Introduction
Esophageal carcinoma is the sixth most common cause
of cancer-related death worldwide. China is one of the
high-incidence countries and esophageal cancer accounts for
approximately 150,000 death each year, nearly a quarter of all
cancer deaths (1) in the country.
Esophageal cancer is usually diagnosed at an advanced stage, making
curative surgical resection, which is initially recommend for early
stage cases, feasible for only 30–40% of patients (2). The outcome of surgery for patients
with such an aggressive tumor is still unsatisfactory, with a
5-year survival rate less than 20% (3). Accumulating evidence from randomized
clinical trials supports the use of neoadjuvant chemoradiotherapy
(CRT), which is shown to improve resectability and survival in
patients with locally advanced esophageal cancer, although mixed
results have been reported (4). The
variations in clinical responses to CRT are most evident for
esophageal squamous cell carcinoma (ESCC), and the survival rates
between responders and non-responders are quite different even with
the same clinical stage (5,6). Therefore, there is a compelling need
to identify novel biomarkers that hold promise of precisely
predicting tumor response to CRT to tailor treatments for different
ESCC patients and enhance survival (7).
MicroRNAs (miRNAs) are endogenous, short, non-coding
RNAs that can regulate the expression of target genes by binding to
RNA-binding proteins to control the occurrence and development of
the disease including cancer. It was first characterized in 1993,
since then, more and more studies have focused on the role of
miRNA-small molecules in the cellular processes and pathways
including the differentiation, progression, apoptosis and
proliferation of different disease. In 2002, Calin et al
(8) first linked miRNAs with cancer
progression. Since then, increased number of studies have reported
the role of miRNAs in carcinogenesis, indicating that miRNAs are
closely related to the process of epithelial-mesenchymal transition
(9,10), characteristic of cancer stem cells
(10,11), the initiation of tumor invasion and
metastasis (12,13), and the therapeutic response to
chemotherapy or radiotherapy (14).
There is growing evidence to indicate miRNAs as biomarkers,
expression profile of miRNAs are different in cancer and normal
tissues, even in different organs or tissues (15,16).
Recent studies have reported that tissue-specific miRNAs are
consistently detected in circulating samples, and cancer
tissue-specific miRNAs also have been found in the circulation at
different stages of the disease (17). In addition, more and more reports
have demonstrated that tumor cells release miRNAs into the
circulation and these circulating miRNAs are in a remarkably
stable, cell-independent form which is protected from endogenous
RNase activity in the bloodstream, suggesting potential
opportunities for using circulating miRNAs as blood-based,
non-invasive biomarkers for molecular diagnostics, physiological
and pathological status, including cancer (18–21).
MicroRNA-7 (miR-7) is an intronic miRNA that resides
in the first intron of the heterogeneous ribonuclear protein K gene
on chromosome 9, and is evolutionarily conserved across all
species. Previous findings suggested that miR-7 participates in
tumorigenesis and progression by several signaling pathways in
various types of tumors (22–27).
Li and Carthew (28) have certified
that 3′-untraslated regions of human EGFR contains miR-7
complementary sites, which enable it to act on EGFR expression.
Furthermore, resent studies have reported that miRNA-7 can affect
sensitivity to chemotherapy by MRP1 (29) and radiotherapy by EGFR (30), providing opportunities for the
development of miRNA-based therapies and/or biomarkers in CRT of
cancer patients.
To investigate the functional roles of miR-7 in
radioresponse of ESCC and its underlying mechanism, we detected the
serum miR-7 expression in ESCC and analyzed its association with
clinical response of ESCC to CRT and evaluated the possibility of
using serum miR-7 as a diagnostic and predicting factor for ESCC.
Furthermore, we investigated the potential miR-7 role in
transfected cellular model of ECA-109, identifying EGFR as a direct
downstream target of miR-7.
Materials and methods
Ethics statement
This research involved human participants, thus
serum-based specimen collection and studies were approved by the
Institutional Review Boards of Shandong Cancer Hospital, the
Shandong Academy of Medical Sciences. All participants provided
written consent and indicated willingness to donate their blood
samples for research.
Samples and treatment regimen
Serum samples were collected from the peripheral
venous blood of 105 patients and 30 healthy volunteers at the
Department of Radiation Oncology, Shandong Cancer Hospital.
Clinical data of enrolled patients including gender, age, tumor
locations, status of lymph node and distant metastases, the maximum
diameter of tumor and tumor differentiation were recorded. Entry
criteria and treatment regimen for the present study have been
described in our published study (31). Immediately after collection, the
serum samples were snap-frozen in liquid nitrogen and then stored
at −80°C for RNA extraction for quantitative RT-PCR (qRT-PCR).
Response Evaluation Criteria In Solid Tumors (RECIST), the
guidelines recommended by the World Health Organization (32) was applied in the present study to
evaluate the response of ESCC patients to CRT (concurrent
radio-chemotherapy): each patient's response was defined as
complete remission (CR), partial remission (PR), stable disease
(SD) or progressive disease (PD).
Cell line and cell culture
ECA-109 cells, a well differentiated human ESCC cell
line, were provided by Tianjin Cancer Hospital. Cells were
maintained in RPMI-1640 medium (HyClone, USA) containing 10%
heat-inactivated fetal bovine serum (FBS; Gibco), 100 U/ml
penicillin, 100 U/ml streptomycin at 37°C in a humidified
atmosphere of 5% CO2. Cells were passaged every 2–3 days
to maintain exponential growth.
miRNA transfection
The mimic of miR-7 was purchased from GenePharma
(Shanghai, China). The ECA-109 cells were transfected with 100 nM
of miR-7 mimic or their corresponding negative controls.
Lipofectamine 2000 (Invitrogen, San Diego, CA, USA) was used for
cell transfection. Transfection complexes were added into the
culture plates and incubated for 4 h, and then replaced by fresh
medium according to the manufacturer's instructions.
Screening and verification of circulating
miRNAs by RT-PCR
Total RNA from human samples, including miRNAs, was
extracted from 400 μl of serum using the mirVana PARIS RNA
isolation kit (Ambion, Austin, TX, USA) according to the
manufacturer's instructions with the cell-miR-39 spike-in (Sangon
Biotech, Shanghai, China). A mirVana miRNA column (Ambion) was used
to collect total RNA. The bound RNA was cleaned with the buffers
provided by the manufacturer to remove impurities and eluted in a
final volume of 50 μl. Total RNA from cells were isolated
using TRIzol reagent (Invitrogen) according to the manufacturer's
instructions.
To analyze the expression miR-7, quantitative
real-time PCR was conducted using TaqMan fluorogenic probes. miR-7
was detected by qRT-PCR using the mirVana™ qRT-PCR primer set and
the mirVana™ qRT-PCR miRNA detection kit (Applied Biosystems, San
Diego, CA, USA) according to the manufacturer's instructions, which
were previously described (31).
Relative levels of EGFR mRNA in cells were examined
by SYBR-Green real-time quantitative reverse transcription-PCR and
normalized to β-actin mRNA. Reverse transcriptions using the
PrimeScript® RT Master Mix (Perfect Real-Time), and
qRT-PCR was performed using SYBR® Premix Ex Taq™ II
(Perfect Real-Time) (both from Takara Bio, Inc., Japan) according
to the manufacturer's instructions.
All qRT-PCRs were performed in duplicate, and the
data are presented as mean ± standard error of the mean. Real-time
polymerase chain reaction (RT-PCR) was carried out on the ABI 7900
Real-Time PCR System (Applied Biosystems). Relative microRNA
expression was calculated with the 2−ΔΔCt method
(31,33), where ΔΔCt = (Ct gene of interest −
Ct normalized gene) of (CR+PR) − (Ct gene of interest − Ct
normalized gene) of (SD+PD).
Western blot analysis
Cell lysates were prepared in lysis buffer [0.15 M
NaCl, 50 mM Tris-Cl (pH 7.5), 2 mM EDTA, 0.5% Triton-100, 5 mM DTT,
0.2 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml
apoptinin] following 48 h transfection. Protein concentrations of
total cell lysates were measured by Bio-Rad protein assay, and 50
μg of total cell lysates/lane was separated by 10% SDS-PAGE.
After electrophoresis, the protein was transferred to
polyvinylidene defluoride (PVDF) membrane, which was blocked with
5% non-fat milk in Tris-buffered saline with Tween-20 (TBST) [50
mmol/l Tris-HCl (pH 7.6), 150 mmol/l NaCl, 0.1% Tween-20] for 1.5 h
at room temperature. Immunoblotting was performed with rabbit
anti-EGFR (1:500; CST), and mouse anti-β-actin (1:500; Santa Cruz)
primary antibodies after washing the membrane 3 times in TBST
buffer. Membranes were subsequently probed with horseradish
peroxidase-conjugated secondary antibody (1:5,000; Zhongshan
Biotechnology, China), developed by chemiluminescence and exposed
to X-ray film. Densitometry was performed with gel imaging system
(AlphaImager 2200; Pharmacia Biotech Co., USA). All experiments
were performed in triplicate.
Statistical analysis
All clinicopathological variables and circulating
miRNA levels were analyzed using PASW Statistics, Windows software
version 17.0 (SPSS, Inc., Chicago, IL, USA). An unpaired t-test was
performed to compare the differences in serum miRNA levels between
groups. Chi-square test and logistic regression analysis were used
to evaluate the association between serum miR-7 relative expression
and clinical pathological variables. All tests were two-sided and
P<0.05 was considered to indicate a statistically significant
result.
Results
Relative levels of serum miR-7
expression
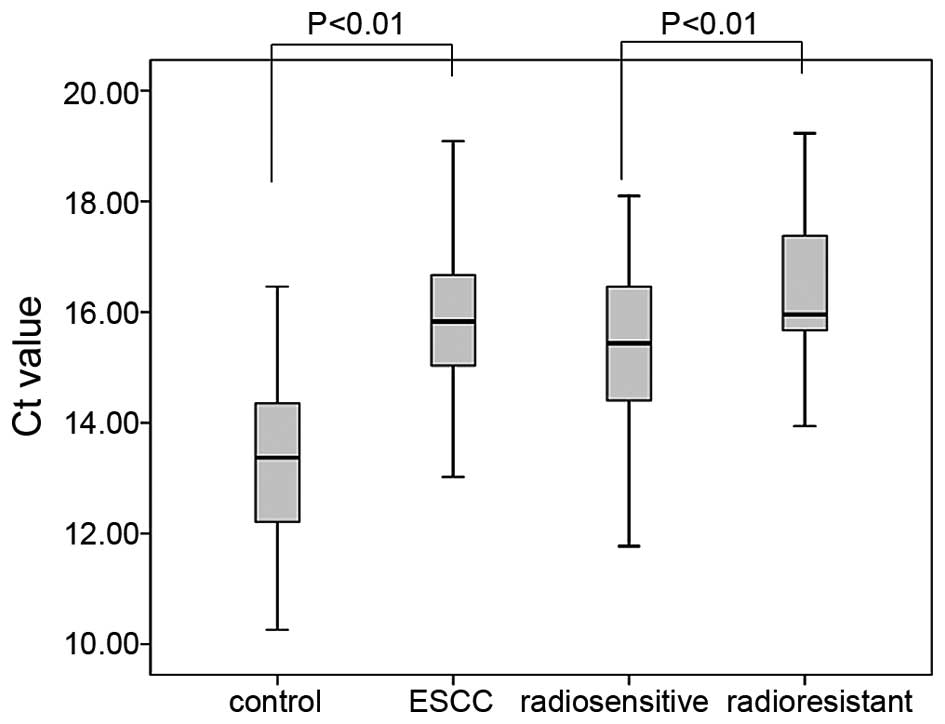
The result analyses revealed that the Ct of miR-7
expression was 15.15±1.80 [95% confidence interval (CI)] for the
radiosensitive group and 16.38±1.44 (95% CI) for the radioresistant
group, as shown in Fig. 1. Based on
statistical analysis, the relative miR-7 serum level was
significantly higher in the SD+PD group when compared with the
CR+PR group (P<0.05). The mean miR-7 serum levels differ by
2.34-fold between these two groups of patients, indicating that
miR-7 may serve as a biomarker for predicting the response of ESCC
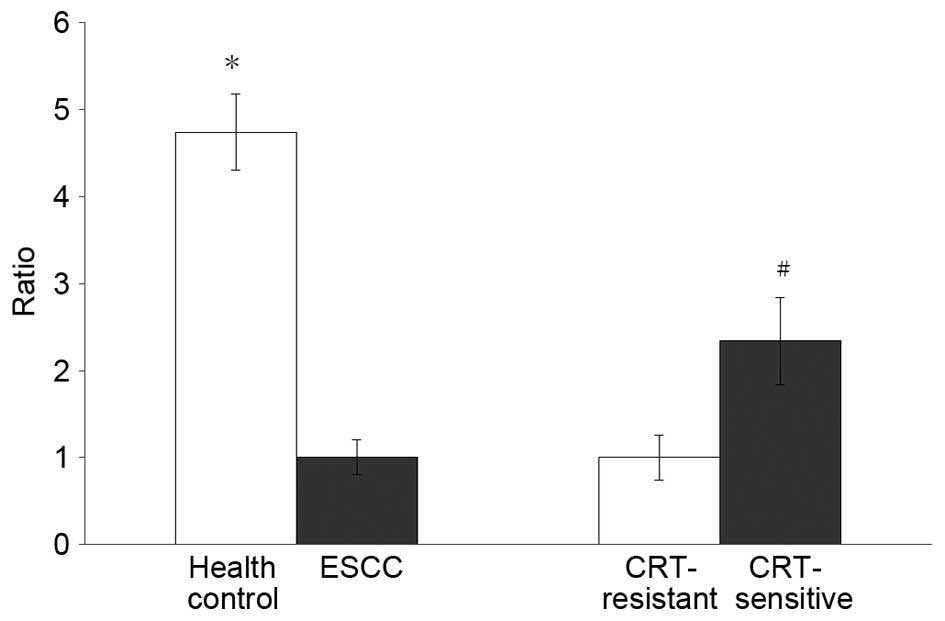
patient to CRT. Fig. 2 presents the
relative expression of serum miR-7 between two different
groups.
The serum miR-7 level of healthy volunteers was
13.41±1.56 (95% CI). As shown by the small range, compared with
ESCC patients, the serum miR-7 level in healthy subjects is 3.35
times higher than that in the radiosensitive group (P<0.01) and
7.84 times higher than that in the radioresistant group
(P<0.01). The comparison between the healthy subjects and ESCC
patients illustrates the possibility that serum miR-7 may also hold
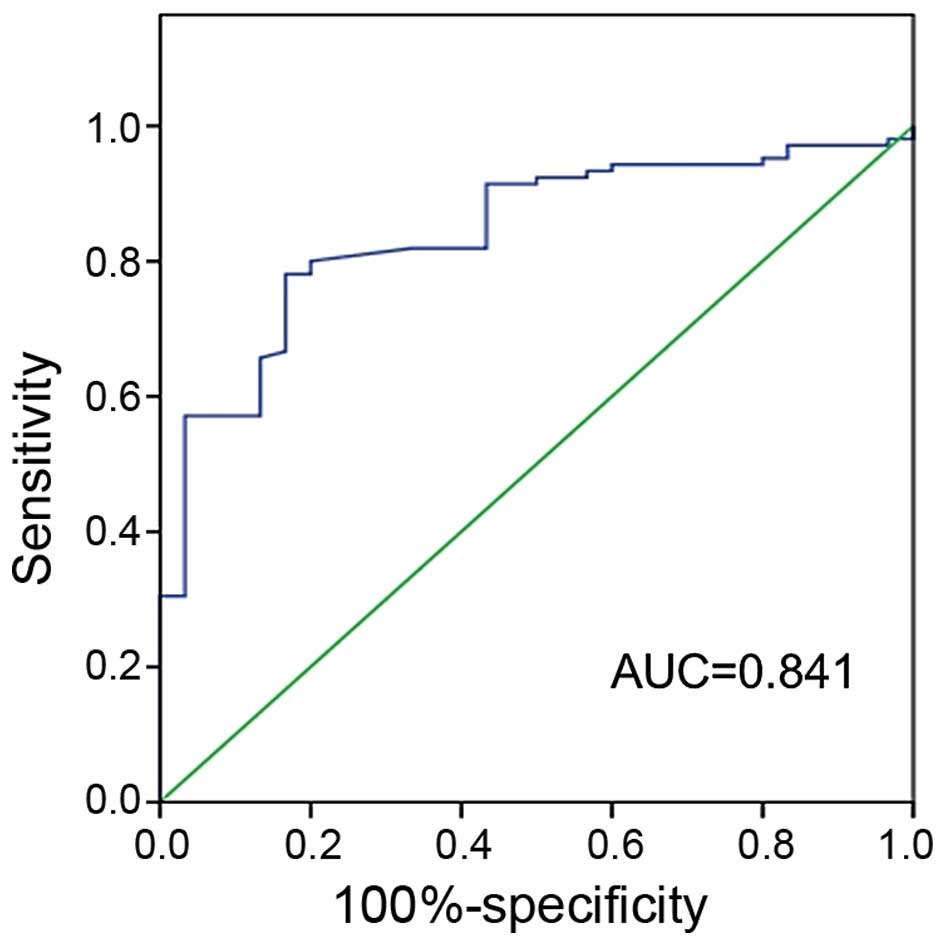
promise as a valuable diagnostic marker. Furthermore, the ROC curve
was plotted to identify a cut-off value that could distinguish ESCC
from healthy control. ROC curve analysis showed that at the optimal
cut-off, serum miR-7 had a 78.1% sensitivity and a 83.3%
specificity in separating ESCC from normal healthy control with an
AUC of 0.841 (Fig. 4).
Relationship between serum miRNA-7
expression and clinicopathological features
In addition to examining the expression of miRNAs in
serum, the relationship between miR-7 expression and
clinicopathological features of enrolled ESCC patients was
examined. By the median value, we demarcate high and low miR-7
levels. As shown in Table I, there
is no correlation between age and gender and relative miR-7 serum
levels. The relative miR-7 serum level is significantly correlated
with tumor length and the status of lymph node metastasis. The
relative miR-7 serum level is significantly lower in patients with
longer tumor compared with patients with shorter ones (P<0.05)
and in patients with lymph node involvement compared with patients
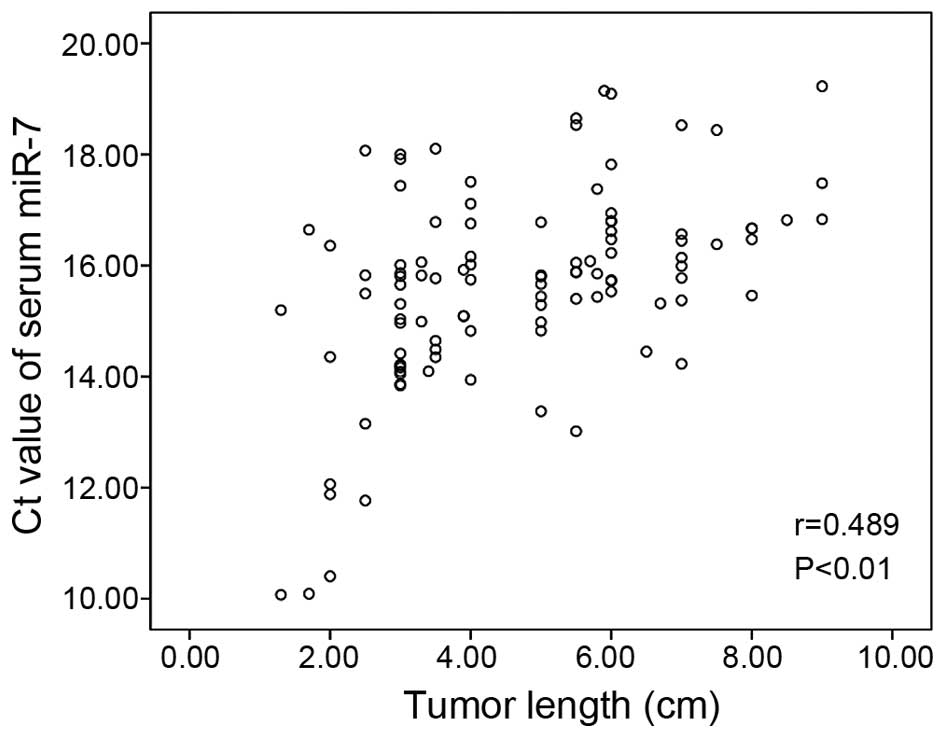
without lymph node involvement (P<0.05). Furthermore, the
relationship between the Ct value of miR-7 and tumor length was
examined by calculating Pearson's correlation coefficient. Our
results showed a correlation (r=0.489; P<0.01) between the serum
miR-7 levels and the tumor length in ESCC patients (Fig. 3).
 | Table IRelationships between effectiveness
of chemoradiotherapy (CRT) and clinicopathological factors as well
as serum levels of tumor markers. |
Table I
Relationships between effectiveness
of chemoradiotherapy (CRT) and clinicopathological factors as well
as serum levels of tumor markers.
| Elements | MicroRNA-7
| χ2 | P-value | Effectiveness
| χ2 | P-value |
|---|
| High
expression | Low expression | CR+PR | SD+PD |
|---|
| Gender | | | 0.116 | 0.733 | | | 0.012 | 0.914 |
| Male | 35 | 34 | | | 41 | 28 | | |
| Female | 17 | 19 | | | 21 | 15 | | |
| Age (years) | | | 0.97 | 0.325 | | | 1.155 | 0.282 |
| ≥60 | 38 | 34 | | | 40 | 32 | | |
| <60 | 14 | 19 | | | 22 | 11 | | |
| Tumor location | | | 0.457 | 0.796 | | | 3.545 | 0.17 |
| Upper third | 16 | 14 | | | 22 | 8 | | |
| Middle third | 28 | 32 | | | 32 | 28 | | |
| Lower third | 8 | 7 | | | 8 | 7 | | |
| Length | | | 7.505 | 0.023 | | | 1.662 | 0.436 |
| ≤4.0 | 33 | 20 | | | 34 | 19 | | |
| 4.1–6.0 | 13 | 19 | | | 16 | 16 | | |
| >6.0 | 6 | 14 | | | 12 | 8 | | |
| Tumor
differentiation | | | 1.157 | 0.561 | | | 1.557 | 0.459 |
| High | 13 | 11 | | | 13 | 11 | | |
| Moderate | 28 | 26 | | | 35 | 19 | | |
| Poor | 11 | 16 | | | 14 | 13 | | |
| Lymph node
metastasis | | | 4.197 | 0.04 | | | 0.264 | 0.607 |
| Positive | 21 | 32 | | | 30 | 23 | | |
| Negative | 31 | 21 | | | 32 | 20 | | |
| Distant
metastasis | | | 0.03 | 0.861 | | | 0.012 | 0.912 |
| Positive | 12 | 13 | | | 15 | 10 | | |
| Negative | 40 | 40 | | | 47 | 33 | | |
| Smoking
history | | | 0.234 | 0.628 | | | 0.367 | 0.545 |
| Positive | 26 | 24 | | | 28 | 22 | | |
| Negative | 26 | 29 | | | 34 | 21 | | |
| Drinking
history | | | 0.106 | 0.745 | | | 0.024 | 0.876 |
| Positive | 19 | 21 | | | 24 | 16 | | |
| Negative | 33 | 32 | | | 38 | 27 | | |
| Family history | | | 0.003 | 0.958 | | | 0.007 | 0.935 |
| Positive | 12 | 12 | | | 14 | 10 | | |
| Negative | 40 | 41 | | | 48 | 33 | | |
| CEA | | | 0.497 | 0.481 | | | 5.237 | 0.021 |
| ≤3.3 | 35 | 39 | | | 49 | 25 | | |
| >3.3 | 17 | 14 | | | 13 | 18 | | |
| Cyfra21-1 | | | 0.293 | 0.589 | | | 5.901 | 0.015 |
| ≤3.4 | 35 | 33 | | | 46 | 22 | | |
| >3.4 | 17 | 20 | | | 16 | 21 | | |
|
Myelosuppression | | | 4.024 | 0.403 | | | 25.076 | 0.000 |
| 0 | 17 | 13 | | | 7 | 23 | | |
| I | 20 | 20 | | | 26 | 14 | | |
| II | 10 | 15 | | | 21 | 4 | | |
| III | 3 | 5 | | | 6 | 2 | | |
| IV | 2 | 0 | | | 2 | 0 | | |
| miR-7 level | | | | | | | 4.418 | 0.036 |
| Higher | | | | | 36 | 16 | | |
| Lower | | | | | 26 | 27 | | |
Moreover, Table I
also presents the relationships between effectiveness of CRT and
clinicopathological factors. As is shown in the table, the
responsiveness of therapy is significantly correlated with
carcinoembryonic antigen (CEA) (P<0.05), Cyfra21-1 (P<0.05),
serum miR-7 level (P<0.05) and the myelosuppression (P<0.01).
Furthermore, by logistic regression analysis, the CR+PR rates of
CRT were significantly associated with the levels of Cyfra21-1
[P=0.038; overall remission (OR)=0.296; 95% CI for OR=0.094–0.934],
CEA (P=0.022; OR=0.276; 95% CI for OR=0.091–0.833), miR-7 (P=0.019;
OR=0.265; 95% CI for OR=0.087–0.800), and myelosuppression
(P=0.000, OR=3.347, 95% CI for OR=1.737–6.449) before treatments
shown in Table II. Thus, ESCC
patients with lower Cyfra21-1 and CEA, higher miR-7 and severe
myelosuppression were much more sensitive to CRT.
 | Table IIMultivariate analysis of the
clinicopathological factors related to responsiveness of
therapy. |
Table II
Multivariate analysis of the
clinicopathological factors related to responsiveness of
therapy.
| Variables | B | SE | Wals | P-value | OR | 95% CI for OR
|
|---|
| Lower U | pper |
|---|
| Gender | −0.105 | 0.743 | 0.020 | 0.888 | 0.901 | 0.210 | 3.862 |
| Age (years) | 0.596 | 0.602 | 0.981 | 0.322 | 1.815 | 0.558 | 5.909 |
| Lymph node
metastasis | −0.098 | 0.571 | 0.029 | 0.864 | 0.907 | 0.296 | 2.778 |
| Distant
metastasis | 0.534 | 0.685 | 0.608 | 0.435 | 1.705 | 0.446 | 6.525 |
| Tumor location | −0.304 | 0.419 | 0.526 | 0.468 | 0.738 | 0.324 | 1.679 |
| Length | −0.186 | 0.350 | 0.283 | 0.595 | 0.830 | 0.418 | 1.648 |
| Tumor
differentiation | 0.284 | 0.370 | 0.591 | 0.442 | 1.329 | 0.644 | 2.744 |
| Smoking
history | 0.390 | 0.748 | 0.272 | 0.602 | 1.477 | 0.341 | 6.397 |
| Drinking
history | −0.245 | 0.686 | 0.127 | 0.721 | 0.783 | 0.204 | 3.003 |
| Family history | −0.114 | 0.612 | 0.035 | 0.852 | 0.892 | 0.269 | 2.961 |
|
Myelosuppression | 1.208 | 0.335 | 13.037 | 0.000 | 3.347 | 1.737 | 6.449 |
| miR-7 level | −1.330 | 0.565 | 5.541 | 0.019 | 0.265 | 0.087 | 0.800 |
| CEA | −1.287 | 0.564 | 5.214 | 0.022 | 0.276 | 0.091 | 0.833 |
| Cyfra21-1 | −1.216 | 0.585 | 4.314 | 0.038 | 0.296 | 0.094 | 0.934 |
| Constant | 4.092 | 2.421 | 2.858 | 0.091 | 59.877 | | |
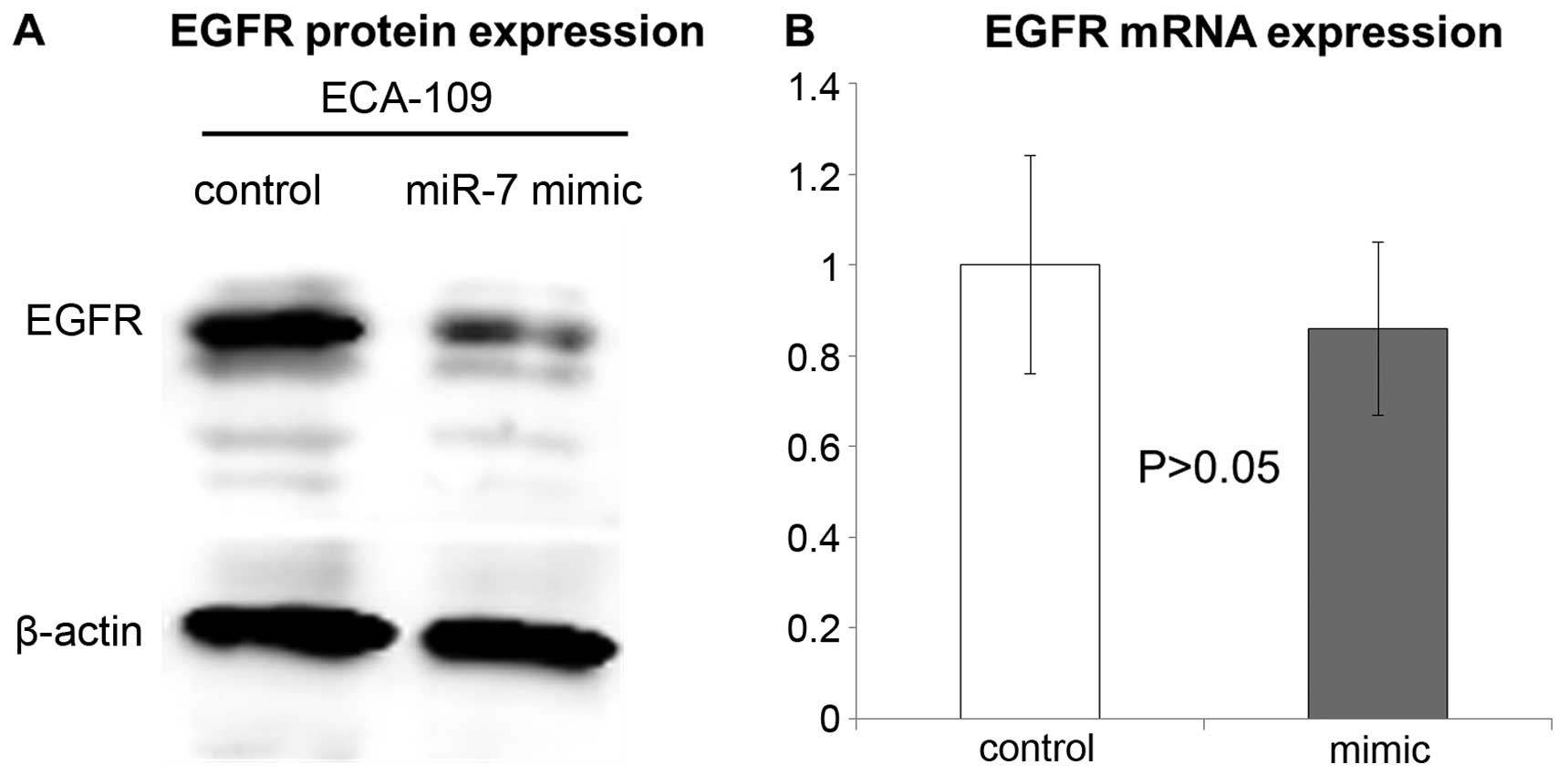
Downregulation of EGFR by miR-7
In previous studies, EGFR has been identified as an
important downstream factor of miR-7 (22,28,30),
therefore, we focused on EGFR for further functional analyses.
Compared with control subjects without any treatment, transfection
of miR-7 into ECA-109 cells suppressed EGFR protein expression
(Fig. 5A). However, no significant
change of EGFR mRNA level was found after transfection of miR-7
mimic (Fig. 5B), suggesting that
miR-7 can interfere with EGFR mRNA translation, but not
degradation.
Discussion
Chemoradiotherapy (CRT) play a very important role
in the treatment of esophageal cancer, however, it remains unclear
on the prediction of treatment responsiveness to CRT. It is
important to identify robust factors that will predict response to
CRT, and will also facilitate appropriate patient selection and
avoid unnecessary delays in patients at high risk of loco-regional
recurrence upon chemoradiation (4).
Wieder et al (34) and
Suzuki et al (35) have
given some clues from uptake value and changes in metabolic
activity in PET/CT that indicating tumor response and patient
survival.
Recently, numerous studies focused on the factors
that affect tumor responses to CRT, such as the presence of tumor
hypoxia (36), tumor
microenvironment (37), DNA damage
repair (38), cell cycle
checkpoint, apoptosis (39) and
radio-related signal transduction pathways (40). Understanding the regulatory
mechanisms of miRNA in tumor radiosensitivity from these diverse
aspects has also become an intense area of interest. Although there
has been great progress in the diagnosis, prediction of response to
treatment and prognosis of ESCC in the past decades, identifying
new biomarkers may greatly benefit the early detection screening
methods.
Great advances concerning miRNA-based therapeutics
have been made in various human diseases, including cancer. Recent
studies have verified that dysregulation of miRNA expression in
human diseases may act as diagnostic, and prognostic factors as
well as predicting response to chemotherapy or/and radiotherapy
biomarkers. miR-7 has previously been characterized as a
tumor-suppressor miRNA in several human cancers by targeting a
number of key signaling molecules (22,25,41).
The present study showed that serum miR-7 levels were downregulated
(4.74-fold-change) in patients with ESCC compared with healthy
controls, indicating that it may be a useful biomarker for early
diagnosis. However, the downregulation is not unique to ESCC. For
example, the level of serum miR-7 is decreased in glioblastoma, and
increased miR-7 inhibits glioma cell proliferation by inhibiting
the EGFR and AKT pathways (24).
Downregulation of miR-7 in breast cancer has also been proven in
the study by Kong et al (42), who demonstrated the inhibition of
epithelial-to-mesenchymal transition and metastasis of breast
cancer cells via targeting FAK expression. Previously studies have
demonstrated that miR-7 is also downregulated in advanced tongue
squamous cell carcinoma cell lines (43,44).
Therefore, our current results are consistent with previous
findings.
The present study shows that the presence of ESCC is
associated with a suppressed level of serum miR-7, and the degree
of suppression is correlated with tumor length and lymph node
status. An interesting feature of our study is the existence of a
statistically significant correlation between lower serum miR-7
expression in ESCC and longer tumor, as well as positive lymph node
metastasis, which are the main prognostic factors for ESCC. ROC
curve analysis showed that at the optimal cut-off, serum miR-7 had
a 78.1% sensitivity and a 83.3% specificity in separating ESCC from
normal healthy control with an AUC of 0.841 (Fig. 3). Such findings imply that miR-7 may
be involved in the initiation and progression of cancer.
Furthermore, the expression level analysis revealed
that serum miR-7 is a valuable biomarker for differentiating the
responsiveness to CRT of ESCC patients. As shown in Table II, the responsiveness to CRT is
significantly associated with the levels of Cyfra21-1, CEA, miR-7
and myelosuppression before treatments. i.e. ESCC patients
with lower Cyfra21-1 and CEA, higher miR-7 and severe
myelosuppression were much more sensitive to CRT, which is similar
to previous studies (45–47). However, this conclusion should be
confirmed in a study with larger and more homogeneous samples.
The higher expression of miR-7 in ESCC may be more
sensitive to CRT, which can be explained with activated EGFR and
AKT associated pathways (24,30).
Recent studies have reported that EGFR can modulate DNA repair
(48–50), consequently influence the
responsiveness to CRT. While the present study showed that miR-7
has the ability to downregulate the expression of EGFR, which is
similar to other studies (24,30,51).
These results suggested that serum miR-7 may serve as a biomarker
for the noninvasive diagnostic and predictive marker for ESCC.
In conclusion, the present study showed that serum
miR-7 levels were significantly suppressed in patients with ESCC
compared with control subjects, and the degree of suppression is
correlated with longer tumor and positive lymph node metastasis.
ESCC patients with lower Cyfra21-1 and CEA, higher miR-7, and
severe myelosuppression were much more sensitive to CRT. These
findings indicate that serum miR-7 may serve as a novel diagnostic
and response predictive marker for ESCC patients, and it can also
play a potential role in selecting CRT. Furthermore, we
demonstrated that miR-7 mediated its function by repressing EGFR,
which could be a novel mechanism of the ESCC patients
responsiveness to CRT, suggesting that miR-7 possibly could be
employed as an effective therapeutic target for ESCC.
Acknowledgments
We are grateful to Professor Cedric X. Yu
(Department of Radiation Oncology, University of Maryland School of
Medicine, Baltimore, MD, USA) for reviewing the manuscript.
References
|
1
|
Zhou ZG, Gao XS, Qiao XY and Zhang P:
Literature analysis of radiotherapy for esophageal cancer in China.
Chin J Cancer. 29:873–881. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schneider BJ and Urba SG: Preoperative
chemoradiation for the treatment of locoregional esophageal cancer:
The standard of care? Semin Radiat Oncol. 17:45–52. 2007.
View Article : Google Scholar
|
|
3
|
Shibata T, Kokubu A, Saito S,
Narisawa-Saito M, Sasaki H, Aoyagi K, Yoshimatsu Y, Tachimori Y,
Kushima R, Kiyono T, et al: NRF2 mutation confers malignant
potential and resistance to chemoradiation therapy in advanced
esophageal squamous cancer. Neoplasia. 13:864–873. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fokas E, Weiss C and Rödel C: The role of
radiotherapy in the multimodal management of esophageal cancer. Dig
Dis. 31:30–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luthra R, Wu TT, Luthra MG, Izzo J,
Lopez-Alvarez E, Zhang L, Bailey J, Lee JH, Bresalier R, Rashid A,
et al: Gene expression profiling of localized esophageal
carcinomas: Association with pathologic response to preoperative
chemoradiation. J Clin Oncol. 24:259–267. 2006. View Article : Google Scholar
|
|
6
|
Wu X, Gu J, Wu TT, Swisher SG, Liao Z,
Correa AM, Liu J, Etzel CJ, Amos CI, Huang M, et al: Genetic
variations in radiation and chemotherapy drug action pathways
predict clinical outcomes in esophageal cancer. J Clin Oncol.
24:3789–3798. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang JX, Tong ZT, Yang L, Wang F, Chai
HP, Zhang F, Xie MR, Zhang AL, Wu LM, Hong H, et al: PITX2: A
promising predictive biomarker of patients' prognosis and
chemoradioresistance in esophageal squamous cell carcinoma. Int J
Cancer. 132:2567–2577. 2013. View Article : Google Scholar
|
|
8
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar
|
|
9
|
Liu Z, Jin ZY, Liu CH, Xie F, Lin XS and
Huang Q: MicroRNA-21 regulates biological behavior by inducing EMT
in human cholangiocarcinoma. Int J Clin Exp Pathol. 8:4684–4694.
2015.PubMed/NCBI
|
|
10
|
Yang X, Ye J, Yan H, Tang Z, Shen 1, Zhang
J and Yang L: MiR-491 attenuates cancer stem cells-like properties
of hepatocellular carcinoma by inhibition of GIT-1/NF-κB-mediated
EMT. Tumour Biol. Jul 19–2015.Epub ahead of print.
|
|
11
|
Chang YL, Zhou PJ, Wei L, Li W, Ji Z, Fang
YX and Gao WQ: MicroRNA-7 inhibits the stemness of prostate cancer
stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21
pathway. Oncotarget. 6:24017–24031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan
M, Jeon YJ, Li B, Vicentini C, Peng Y, et al: MicroRNA-224 promotes
tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci
USA. 112:E4288–E4297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Shen S, Wu S, Chen Z, Hu C and Yan
R: Regulation of tumorigenesis and metastasis of hepatocellular
carcinoma tumor endothelial cells by microRNA-3178 and underlying
mechanism. Biochem Biophys Res Commun. 464:881–887. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao L, Bode AM, Cao Y and Dong Z:
Regulatory mechanisms and clinical perspectives of miRNA in tumor
radiosensitivity. Carcinogenesis. 33:2220–2227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Erson AE and Petty EM: MicroRNAs in
development and disease. Clin Genet. 74:296–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
21
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Webster RJ, Giles KM, Price KJ, Zhang PM,
Mattick JS and Leedman PJ: Regulation of epidermal growth factor
receptor signaling in human cancer cells by microRNA-7. J Biol
Chem. 284:5731–5741. 2009. View Article : Google Scholar
|
|
23
|
Buckley AF, Burgart LJ, Sahai V and Kakar
S: Epidermal growth factor receptor expression and gene copy number
in conventional hepatocellular carcinoma. Am J Clin Pathol.
129:245–251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, et
al: microRNA-7 inhibits the epidermal growth factor receptor and
the Akt pathway and is down-regulated in glioblastoma. Cancer Res.
68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reddy SD, Ohshiro K, Rayala SK and Kumar
R: MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase
1 and regulates its functions. Cancer Res. 68:8195–8200. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok
MT, Wang H, Chen J, Ng SS, Chen M, et al: microRNA-7 is a novel
inhibitor of YY1 contributing to colorectal tumorigenesis.
Oncogene. 32:5078–5088. 2013. View Article : Google Scholar
|
|
28
|
Li X and Carthew RW: A microRNA mediates
EGF receptor signaling and promotes photoreceptor differentiation
in the Drosophila eye. Cell. 123:1267–1277. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee KM, Choi EJ and Kim IA: microRNA-7
increases radiosensitivity of human cancer cells with activated
EGFR-associated signaling. Radiother Oncol. 101:171–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong W, Li B, Wang Z, Zhang Z and Wang J:
Clinical significance of microRNA-24 expression in esophageal
squamous cell carcinoma. Neoplasma. 62:250–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miller AB, Hoogstraten B, Staquet M and
Winkler A: Reporting results of cancer treatment. Cancer.
47:207–214. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmittgen TD, Lee EJ and Jiang J:
High-throughput real-time PCR. Methods Mol Biol. 429:89–98. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wieder HA, Brücher BL, Zimmermann F,
Becker K, Lordick F, Beer A, Schwaiger M, Fink U, Siewert JR, Stein
HJ, et al: Time course of tumor metabolic activity during
chemoradiotherapy of esophageal squamous cell carcinoma and
response to treatment. J Clin Oncol. 22:900–908. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Suzuki A, Xiao L, Hayashi Y, Macapinlac
HA, Welsh J, Lin SH, Lee JH, Bhutani MS, Maru DM, Hofstetter WL, et
al: Prognostic significance of baseline positron emission
tomography and importance of clinical complete response in patients
with esophageal or gastroesophageal junction cancer treated with
definitive chemoradiotherapy. Cancer. 117:4823–4833. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vaupel P and Harrison L: Tumor hypoxia:
Causative factors, compensatory mechanisms, and cellular response.
Oncologist. 9(Suppl 5): S4–S9. 2004. View Article : Google Scholar
|
|
37
|
Jamal M, Rath BH, Williams ES, Camphausen
K and Tofilon PJ: Microenvironmental regulation of glioblastoma
radioresponse. Clin Cancer Res. 16:6049–6059. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Thoms J and Bristow RG: DNA repair
targeting and radiotherapy: A focus on the therapeutic ratio. Semin
Radiat Oncol. 20:217–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Langerak P and Russell P: Regulatory
networks integrating cell cycle control with DNA damage checkpoints
and double-strand break repair. Philos Trans R Soc Lond B Biol Sci.
366:3562–3571. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chou CH, Chen SU and Cheng JC:
Radiation-induced interleukin-6 expression through
MAPK/p38/NF-kappaB signaling pathway and the resultant
antiapoptotic effect on endothelial cells through Mcl-1 expression
with sIL6-Ralpha. Int J Radiat Oncol Biol Phys. 75:1553–1561. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder
CE, Kolokythas A, Wang A, Dai Y and Zhou X: MicroRNA-7 targets
IGF1R (insulin-like growth factor 1 receptor) in tongue squamous
cell carcinoma cells. Biochem J. 432:199–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W,
Wu Z, Chen T, Wu W, Lobie PE, et al: MicroRNA-7 inhibits
epithelial-to-mesenchymal transition and metastasis of breast
cancer cells via targeting FAK expression. PLoS One. 7:e415232012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu X, Jiang L, Wang A, Yu J, Shi F and
Zhou X: MicroRNA-138 suppresses invasion and promotes apoptosis in
head and neck squamous cell carcinoma cell lines. Cancer Lett.
286:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H
and Zhou X: MicroRNA-222 regulates cell invasion by targeting
matrix metalloproteinase 1 (MMP1) and manganese superoxide
dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines.
Cancer Genomics Proteomics. 6:131–139. 2009.PubMed/NCBI
|
|
45
|
Kunisaki C, Imada T, Yamada R, Hatori S,
Kinbara K, Watai K, Akiyama H, Nomura M, Matsuda G, Otsuka Y, et
al: Prognostic factors after chemoradiotherapy for patients with
inoperable esophageal squamous cell carcinoma.
Hepatogastroenterology. 53:366–371. 2006.PubMed/NCBI
|
|
46
|
Wakatsuki M, Suzuki Y, Nakamoto S, Ohno T,
Ishikawa H, Kiyohara H, Kiyozuka M, Shirai K, Nakayama Y and Nakano
T: Clinical usefulness of CYFRA 21-1 for esophageal squamous cell
carcinoma in radiation therapy. J Gastroenterol Hepatol.
22:715–719. 2007.PubMed/NCBI
|
|
47
|
Yi Y, Li B, Wang Z, Sun H, Gong H and
Zhang Z: CYFRA21-1 and CEA are useful markers for predicting the
sensitivity to chemoradiotherapy of esophageal squamous cell
carcinoma. Biomarkers. 14:480–485. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liccardi G, Hartley JA and Hochhauser D:
EGFR nuclear translocation modulates DNA repair following cisplatin
and ionizing radiation treatment. Cancer Res. 71:1103–1114. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Szumiel I: Epidermal growth factor
receptor and DNA double strand break repair: The cell's
self-defence. Cell Signal. 18:1537–1548. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kriegs M, Kasten-Pisula U, Rieckmann T,
Holst K, Saker J, Dahm-Daphi J and Dikomey E: The epidermal growth
factor receptor modulates DNA double-strand break repair by
regulating non-homologous end-joining. DNA Repair. 9:889–897. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kalinowski FC, Giles KM, Candy PA, Ali A,
Ganda C, Epis MR, Webster RJ and Leedman PJ: Regulation of
epidermal growth factor receptor signaling and erlotinib
sensitivity in head and neck cancer cells by miR-7. PLoS One.
7:e470672012. View Article : Google Scholar : PubMed/NCBI
|