy5 mono-reactive dyes (GE
Healthcare, Buckinghamshire, UK). Labeling reactions were performed
using 1 µg of total RNA and an Amino Allyl aRNA kit (Life
Technologies). Microarray analyses were performed using a Toray
mRNA microarray system (3D-Gene Scanner 3000 system; Toray, Tokyo,
Japan).
EV isolation and RNA extraction
Cell culture media were centrifuged at 2,000 × g for
15 min and 4°C to remove cell debris. The supernatants were then
passed through a 0.22-µm filter. The filtrates were
ultracentrifuged at 120,000 × g for 70 min and 4°C on an Optima TLX
Ultracentrifuge (Beckman Coulter, Brea, CA, USA) to collect EVs.
Total RNA was extracted from EVs or cells using an ISOGEN II
(Nippon Gene, Tokyo, Japan) according to the manufacturer's
instructions.
Reverse transcription-polymerase chain
reaction (RT-PCR)
To synthesize cDNAs from cells or EVs, high-capacity
cDNA reverse transcriptase kits (Life Technologies) were used. The
synthesized cDNAs were then subjected to PCR in a 15 µl
reaction mixture containing 1X Power SYBR Green Master Mix (Life
Technologies), 0.5 µM concentrations of the primer pairs
described in Table I, and cDNA
template. Primer pairs were designed from human-specific sequence
regions. Therefore, potential bovine mRNA contamination in FBS was
not detected. Quantitative PCR was performed using real-time PCR
system (StepOne Plus; Life Technologies) under the following
conditions: 10 min at 95°C, followed by 40 cycles each of 95°C for
15 sec, and 60°C for 60 sec. GAPDH mRNA was used as an internal
control. Cellular expression values and standard deviations were
calculated by the comparative Ct method, and values were
normalized according to the values from Wt-HL60 cells, which were
set at 1.0. PCR products derived from the mRNA of EVs were
electrophoresed on 4% agarose gels. Detection of amplified
fragments was achieved via ethidium bromide staining using a
ChemiDoc xRS and Quantity One software (both from Bio-Rad).
 | Table IPrimers and accession numbers in the
focused genes. |
Table I
Primers and accession numbers in the
focused genes.
| Primer name | Accession no. | Sequence (5′-3′) | Size (nt) | Amplication size
(bp) |
|---|
|
SEPT11-forward | NM_018243 |
GAAAGCAGCGGCTCAGTTA | 19 | 110 |
|
SEPT11-reverse | |
GGCTTGCCAGGCTTTATGT | 19 | |
|
MAD2L1-forward | NM_002358 |
GCGTGCTTTTGTTTGTGTC | 19 | 122 |
|
MAD2L1-reverse | |
TAAAATGCTGTTGATGCCG | 19 | |
|
VASP-forward | NM_003370 |
ACCTGGTCGGTCCCGAAC | 18 | 96 |
|
VASP-reverse | |
GGAGACCCGGCGCTCTATG | 19 | |
|
MXD1-forward | NM_002357.2 |
AGCTGGGCATTGAGAGGAT | 19 | 96 |
|
MXD1-reverse | |
CCACGTCAACGTCGATTT | 18 | |
|
RNF2-forward | NM_001846.2 |
GCGTCCGCGGCAGCTGATA | 19 | 77 |
|
RNF2-reverse | |
ATTGCGGCTCCTGCCCCAG | 19 | |
|
CCND1-forward | NM_001725.2 |
CGAGAAGCTGTGCATCTACACC | 22 | 86 |
|
CCND1-reverse | |
ACTTGAGCTTGTTCACCAGGAG | 22 | |
|
CSE1L-forward | NM_001846.2 |
TTCAGAAGCAGTTAAGTGATGCA | 23 | 72 |
|
CSE1L-reverse | |
GCAAGTCAGGCCATTTCTGT | 20 | |
|
ITPKA-forward | NM_001093772.1 |
CGACCTGCTGAGCGACAGT | 19 | 96 |
|
ITPKA-reverse | |
CGGATCTTCTGCCAGTGGT | 19 | |
|
TNF-forward | NM_000860 |
CAGCCTCTTCTCCTTCCTGA | 20 | 124 |
|
TNF-reverse | |
GGCCAGAGGGCTGATTAGA | 19 | |
|
HPGD-forward | NM_001725.2 |
GAACCTCAGAAGACTCTGTTCATC | 24 | 115 |
|
HPGD-reverse | |
CATTATTGACCAAAATGTCCAGTC | 24 | |
|
GAPDH-forward | NM_002046 |
GCCACATCGCTCAGACACC | 19 | 69 |
|
GAPDH-reverse | |
AGGCGCCCAATACGACCA | 18 | |
Statistics
Statistical analysis was performed using the Origin
software package (OriginLab® Pro version 9.0; OriginLab
Co., Northampton, MA, USA) and SPSS version 17.0 for Windows (IBM,
Chicago, IL, USA). Statistical analysis of the cDNA microarray was
performed using Ingenuity® Pathway Analysis tools
(Qiagen Silicon Valley, Redwood City, CA, USA); a P-value <0.05
was considered statistically significant.
Results
Expression network of mRNA in human
Res-HL60
In order to clarify the mRNA expression profile in
Res-HL60 cells, a cDNA microarray analysis and
Ingenuity® statistical analysis were performed. Res-HL60
cells, which show resistance to radiation, were extracted from
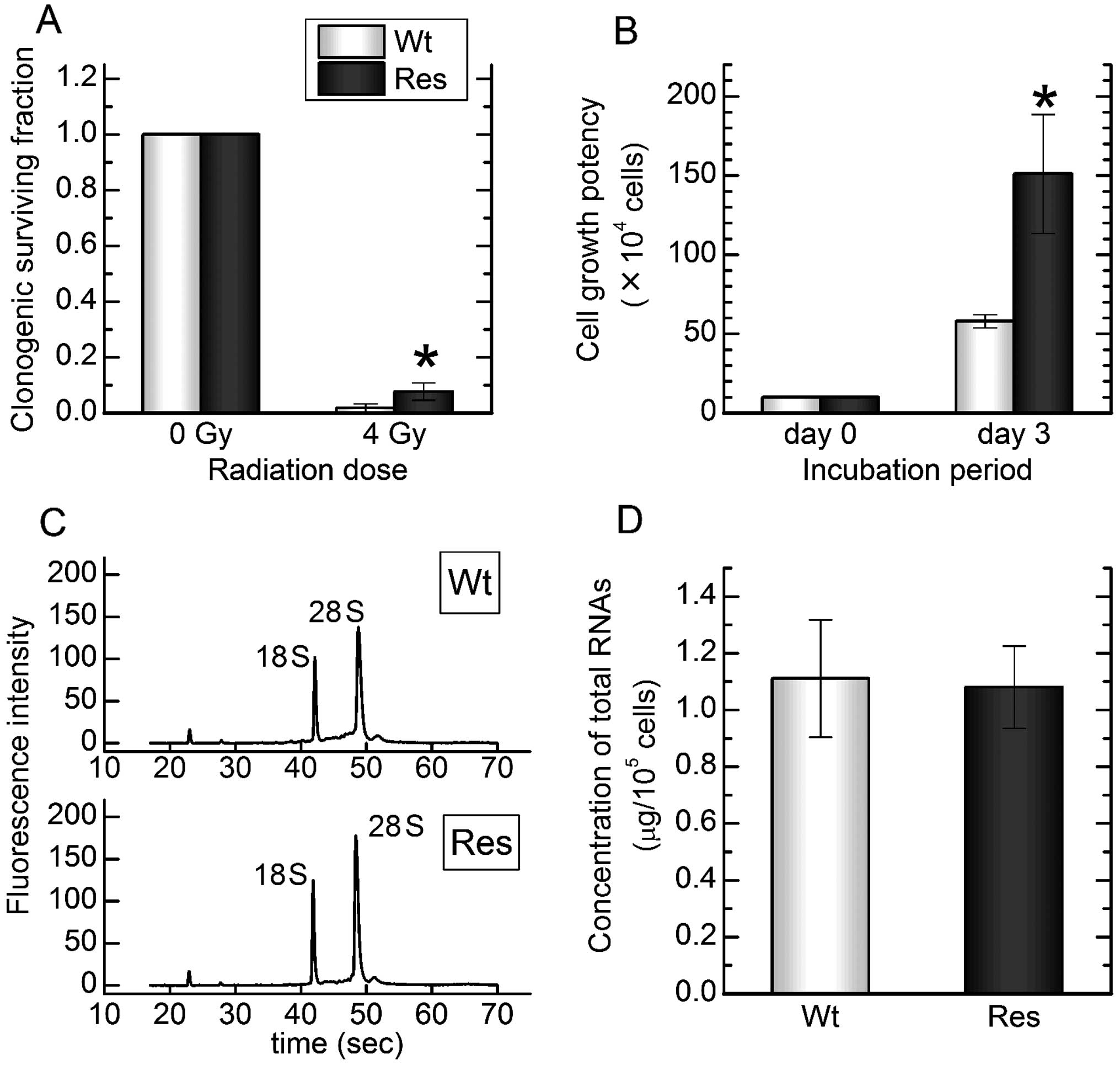
total RNAs at high quality and sufficient concentration (Fig. 1). A total of 25 k target molecules
were exported, and 7,309 known molecules were identified as
significantly upregulated or downregulated in comparison to
wild-type control cells (Wt-HL60) (Table II). Of these molecules, 4,268 were
uploaded to Ingenuity® for functional analysis.
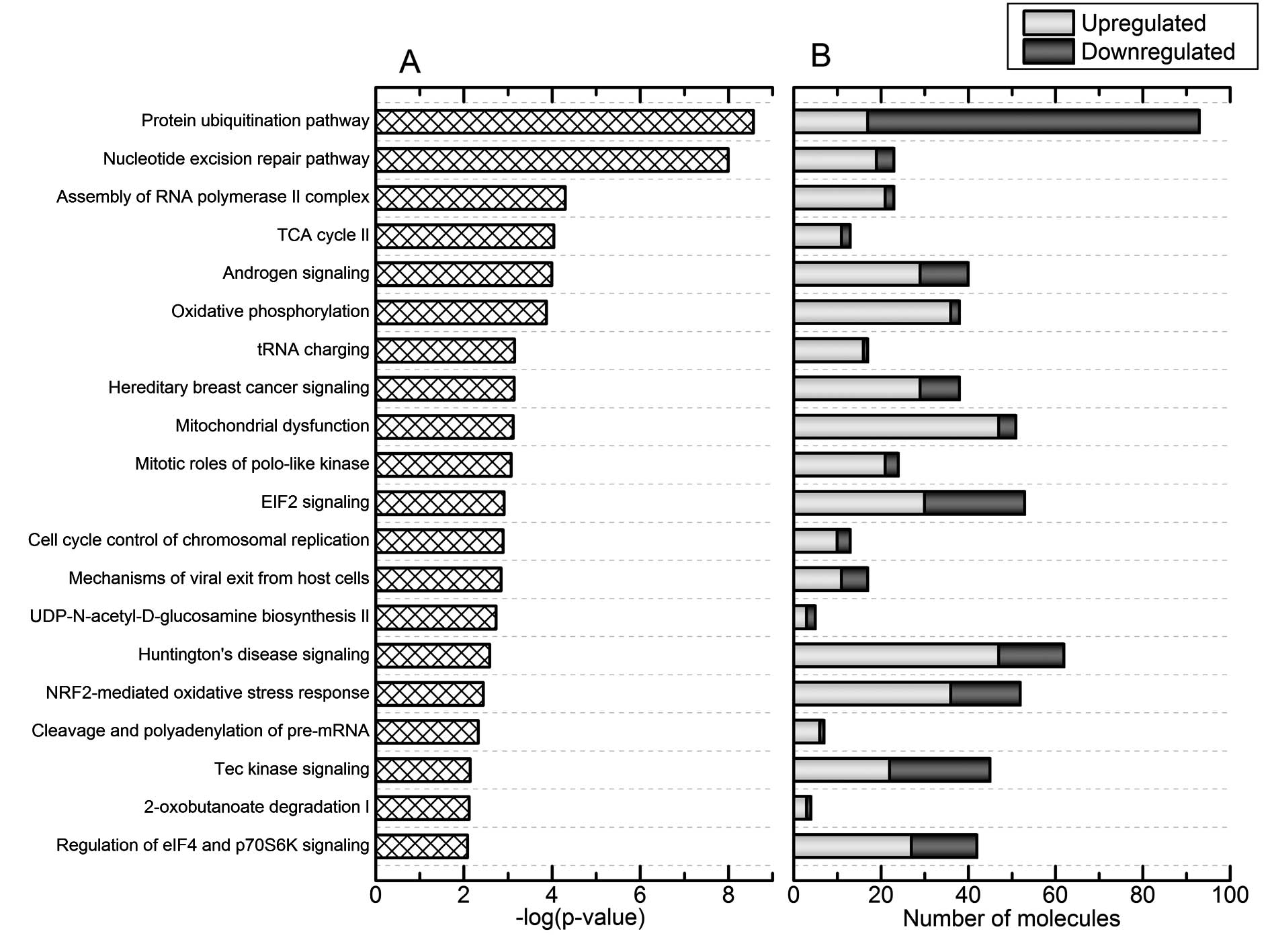
According to the canonical pathway analysis of Res-HL60 by
Ingenuity®, significant changes were observed in the top
4 categories of 'protein ubiquitination pathway', 'nucleotide
excision repair pathway', 'assembly of RNA polymerase II complex',
and 'TCA cycle II' relative to Wt-HL60 [the-log(p-value) for each
pathway was 8.6, 8.0, 4.3, and 4.0, respectively] (Fig. 2A). In addition, the 'protein
ubiquitination pathway' category featured the greatest number of
affected molecules (Fig. 2B).
 | Table IISignificant differences in all known
mRNAs in the cDNA microarray. |
Table II
Significant differences in all known
mRNAs in the cDNA microarray.
| Contents | No. of genes |
|---|
| Total number of
target genes in collected data | 25,000 |
| Number of known
genesa in target genes | 12,245 |
| Significantly
changing genes in known genes (Res vs. Wt)b | 7,309 |
| Mapped by
Ingenuity® in significant genes | 4,268 |
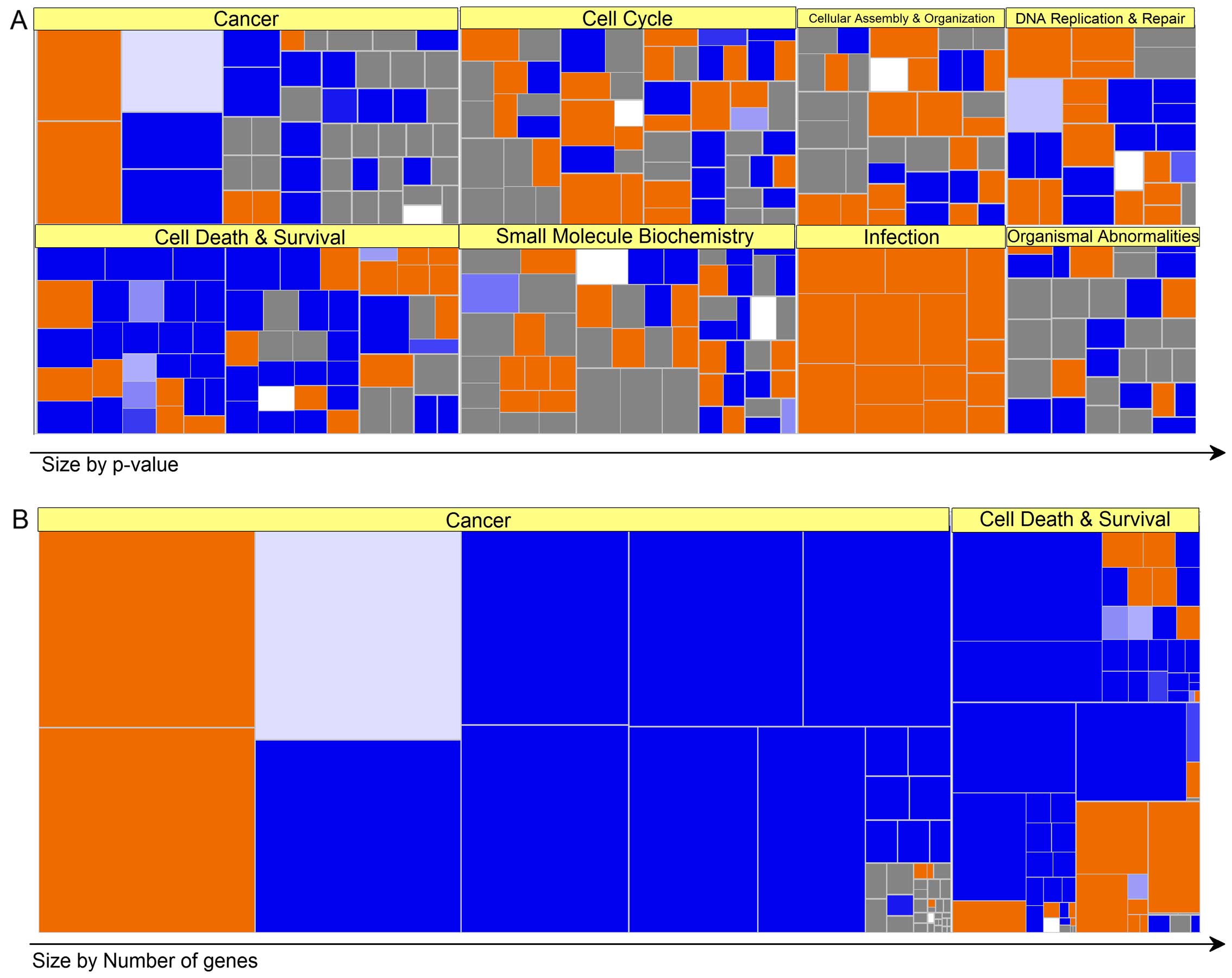
Based on the above information, a biofunctional
analysis of the Ingenuity® heat map revealed that the
genetic categories of 'cancer', 'cell death and survival', 'cell
cycle', 'small molecule biochemistry', 'cellular assembly', and
'organization and infection' correlated strongly in this order
(Fig. 3A). In addition, various
molecules in the categories of 'cancer,' 'cell death and survival',
were particularly observed in Res-HL60 relative to Wt-HL60
(Fig. 3B).
Quantitative mRNA expression
analysis
To identify the target mRNA(s) related to
radio-resistance, a quantitative expression analysis of mRNAs in
the top 4 categories (e.g., 'cell cycle', 'DNA replication', 'cell
death and survival', and 'infection') was performed. As shown in
Table III, Res-HL60 exhibited
significantly increased expression of the cell cycle-related mRNAs
SEPT11, MAD2L1, and CHFR (6.97-, 5.05- and
3.54-fold, respectively) and significantly decreased expression of
MCPH1, VASP, and MXD1 (0.59-, 0.54- and
0.21-fold, respectively) relative to Wt-HL60. expression of the DNA
replication-related mRNAs RNF2, SIRT1, and
RNF4 was significantly higher (5.41-, 4.24- and 3.86-fold,
respectively) and that of LIG1, TDRD7, and
CCND1 significantly lower (0.55-, 0.53- and 0.15-fold,
respectively) in Res-HL60 relative to Wt-HL60. Expression of the
cell death and survival-related mRNAs COL4A2, KIT,
and IGFBP7 was higher (35.2-, 24.7- and 23.2-fold,
respectively) and that of PRG2, HPGD, and BPI
significantly lower (0.06-, 0.05- and 0.04-fold, respectively) in
Res-HL60 than in Wt-HL60. Expression of the infection-related mRNAs
CSE1L, ITPKA, and GBAS was significantly
higher (10.9-, 7.30- and 6.43-fold, respectively) and that of
CRIPAK, EGR1, and TNF significantly lower
(0.27-, 0.19- and 0.19-fold, respectively) in Res-HL60 than in
Wt-HL60.
 | Table IIIAnalysis of significant expression of
mRNA in cDNA microarray. |
Table III
Analysis of significant expression of
mRNA in cDNA microarray.
| Gene name | Accession no. | Predictiona | Ratio (Res/Wt) |
|---|
| Cell cycle-related
mRNAs | | | |
| SEPT11 | NM_018243 | Activated | 6.97±2.14 |
| MAD2L1 | NM_002358 | Activated | 5.05±0.81 |
| CHFR | NM_018223.1 | Activated | 3.54±1.61 |
| MCPH1 | NM_024596.2 | Inhibited | 0.59±0.02 |
| VASP | NM_003370 | Inhibited | 0.54±0.02 |
| MXD1 | NM_002357.2 | Inhibited | 0.21±0.04 |
| DNA
replication-related mRNAs | | | |
| RNF2 | NM_007212.3 | Activated | 5.41±2.44 |
| SIRT1 | NM_012238.4 | Activated | 4.24±2.26 |
| RNF4 | NM_002938.3 | Activated | 3.86±1.63 |
| LIG1 | NM_000234 | Inhibited | 0.55±0.05 |
| TDRD7 | NM_014290 | Inhibited | 0.53±0.04 |
| CCND1 | NM_053056 | Inhibited | 0.15±0.02 |
| Cell death and
survival-related mRNAs | | | |
| COL4A2 | NM_001846.2 | Activated | 35.2±8.43 |
| KIT | NM_001093772.1 | Activated | 24.7±11.8 |
| IGFBP7 | NM_001553 | Activated | 23.2±9.46 |
| PRG2 | NM_002728 | Inhibited | 0.06±0.03 |
| HPGD | NM_000860 | Inhibited | 0.05±0.02 |
| BPI | NM_001725.2 | Inhibited | 0.04±0.01 |
| Infection-related
mRNAs | | | |
| CSE1L | NM_001316.2 | Activated | 10.9±5.37 |
| ITPKA | NM_002220 | Activated | 7.30±1.72 |
| GBAS | NM_001483 | Activated | 6.43±1.61 |
| CRIPAK | NM_175918 | Inhibited | 0.27±0.02 |
| EGR1 | NM_001964 | Inhibited | 0.19±0.04 |
| TNF | NM_000594 | Inhibited | 0.19±0.07 |
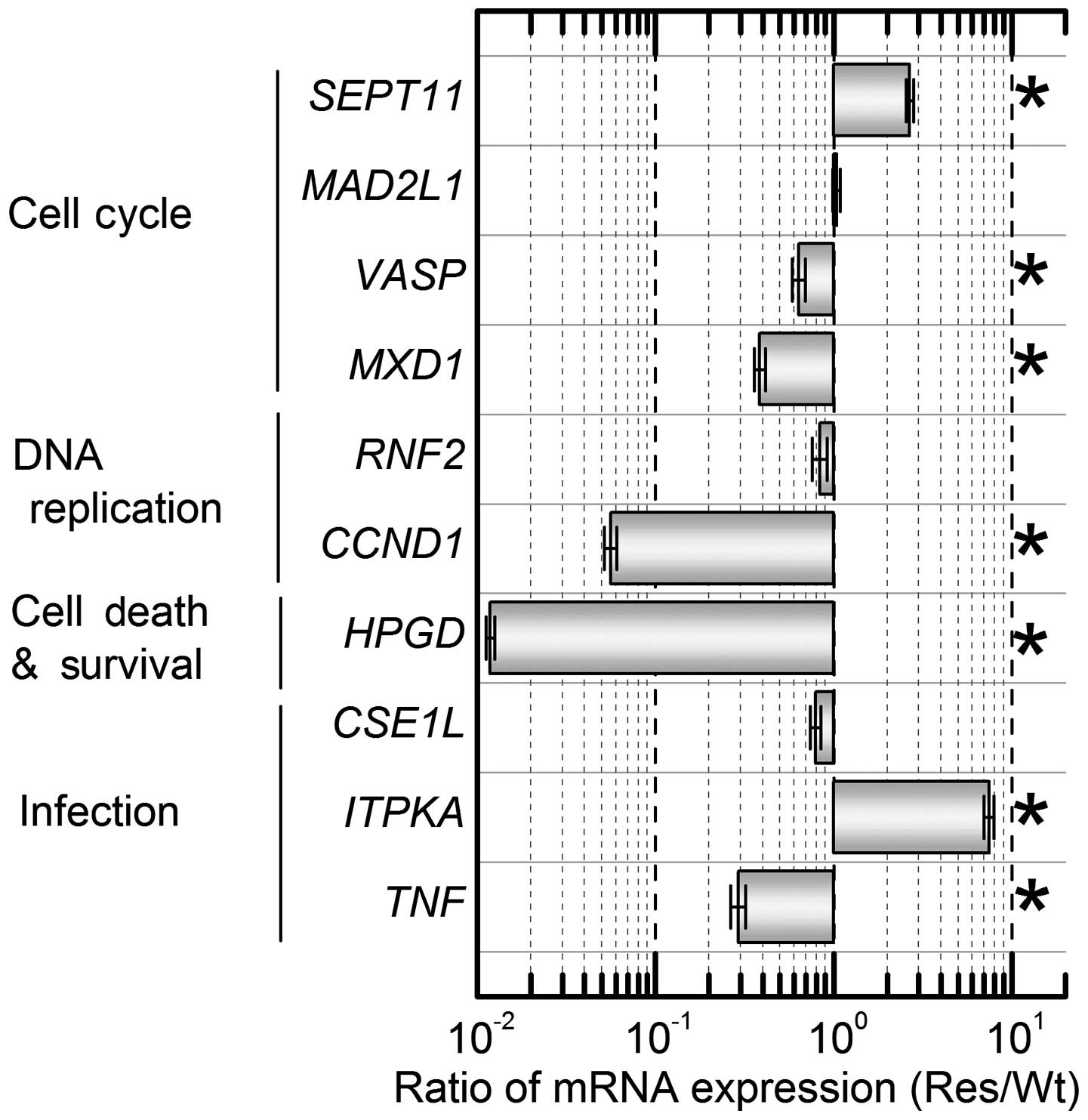
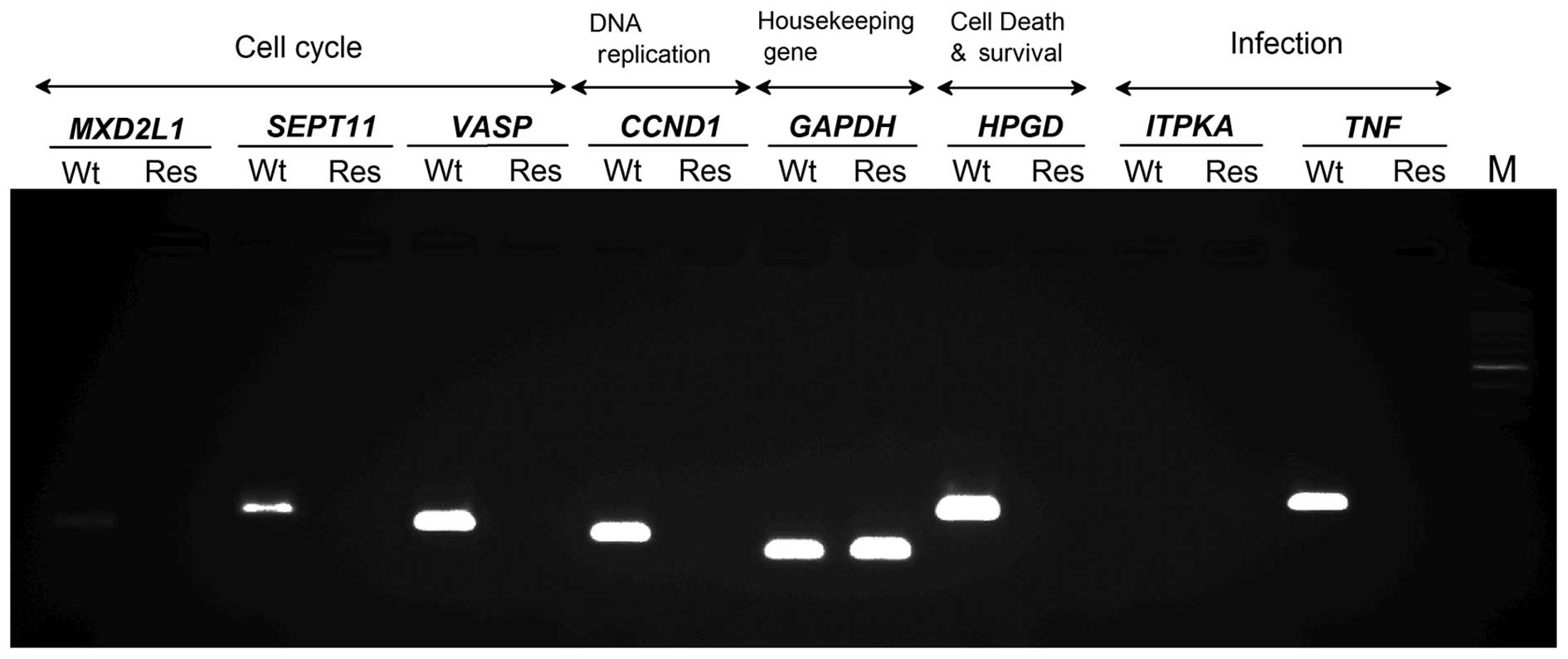
The reproducibility of these mRNA expression results
was confirmed using real-time RT-PCR. Ten primers for verification
of sufficient accuracy were prepared (Table I). RT-PCR detected upregulation of
SEPT11 and ITPKA and down-regulation of VASP,
MXD1, CCND1, HPGD, and TNF (Fig. 4). Therefore, similar expression
patterns of these 7 mRNAs in Res-HL-60 were detected using both
cDNA microarray and real-time RT-PCR.
Analysis of EVs in Res-HL60
To clarify whether these mRNAs were expressed in
EVs, a mode of intercellular communication, EVs from Res-HL60 were
analyzed. As EVs released from cells can be detected and harvested
from cell culture supernatants, fetal EVs (i.e., from FBS) in cell
culture must be eliminated before collecting EVs derived from
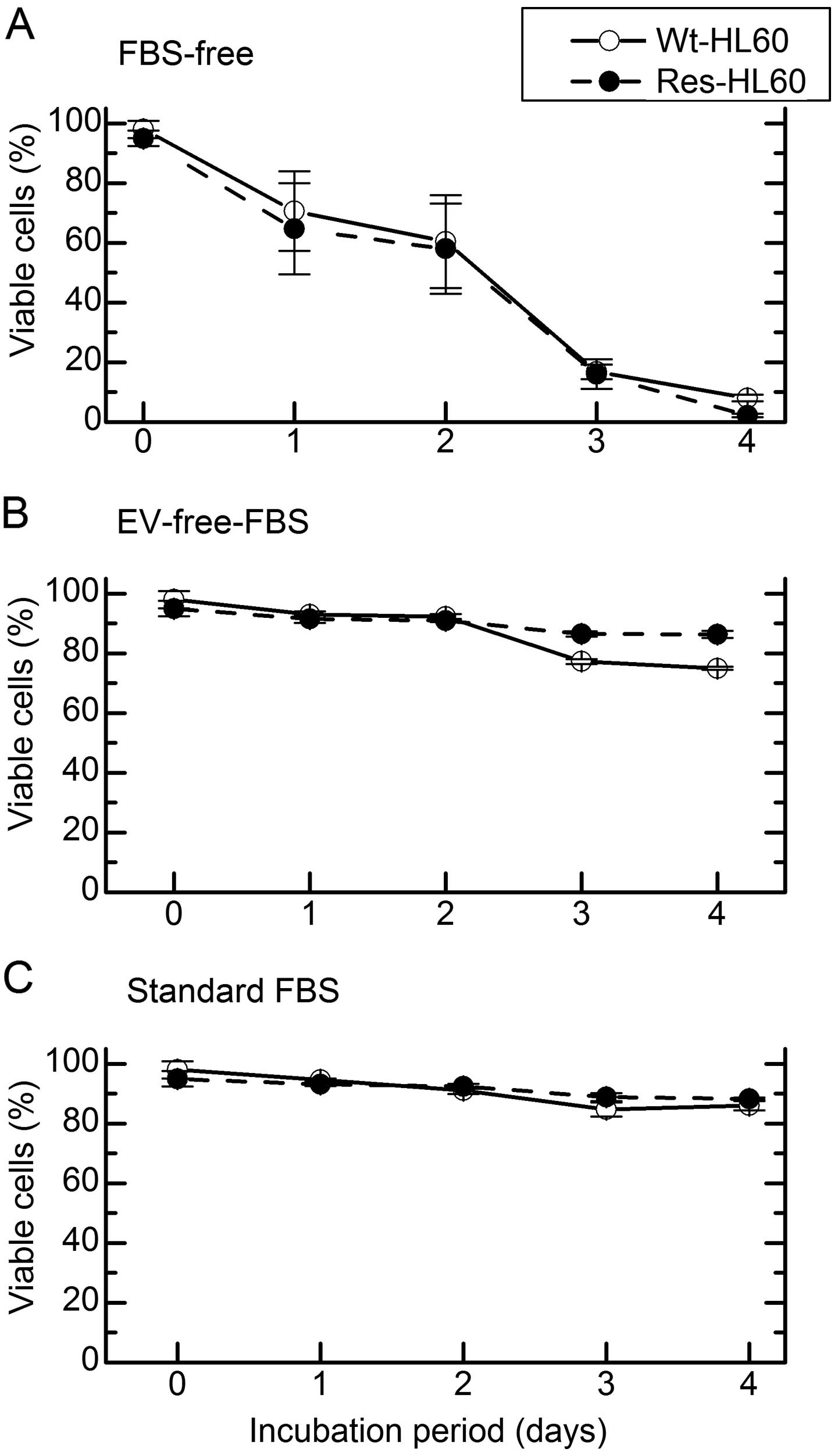
Res-HL60 cells in vitro. The cell viability in FBS-free
media and media with EV-free-FBS was analyzed to determine the
optimal condition of cell culture which performs normal cellular
metabolism without fetal EVs. Compared to standard FBS media,
Res-HL60 and Wt-HL60 fared similarly with EV-free FBS media;
however, the viability of HL60 decreased to <20% by day 3 in
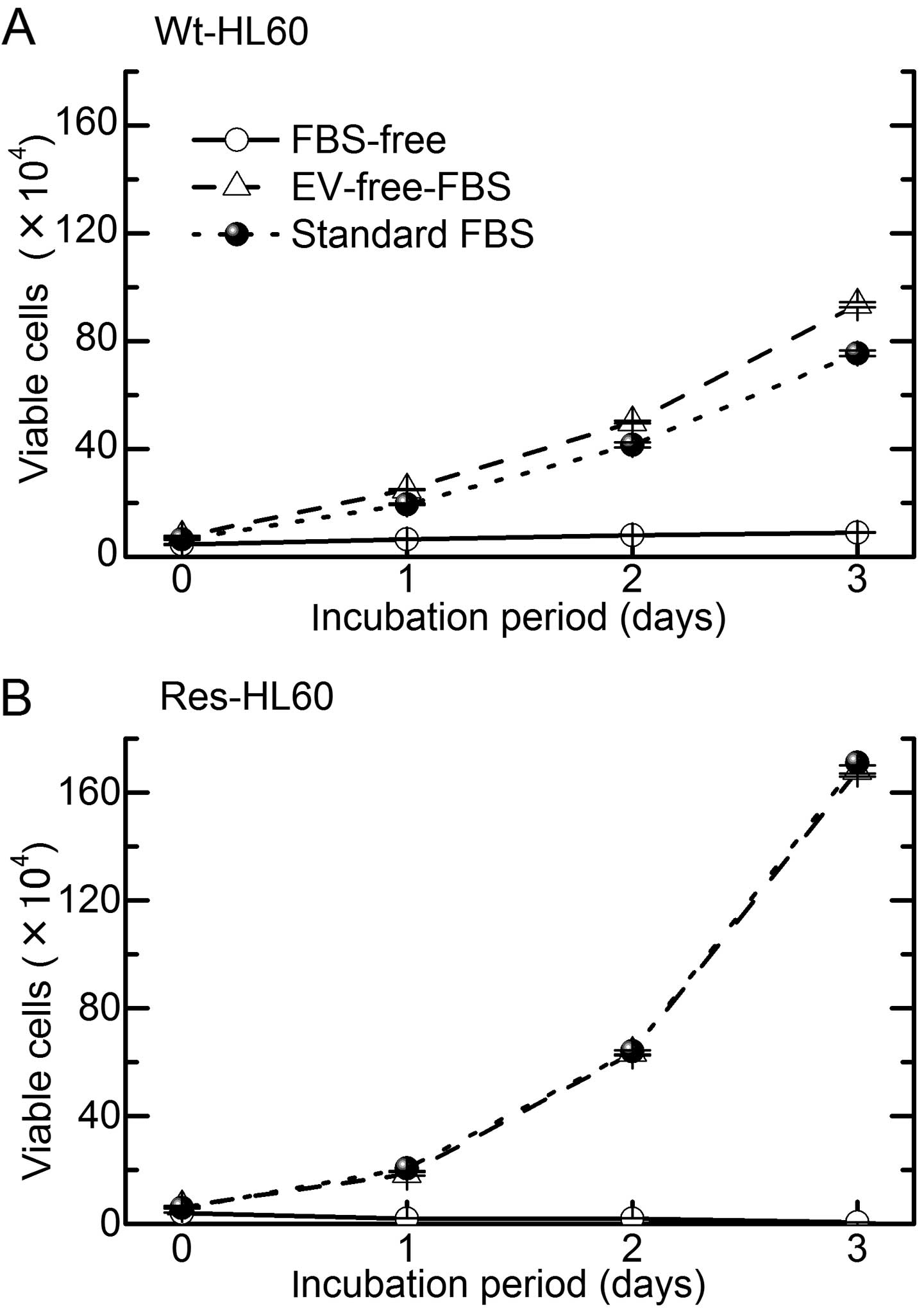
FBS-free medium (Fig. 5). In
addition, the cell growth abilities of Res-HL-60 and Wt-HL60 cells
in EV-free FBS media were similar to that in standard FBS media
(Fig. 6). Therefore, an analysis of
mRNA expression in EVs from Res-HL60 was performed using cellular
debris collected from cell culture supernatants on day 2. Five
mRNAs, SEPT11, VASP, CCND1, HPGD and
TNF, were detected in EVs from Wt-HL60; however, these
molecules were not detected in EVs from Res-HL60 by either
real-time RT-PCR or electropherogram (Fig. 7).
Discussion
In the present study, an analysis of the
intracellular genetic network and transference potency of
radio-resistant specific mRNAs between intercellular communicating
EVs was performed via quantitative RNA analysis. Significantly
changes in the expression of 7,309 known mRNAs were observed in
Res-HL60 cells relative to Wt-HL60 cells; in particular, changes in
the expression of protein ubiquitination pathway-related molecules
was observed in Res-HL60 (Fig. 2).
Protein ubiquitination, which serves as a cell signaling activator
or suppressor in acute leukemia cells, maintaining DNA damage
responses and tumorigenesis (10,11).
Res-HL60 may regulate tumorigenesis to a greater extent than
Wt-HL60. Generally, reactive oxygen species or free radicals
produced by IR, such as X-rays and gamma-rays, are known to
indirectly and/or directly induce DNA strand breaks and to exert
various cytotoxic effects (12,13).
Ai et al reported that ubiquitination of the cytokine
receptor G-CSFR regulates myeloid cell survival and proliferation
(14). Therefore, activation of the
protein ubiquitination system via radiation reiteration exposure
may affect the potency of radiation protection. In the category of
ubiquitination-related biofunction, gene expression in the
categories of 'cell cycle', 'DNA replication/repair', and
'infection' were found to correlate closely with Res-HL60. Among
the 7 reproducibly identified mRNAs, SEPT11 and VASP
are necessary for developing microtubules and cytoskeleton
structures and are related to the G2/M transition (15-17).
The Max protein, encoded by MXD1, activates the
transcription factor myc to form a Myc-Max heterodimer and thus
promotes cell proliferation and/or transformation (18,19).
Therefore, our present data suggest that the behavior of cell
cycle-related genes (up of SEPT11, down of VASP/MXD1)
in Res-HL60 modify the intracellular environment, including
cytoskeletal formation, whereas repeated exposure to IR suppresses
Myc signaling.
On the contrary, the downregulation of CCND1,
which encodes cyclin D1 and affects DNA replication and cell
proliferation, was an unexpected phenomenon (5,8,20).
Shimura et al recently reported that repeated exposure to
low-dose fractionated radiation abrogates cell cycle-dependent
cyclin D1 degradation via the constitutive activation of AKT
survival signaling in normal human fibroblasts (21). High- and low-dose radiation rates
may induce different behaviors of some CCND1 gene
regulators.
Xun et al reported that the rapid turnover of
15-PGDH, which is encoded by HPGD, in HL60 indicates that
enzymatic activity depends on continued enzyme synthesis, which
could be susceptible to hormone- and drug-controlled mechanisms.
Upregulation of ITPKA, which promotes stem cell
differentiation, and downregulation of TNF, which encodes a
pro-inflammatory cytokine, were also observed (22-24).
Therefore, these regulatory mechanisms may indicate the mechanism
underlying radio-resistant APL. Interestingly, none of our target
molecules were transferred among Res-HL60 cells via EVs.
Accordingly, radio-resistant regulation in APL may be restricted to
an intracellular phenomenon and may affect other cells. Szabó et
al reported that in leukemic cells, the transfer of EVs and
stimulating cytokines through intercellular interactions differs
from inflammatory processes (25).
It is necessary to confirm whether radio-resistant behavior can be
countered by targeting the molecules identified in the present
study. More precise approaches are required to elucidate the role
of a genetic network in radio-resistant APL induced by exposure to
repeated IR.
In conclusion, the specific phenomenon of
radio-resistance acquisition is induced through changes of
intracellular gene expression networks, but is not affected by the
intercellular transfer of molecules.
Acknowledgments
This study was supported by a grant for Hirosaki
University Young Institutional Research (2013–2015), the Takeda
Science Foundation (2013 S.M.), and a KAKENHI Grant-in-Aid for
Young Scientists (B) (no. 25861054 S.M.).
References
|
1
|
Russell JA, Irish W, Balogh A, Chaudhry
MA, Savoie ML, Turner AR, Larratt L, Storek J, Bahlis NJ, Brown CB,
et al: The addition of 400 cGY total body irradiation to a regimen
incorporating once-daily intravenous busulfan, fludarabine, and
antithymocyte globulin reduces relapse without affecting nonrelapse
mortality in acute myelogenous leukemia. Biol Blood Marrow
Transplant. 16:509–514. 2010. View Article : Google Scholar
|
|
2
|
Termuhlen AM, Klopfenstein K, Olshefski R,
Rosselet R, Yeager ND, Soni S and Gross TG: Mobilization of
PML-RARA negative blood stem cells and salvage with autologous
peripheral blood stem cell transplantation in children with
relapsed acute promyelocytic leukemia. Pediatr Blood Cancer.
51:521–524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mikell JL, Waller EK, Switchenko JM,
Rangaraju S, Ali Z, Graiser M, Hall WA, Langston AA, Esiashvili N,
Khoury HJ, et al: Similar survival for patients undergoing
reduced-intensity total body irradiation (TBI) versus myeloablative
TBI as conditioning for allogeneic transplant in acute leukemia.
Int J Radiat Oncol Biol Phys. 89:360–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rashidi A and Fisher SI: Therapy-related
acute promyelocytic leukemia: A systematic review. Med Oncol.
30(625)2013. View Article : Google Scholar
|
|
5
|
Hazawa M, Hosokawa Y, Monzen S, Yoshino H
and Kashiwakura I: Regulation of DNA damage response and cell cycle
in radiation-resistant HL60 myeloid leukemia cells. Oncol Rep.
28:55–61. 2012.PubMed/NCBI
|
|
6
|
Stein EM and Tallman MS: Acute
promyelocytic leukemia in children and adolescents. Acta Haematol.
132:307–312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dalia SM, Horna P and Zhang L: Tetraploidy
acute promyelocytic leuemia with double t(15;17)/PML-RARA, a case
report with review of literature. Int J Clin Exp Pathol.
7:5363–5368. 2014.PubMed/NCBI
|
|
8
|
Monzen S, Takimura K, Kashiwakura I and
Hosokawa Y: Acute promyelocytic leukemia mutated to radioresistance
suppressed monocyte lineage differentiation by phorbol 12-myristate
13-acetate. Leuk Res. 37:1162–1169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Turturici G, Tinnirello R, Sconzo G and
Geraci F: Extracellular membrane vesicles as a mechanism of
cell-to-cell communication: Advantages and disadvantages. Am J
Physiol Cell Physiol. 306:C621–C633. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao H, Zhu M, Dou G, Zhao H, Zhu B, Li J,
Liao J and Xu X: BCL10 regulates RNF8/RNF168-mediated
ubiquitination in the DNA damage response. Cell Cycle.
13:1777–1787. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guan D, Factor D, Liu Y, Wang Z and Kao
HY: The epigenetic regulator UHRF1 promotes ubiquitination-mediated
degradation of the tumor-suppressor protein promyelocytic leukemia
protein. Oncogene. 32:3819–3828. 2013. View Article : Google Scholar :
|
|
12
|
Bajinskis A, Natarajan AT, Erixon K and
Harms-Ringdahl M: DNA double strand breaks induced by the indirect
effect of radiation are more efficiently repaired by non-homologous
end joining compared to homologous recombination repair. Mutat Res.
756:21–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vignard J, Mirey G and Salles B:
Ionizing-radiation induced DNA double-strand breaks: A direct and
indirect lighting up. Radiother Oncol. 108:362–369. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ai J, Druhan LJ, Loveland MJ and Avalos
BR: G-CSFR ubiquiti-nation critically regulates myeloid cell
survival and proliferation. PLoS One. 3:e34222008. View Article : Google Scholar
|
|
15
|
Hanai N, Nagata K, Kawajiri A, Shiromizu
T, Saitoh N, Hasegawa Y, Murakami S and Inagaki M: Biochemical and
cell biological characterization of a mammalian septin, Sept11.
FEBS Lett. 568:83–88. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tao Y, Chen YC, Sang JR and Xu WR:
Phosphorylation of vasodilator stimulated phosphoprotein is
correlated with cell cycle progression in HeLa cells. Mol Med Rep.
3:657–662. 2010.
|
|
17
|
Zhang YT, xu LH, Lu Q, Liu KP, Liu PY, Ji
F, Liu XM, Ouyang DY and He XH: VASP activation via the
Gα13/RhoA/PKA pathway mediates cucurbitacin-B-induced actin
aggregation and cofilinactin rod formation. PLoS One. 9:e935472014.
View Article : Google Scholar
|
|
18
|
Nair SK and Burley SK: X-ray structures of
Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation
by proto-oncogenic transcription factors. Cell. 112:193–205. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garcia-Sanz P, Quintanilla A, Lafita MC,
Moreno-Bueno G, García-Gutierrez L, Tabor V, Varela I, Shiio Y,
Larsson LG, Portillo F, et al: Sin3b interacts with Myc and
decreases Myc levels. J Biol Chem. 289:22221–22236. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pagano M, Theodoras AM, Tam SW and Draetta
GF: Cyclin D1-mediated inhibition of repair and replicative DNA
synthesis in human fibroblasts. Genes Dev. 8:1627–1639. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shimura T, Kobayashi J, Komatsu K and
Kunugita N: DNA damage signaling guards against perturbation of
cyclin D1 expression triggered by low-dose long-term fractionated
radiation. Oncogenesis. 3:e1322014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sonnet M, Claus R, Becker N, Zucknick M,
Petersen J, Lipka DB, Oakes CC, Andrulis M, Lier A, Milsom MD, et
al: Early aberrant DNA methylation events in a mouse model of acute
myeloid leukemia. Genome Med. 6(34)2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Obeid LM, Linardic CM, Karolak LA and
Hannun YA: Programmed cell death induced by ceramide. Science.
259:1769–1771. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xun CQ, Tian ZG and Tai HH: Stimulation of
synthesis de novo of NAD(+)-dependent 15-hydroxyprostaglandin
dehydrogenase in human promyelocytic leukaemia (HL-60) cells by
phorbol ester. Biochem J. 279:553–558. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szabó GT, Tarr B, Pálóczi K, Éder K, Lajkó
E, Kittel Á, Tóth S, György B, Pásztói M, Németh A, et al: Critical
role of extracellular vesicles in modulating the cellular effects
of cytokines. Cell Mol Life Sci. 71:4055–4067. 2014. View Article : Google Scholar : PubMed/NCBI
|