1. Introduction
Whereas surgical resection and adjuvant therapy can
cure well-confined primary tumors, metastatic disease is largely
incurable due to its systemic nature and the resistance of
disseminated tumor cells to existing therapeutic agents. This
explains why above 90% of mortality from cancer is attributable to
metastases, but not the primary tumors from which these malignant
lesions arise (1,2). Thus, our ability to effectively treat
cancer is largely dependent on our capacity to interdict, and
perhaps even reverse, the process of metastasis.
Metastasis is commonly viewed as a linear chain of
events resulting in the re-localization of tumorigenic cells from
the primary tumor site to a distant location. The stages of
metastasis may be divided into the following categories: i)
invasion locally through surrounding extracellular matrix (ECM) and
stromal cell layers; ii) intravasation into the lumina of blood
vessels; iii) survival of the transport through the vasculature;
iv) arrest at distant organ sites; v) extravasation into the
parenchyma of distant tissues; vi) initial survival in these
foreign microenvironments in order to form micrometastases; and
vii) re-initiation of their proliferative programs at metastatic
sites, thereby generating macroscopic, clinically detectable
neoplastic growths (the step often referred to as 'metastatic
colonization') (3). Molecules
involved in the metastasis process are potential therapeutic target
for cancer metastasis (4).
The RUNX factors describe a family of evolutionarily
conserved metazoan proteins [RUNX1, RUNX2 and runt-related
transcription factor 3 (RUNX3)], each of which is capable of
forming heterodimers with the common β-subunit CBFβ. RUNX families
share the highly conserved homologous N-terminal runt domain, a
DNA-binding and heterodimerization region of ~120 amino acids.
Although their strong homology indicates a certain degree of
redundancy as observed during mouse embryogenesis (5) and transcription regulation of various
genes (for example Sgpp1, Ncam1 and p21WAF1) (6), the RUNX proteins have been shown to
perform distinct functions that are dependent on tissue type
(7).
Among the RUNX families, RUNX3 was demonstrated to
be the oldest of the three genes by sequence analysis, and is found
in cnidarians, the most primitive animals, where it regulates the
growth of their primitive gastrointestinal system. RUNX3 is
involved in a variety of biological activities, including the
development of the gastrointestinal tract (8), neurogenesis of dorsal root ganglia
(9) and T-cell differentiation
(10). The RUNX3 gene is located on
1p.13-p36.11, where loss of heterozygosity (LOH) is often reported
in many types of cancers, indicating its potential role of tumor
suppressor in cancers (11). RUNX3
was first claimed to be a tumor suppressor in gastric cancer more
than a decade ago. This assertion was based on the following
observations: i) high expression of Runx3 in normal mouse
gastrointestinal tract (GIT) epithelium; ii) gastric hyperplasia in
Runx3−/− newborn mice; iii) RUNX3 LOH in gastric cancer
patients; iv) DNA hypermethylation of RUNX3 promoter in gastric
cancer; and v) reduced RUNX3 mRNA in gastric cancer vs. normal
gastric tissue in 60% of gastric cancer patients assayed by RNA
in situ hybridization (RISH). Since then, RUNX3 has emerged
as a candidate tumor suppressor that is epigenetically inactivated
in a wide spectrum of invasive and pre-invasive epithelial and
mesenchymal neoplasms (12).
Hypermethylation of RUNX3 promoter is the leading cause which
results in RUNX3 inactivation in cancer tissues (13–15).
Besides hypermethylation, inactivation of RUNX3 also occurs by
mislocalization of RUNX3 protein from the nucleus to the cytoplasm
(16). RUNX3 can function as a
tumor suppressor and is involved in tumor development and
metastasis at different levels, such as EMT (17), adhesion (18), invasion (18,19),
apoptosis (20) and angiogenesis
(21). However, it is not clear how
and where RUNX3 interacts with pathways that regulate tumor
development and metastasis. Investigations of the factors
controlling RUNX3 function and its downstream effectors have begun
to unravel this biological mystery. The present review will focus
on the existing knowledge of the role of RUNX3 gene in regulating
cancer metastasis, and provide some considerations for future
investigations.
2. RUNX3 inactivation in human cancers
The RUNX3 gene is located on 1p.13-p36.11, a
deletion hotspot in diverse cancers of epithelial, hematopoietic
and neural origins, indicating its potential role of tumor
suppressor in cancers (11). RUNX3
was first found lost in gastric cancer, since then, more and more
studies have reported that the loss or reduction of the RUNX3
protein can be detected in various cancers, such as colorectal
cancer, glioma and melanoma (15,22,23).
In our previous studies, we showed the reduction of RUNX3 was
correlated with tumor stage and grade in prostate cancer and renal
cell carcinoma (18,24). Analysis of clinical tissue samples
from peritoneal metastases arising from gastric cancers provides
evidence that RUNX3 expression significantly decreased in the
metastatic tissue, compared to normal gastric mucosa or primary
main tumors (19). Conversely,
RUNX3 overexpression correlates with reduced invasive potential of
breast cancer cells (25).
Similarly, RUNX3 expression in the breast stroma is associated with
low rate of recurrence, and thus, a good clinical outcome in breast
cancer (26). Furthermore, the
presence of nuclear RUNX3 protein in esophageal squamous cell
carcinomas correlates with increased sensitivity to radiotherapy,
whereas RUNX3 inactivation is linked to radioresistance and poor
prognosis (27). The above suggests
that RUNX3 inactivation is a significant risk factor for
tumorigenesis and metastasis.
Aberrant methylation is the main reason that results
in RUNX3 inactivation in human cancers. Although it is unclear what
causes aberrant methylation of RUNX3, hypermethylation of the CpG
islands at the RUNX3 promoter is one of the most common aberrant
methylation events in cancer. Diverse tumor tissues and cancer cell
lines from human patients have RUNX3 promoter hypermethylation and
inactivation, as identified by methylation-specific PCR or
bisulfate genomic sequencing analysis. These tissues and cells
include those derived from gastric, bladder, colorectal, breast,
lung, pancreatic and brain cancers, and hepatocellular carcinoma
(12). RUNX3 hypermethylation in
these cancer tissues is strongly correlated with poor prognosis and
a low patient survival rate (28).
Recently, RUNX3 hypermethylation status has been used not only as a
helpful and valuable biomarker for diagnosis of breast cancer,
particularly in ER-negative breast cancer (29), but also a factor in predicting or
monitoring postoperative recurrence of papillary thyroid cancer
patients (30). These reports
suggest that RUNX3 methylation occurs early in tumorigenesis and
may increase with age. To date, only three contributors to the
increase in RUNX3 promoter hypermethylation have been identified.
First, Helicobacter pylori, a well-known factor involved in
gastric cancer, induces nitric oxide production, liposaccharide
production and inflammation, leading to hypermethylation and
silencing of RUNX3 (31). Second,
inflammatory insults such as that caused by lipopolysaccharide
(LPS) enhance RUNX3 methylation at the promoter through increased
nitric oxide production (31).
Third, estrogen induces hypermethylation and silencing of RUNX3 in
mammosphere-derived cells (32).
Enhancer of zeste homologue 2 (EZH2)-mediated
histone methylation is another epigenetic mechanism that is
involved in RUNX3 inactivation. Overexpression of EZH2 caused H3K27
trimethylation of the RUNX3 promoter, which led to repression of
RUNX3 expression in the absence of DNA methylation. EZH2 binds to
the RUNX3 promoter, resulting in upregulation of H3K27 methylation
and concomitant down-regulation of RUNX3 expression (33). Lee et al extended this
finding by showing that hypoxia-induced upregulation of G9a histone
methyltransferase and histone deacetylase 1 (HDAC1) expression also
cause epigenetic RUNX3 silencing through H3K9 methylation and
decreased H3 acetylation at RUNX3 promoter. This proposed that
hypoxia silences RUNX3 by epigenetic histone regulation during the
progression of gastric cancer (34).
MicroRNAs are the third epigenetic mechanism
responsible for silencing of RUNX3. MicroRNAs have been recently
highlighted as regulators of gene expression at the
post-transcriptional level. A recent study found that frequent
overexpression of EZH2 in aggressive solid tumors is largely due to
the loss of its regulator miR-101. Specifically, reintroduction of
miR-101 or knockdown of EZH2 expression was sufficient to reduce
H3K27 trimethylation and increase RUNX3 mRNA levels (35). Recently, miR130b was proposed as a
candidate for silencing RUNX3 in gastric cancer cells and tissues.
Overexpression of miR-130b increases cell viability and decreases
TGF-β-induced apoptosis, resulting in RUNX3 protein expression
(36). In melanoma tissues, RUNX3
silencing is linked to miR-532-5p (37). The role of these miRNAs in silencing
RUNX3 should be explored further to evaluate the restoration of
RUNX3 expression for therapeutic approaches.
Mislocalization of RUNX3 protein from the nucleus to
the cytoplasm also contributes to inactivation of RUNX3. RUNX3 was
reported primarily confined to the cytoplasm of cells in RUNX3
induced tumors, whereas a substantial fraction of RUNX3 was present
in the nucleus of cells in control tumors. Cytoplasmic RUNX3
protein does not elicit tumor suppressive activity, and is
therefore a novel mechanism of RUNX3 inactivation (16). Several mechanisms for excluding
RUNX3 from the nucleus have been suggested. Src kinase
phosphorylates tyrosine residues in RUNX3, and overexpression of
activated Src correlates with RUNX3 cytoplasmic localization
(38). In contrast, Pim-1
phosphorylates four Ser/Thr residues within the runt domain and
thereby stabilizes the RUNX3 protein. It alters the cellular
localization of RUNX3 from the nucleus to the cytoplasm (39). Phosphorylation of the RUNX3 by
Jun-activation domain-binding protein 1 (Jab1/CSN5) is also a major
cause of cytoplasmic sequestration (40).
In summary, RUNX3 inactivation can be regulated by
several epigenetic mechanisms, including DNA and histone
methylation, and some types of miRNAs. Another mode of RUNX3
inactivation is the mislocalization of RUNX3.
3. Overview of RUNX3 signaling in cancer
metastasis
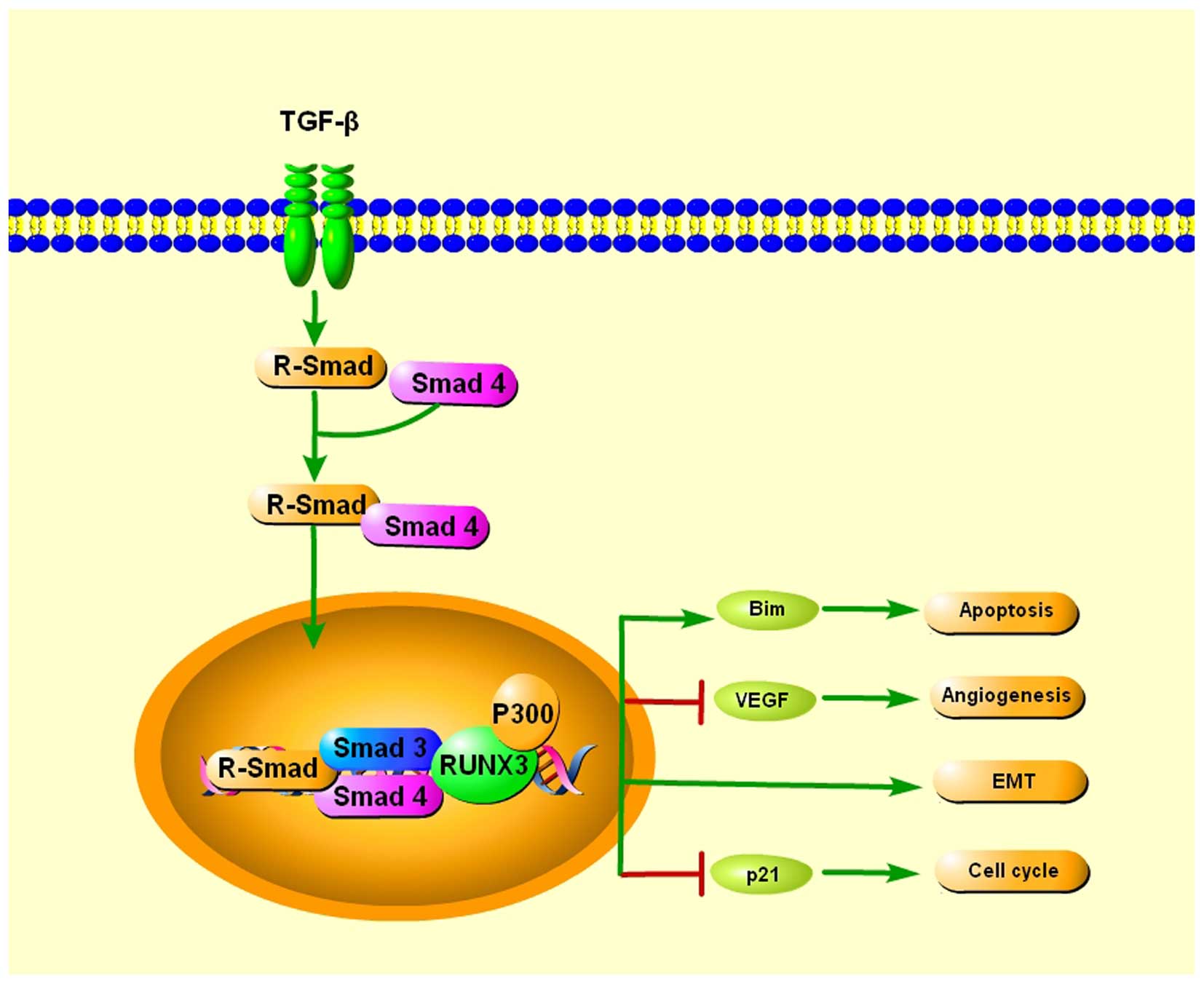
TGF-β is a ubiquitously expressed cytokine that
plays very important roles in many cellular functions. When TGF-β
binds to the respective cognate receptors, the serine kinases are
activated, which then phosphorylate signal transducers called
R-Smad. TGF-β activates R-Smad, which associate with a common Smad4
and enter the nucleus, where the R-Smad/Smad4/Smad3 complexes bind
to transcription factors in order to regulate the transcription of
target genes (41).
Runx3−/− gastric epithelial cells were insensitive to
the growth inhibitory effects of TGF-β, indicating a potential role
of RUNX3 as a downstream effector of TGF-β (42), providing a possible explanation for
its broad involvement in human tumorigenesis and metastasis. Chi
et al showed that RUNX3 inhibits gastric epithelial cell
growth by inducing p21 gene expression in response to TGF-β
(43). Yamamura et al
reported that RUNX3 works together with FoxO3a/FKHRL1 in the
induction of apoptosis by activating Bim when exposed to TGF-β and
may play an important role in tumor suppression of cancer (44). Peng et al found three RUNX3
binding sites in the vascular endothelial growth factor (VEGF)
promoter. Restoration of RUNX3 expression inhibited the angiogenic
potential of gastric cancer cells, and correlated with
downregulated VEGF expression via promoter repression (21). Furthermore, Runx3-null gastric
epithelial lines are unexpectedly sensitized to TGF-β-induced EMT,
indicating a potential role of RUNX3 in inhibiting EMT in TGF-β
signaling (17) (Fig. 1).
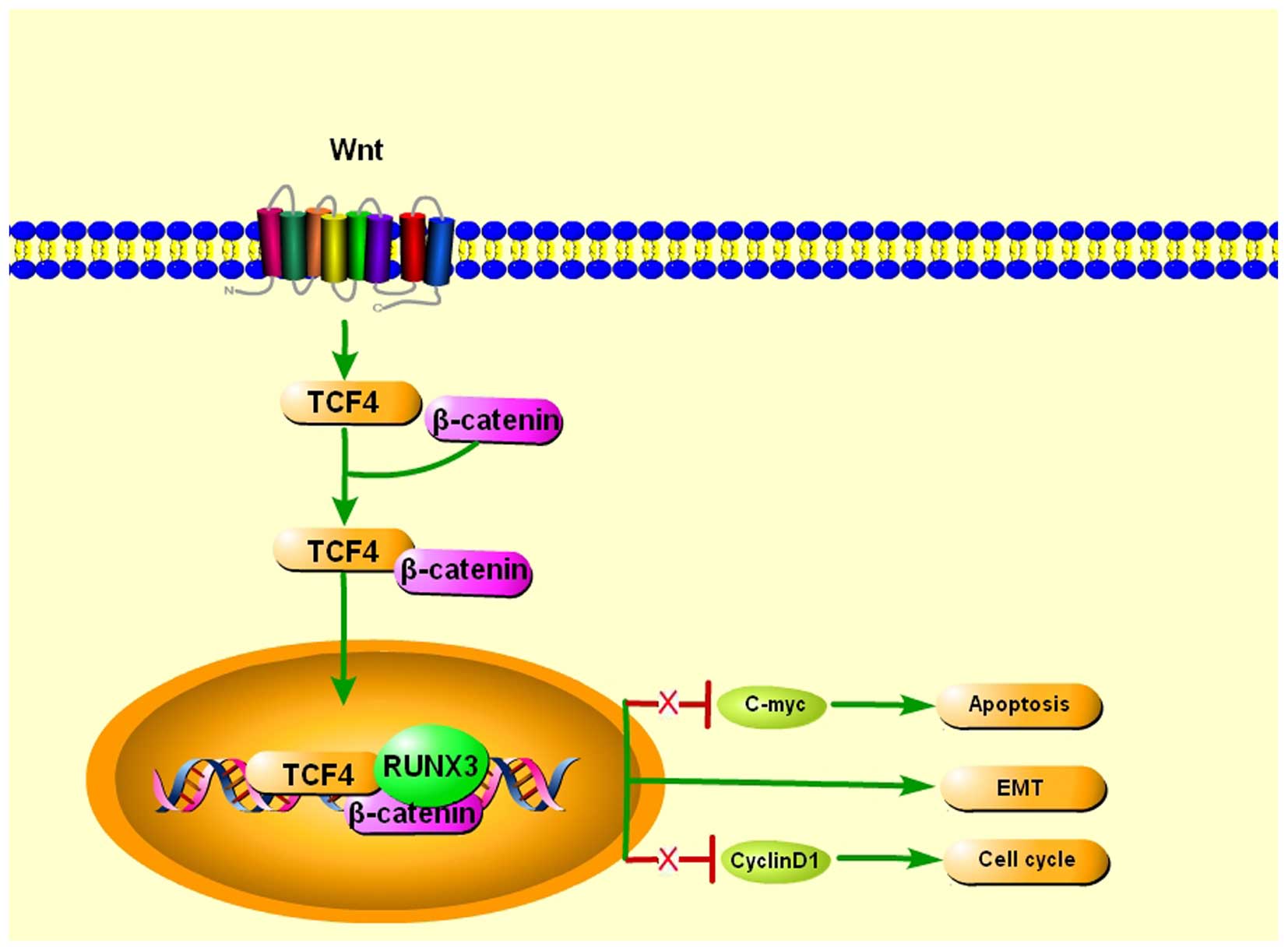
The Wnt/β-catenin signaling pathway has a
well-established role in the regulation of cell invasion, EMT, cell
growth and proliferation, as well as in cancer stem cell
differentiation, and its constitutive activation is commonly found
in human cancers (45). Notably,
RUNX3 was demonstrated to inhibit EMT, which promotes metastasis,
through the regulation of Wnt signaling pathways (17,46).
The detection of Runx3 in the epithelial cells of small and large
intestines in mice hinted that RUNX3 may function as a tumor
suppressor in the intestinal epithelium. Indeed, the intestinal
epithelia of Runx3−/− mice exhibit hyperplasia that is
associated with robust proliferation. Further examination revealed
that this phenomenon is the likely consequence of enhanced Wnt
signaling activity and transcriptional upregulation of Wnt target
genes such as c-Myc and cyclin D1. Further discovery
indicated that RUNX3 forms a ternary complex with β-catenin/TCF4,
and attenuates Wnt signaling activity and reduced ability to bind
DNA, and activate transcription of Wnt target genes. RUNX3,
therefore, antagonizes aberrant Wnt signaling, which substantial
evidence has shown, provides an oncogenic impetus to sustain
colorectal cancer growth (47).
However, RUNX3 stabilizes the TCF4/β-catenin complex on the Wnt
target gene promoter in the gastric cancer cells Kato III, leading
to activation of Wnt signaling, and its expression resulted in
suppression of tumorigenesis of Kato III cells, indicating that
RUNX3 plays a tumor-suppressing role in Kato III cells through a
Wnt-independent mechanism, suggesting RUNX3 can either suppress or
activate the Wnt signaling pathway through its binding to the
TCF4/β-catenin complex by cell context-dependent mechanisms
(48) (Fig. 2).
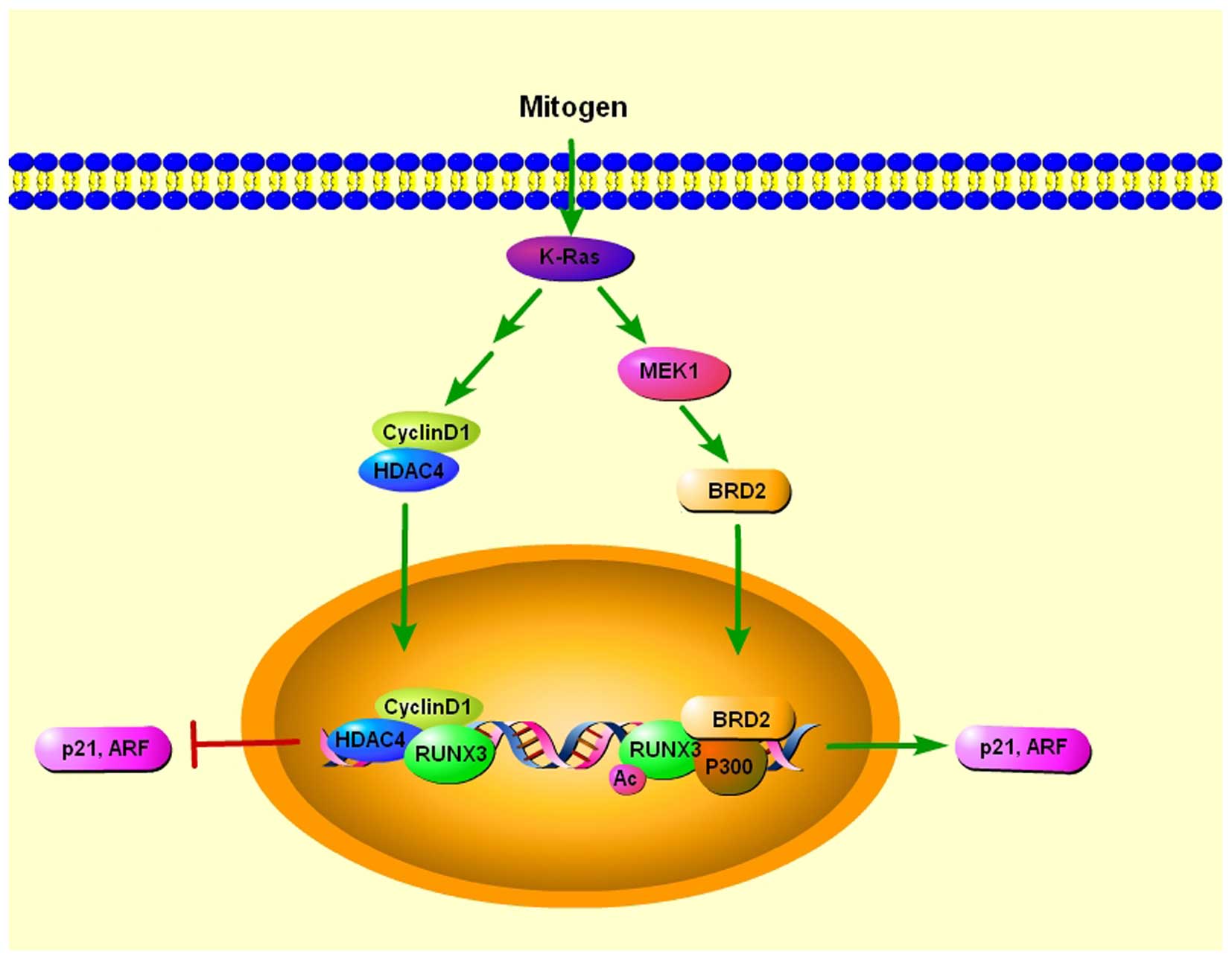
Oncogenic Ras induces premature senescence in
primary cells, except when its expression is accompanied by
coincident loss of tumor-suppressor activity, suggesting the
existence of a cellular defense mechanism against oncogenic insults
(49). RUNX3 is preferentially
inactivated in K-RAS-induced lung adenocarcinoma, indicating its
potential role of tumor suppressor in lung adenocarcinoma (50). ARF and p21 play essential roles in
the p53-mediated cellular defense against oncogenic insult
(51), and both genes are targets
of Runx proteins (43,52). Transcriptional activation of ARF is
essential for oncogenic RAS-induced stabilization of p53, which
plays major roles in tumor suppression (53). In addition, p21, which is induced in
both p53-dependent and -independent manner, also plays a key role
in defense against oncogenic RAS (51). Lee et al found that both p21
and ARF were transiently induced by serum stimulation, and the
induction of both genes was dependent on both K-RAS activity and
RUNX3 (54). BRD2 is a member of
the bromodomain extra terminal (BET) protein family, which consists
of four chromatin-interacting proteins that regulate gene
expression (55). They further
found that serum stimulation induces K-RAS activation, which
triggered the formation of RUNX3-BRD2 complex which is responsible
for oncogenic K-RAS-induced expression of p21 and ARF. When K-RAS
activity was reduced and cyclin D1 was induced, and subsequently
bound to RUNX3. At the same time, RUNX3 interacted with histone
deacetylase 4 (HDAC4), resulting in deacetylation of RUNX3
(56) and RUNX3 was released from
both p300 and BRD2, converting the RUNX3-BRD2 complex to a
RUNX3-HDAC4 complex. As a result, the expression of ARF and p21 is
switched off. When K-Ras is constitutively activated, the oncogenic
Ras-activated MEK1 pathway inhibits conversion of the RUNX3-BRD2
complex into the RUNX3-HDAC4 complex, which switches off ARF and
p21 expression in lung adenocarcinoma (54) (Fig.
3).
4. RUNX3 in cancer metastasis
RUNX3 in EMT
EMT is a process whereby epithelial cells undergo a
shift in plasticity and acquire the ability to disseminate, invade
and cause metastasis. Established as a central process during the
early stages of development, it is now clear that EMT has
implications on cancer metastasis by triggering the loss of
cell-cell adhesion to facilitate tumor cell invasion and remodeling
of the extracellular matrix. While epithelial cells express high
levels of E-cadherin and are closely connected to each other by
tight junctions, mesenchymal cells express N-cadherin, fibronectin
and vimentin, have a spindle-shaped morphology and less tight
junctions (57).
Many of the pathways known to be involved in
metastasis also have close connections with EMT. Prominent among
these extracellular signals are the Wnt and TGF-β/BMP signaling
pathways, which contribute to the EMT activation network via
β-catenin-mediated activation of EMT inducing factors and SMAD
protein interactions, respectively (58). The Wnt signaling pathway
participates in EMT by inhibiting glycogen synthase kinase-3β
(GSK3β) to stabilize β-catenin, promoting a gene expression program
that favors EMT (59). During the
later stages of carcinogenesis, TGF-β signaling plays a paradoxical
role in the promotion of the invasion and metastasis of cancer
through its induction of EMT (57).
However, the convergence of signaling pathways is necessary for
EMT. While RUNX3 acts concurrently as a mediator of TGF-β signaling
and an antagonist of Wnt, RUNX3 was demonstrated to inhibit EMT,
which promotes metastasis, through the regulation of these
signaling pathways (17,46). Gastric epithelial cells undergo
spontaneous EMT in the absence of Runx3 and p53, which induces the
formation of a tumorigenic subpopulation in immortalized
Runx3−/−p53−/− gastric epithelial (GIF)
cells. Remarkably, the mesenchymal-like subpopulation also
possesses stem-like properties, as reflected in the enrichment of
the gastrointestinal stem cell marker Lgr5 and greater
sphere-initiating and colony-forming abilities. Contrary to their
resistance to TGF-β-mediated apoptosis, Runx3-null GIF lines are
unexpectedly sensitized to TGF-β-induced EMT, indicating a
rerouting of the TGF-β signal. In addition, a greatly increased
Wnt-responsiveness was observed in Runx3-null GIF cells, which acts
synergistically with TGF-β to induce Lgr5 expression. These
findings point to the possibility that RUNX3 acts as a tumor
suppressor by safeguarding gastric epithelial differentiation and
phenotypes; in its absence, gastric epithelial cells are prone to
spontaneous EMT and aberrant TGF-β and Wnt signaling, giving rise
to a tumorigenic stem cell-like subpopulation (17). Lee et al used Runx3-null
mouse to explore the role of RUNX3 in lung tumorigenesis. During
lung development, various cellular and molecular events occur such
as cell proliferation, cell death, differentiation and EMT.
Excessive EMT was observed in lungs in Runx3-null mice and PN18 in
Runx3 heterozygous mice. Pharmacologic inhibition of EMT resulted
in increased life spans of newborn mice were and lung hyperplasia
was partially rescued by downregulated EMT (60). In hepatocellular carcinoma, ectopic
RUNX3 expression had an anti-EMT effect in low-EMT HCC cell lines
characterized by increased E-cadherin expression and decreased
N-cadherin and vimentin expression (46).
Numerous studies have suggested that miRNAs
contribute to the invasion and metastasis of various types of human
cancers by regulating the EMT (61,62).
Vimentin is one of the targets that can be regulated by miRNAs.
Yamasaki et al showed that miR-138 targeted vimentin and
inhibited cell migration and invasion in renal cell carcinoma
(63). Cheng et al showed
that miR-30a could directly bind to the 3′UTR of vimentin and
inhibited its translation, then reduced the protein level of
vimentin in breast cancer (64). In
addition, overexpression of RUNX3 increased the expression of
miR-30a, which directly targeted the 3′UTR of vimentin and
decreased its protein level. The miR-30a inhibitor abrogated
RUNX3-mediated inhibition of cell invasion and downregulated
vimentin. In gastric cancer patients, levels of RUNX3 were
positively correlated with miR-30a and negatively associated with
vimentin. Thus, RUNX3 plays a significant role in regulating the
EMT by activating miR-30a (65).
RUNX3 in angiogenesis
Folkman, proposed the hypothesis that tumor growth
is dependent on the formation of new blood vessels (66). Tumor vessels eventually formed are
different when compared with normal vasculature: they are
disorganized with irregular structure and with altered interaction
between endothelial cells (67).
Once cancer cells generate their own blood supply, they are capable
of further invasion and have the capacity to metastasize.
Consequently, angiogenesis is essential for the development and
evolution of neoplastic disease, as both tumor growth and
metastasis require persistent new blood vessels and ongoing
angiogenesis is necessary for rapid expansion of a tumor mass
(68). Targeting of tumor-related
vessels has been regarded as an effective strategy to treat cancer.
The vast majority of anti-angiogenic agents being tested in the
clinic are based on the strategies that either interfere with
pro-angiogenic ligands or block the signaling of pro-angiogenic
receptor tyrosine kinases (69).
The most well-described angiogenic factor is VEGF.
The VEGF pathway holds the key in regulating angiogenesis,
vasopermeability as well as the proliferation and migration of
endothelial cells. Targeted therapies against this pathway have
been established in the clinic. In several types of cancer (colon
carcinoma, soft tissue sarcomas and gastric cancer), serum VEGF
level is a marker for disease stage and an indicator of metastasis.
VEGF level is significantly elevated in uveal melanoma patients
with metastatic disease compared to patients without metastases
(70). In our previous studies, we
injected nude mice with RUNX3 stably expressed prostate cancer
cells and our results showed reduced angiogenesis and metastasis
in vivo. The VEGF secretion was inhibited by RUNX3 in
vitro (24). Lee et al
used Runx3 knock-out mice to detect angiogenesis and liver
differentiation. The staining and real-time quantitative polymerase
chain reaction (RT-qPCR) displayed that VEGF were markedly
upregulated by the loss of RUNX3. Clarifying the mechanisms of
angiogenesis and liver differentiation may aid in the design of
efficient and safe anti-angiogenic and gene therapies for liver
disorders (71). Similarly, Runx3
knockout mice displayed vasculogenesis and angiogenesis by
increased CD31, VEGF and vWF (72).
Furthermore, Peng et al found three RUNX3 binding sites in
the VEGF promoter. Restoration of RUNX3 expression inhibited the
angiogenic potential of gastric cancer cells, and correlated with
downregulated VEGF expression via promoter repression in
vitro, and attenuation of tumorigenicity and abrogation of
metastasis in animal models (21).
In solid tumors >1 mm3, tumor cells
removed from blood vessels experience hypoxia (34,73).
Hypoxia stabilizes hypoxia-inducible factor (HIF)-1α, which
dimerizes with aryl hydrocarbon receptor nuclear translocator or
HIF-1β, a constitutively expressed binding partner of several
bHLH-PAS domain proteins, to increase the HIF-1-mediated gene
transcriptions involved in tumor angiogenesis (74). Hypoxic conditions also contribute to
RUNX3 silencing by the induction of histone methylation and the
deacetylation of RUNX3 promoter by G9a histone methyl transferase
and HDAC1. Thus, this decreased expression of RUNX3 under hypoxia
may potentiate tumor progression and metastasis in gastric cancer
by aggravating the angiogenic microenvironment (34). Lee et al further investigated
cross-talk between RUNX3 and HIF-1α under hypoxia. They
demonstrated that RUNX3 overexpression significantly inhibited
hypoxia-induced angiogenesis in vitro and in vivo.
RUNX3 directly interacted with the C-terminal activation domain of
HIF-1α and prolyl hydroxylase (PHD) 2 and enhanced the interaction
between HIF-1α and PHD2, which potentiated proline hydroxylation
and promoted the degradation of HIF-1α. RUNX3 appears to be a novel
suppressor of HIF-1α and of hypoxia-mediated angiogenesis in
gastric cancer cells (75).
miRNAs are key regulators of diverse cellular
processes, including angiogenesis. In has been reported that
downregulated miR-130a in patients with type 2 diabetes mellitus
(DM) results in endothelial progenitor cells (EPC) dysfunction, via
its target RUNX3 (76). However,
the relationship between miRNAs and RUNX3 in regulating tumor
angiogenesis has not been reported. Therefore, further
investigations need to focus on miRNAs that may possess potential
ability to regulate angiogenesis through target RUNX3 in human
cancers.
RUNX3 in programmed cell death
During the process of metastasis, any mistakes made
by a metastatic cell during cellular events may lead to cell death.
Therefore, the regulation of cell death is critical for cancer
cells to survive during metastasis. Programmed cell death is
defined as regulated cell death mediated by an intracellular
program. Stresses include the loss of cell-cell contacts, the
recognition and destruction of the cancer cells by the immune
system, and the lack of necessary growth factors, all of which may
trigger programmed cell death (77).
RUNX3 and apoptosis
Apoptosis is a type of programmed cell death that
characterized by cell membrane blebbing, cell shrinkage, nuclear
fragmentation, chromatin condensation, and chromosomal DNA
fragmentation (78,79). Apoptosis may block metastatic
dissemination by killing misplaced cells. Thus, apoptosis serves as
an important process for inhibiting metastasis. It has been
reported that lack of endogenous RUNX3 helps HCC to escape
apoptosis (80). The combination of
demethylation drugs 5-aza-CdR and TSA induced the translocation of
RUNX3 from the cytoplasm into the nucleus, induced
caspase-3-dependent apoptosis in leukaemia cell lines (81). Apoptosis is notably reduced in
Runx3−/− gastric epithelial cells. Further study showed
that in Runx3−/− mouse gastric epithelium, pro-apoptotic
gene Bim was downregulated, and apoptosis was reduced to the same
extent as that in Bim-/- gastric epithelium. RUNX3 is responsible
for transcriptional upregulation of Bim via binding to Bim promoter
in TGF-β-induced apoptosis (20).
Notably, both RUNX3 and FoxO3a/FKHRL1 were required for induction
of Bim expression. RUNX3 cooperates with FoxO3a/FKHRL1 to
participate in the induction of apoptosis by activating Bim and
play an important role in tumor suppression in gastric cancer
(44). Survivin is a member of the
inhibitor of apoptosis (IAP) family and has been shown to inhibit
cell apoptosis and promote cell proliferation. RUNX3 binds to
survivin promoter, leads to significantly inhibition of survivin
expression (82). Therefore, as a
transcriptional factor, RUNX3 is involved in regulating apoptosis
via its target gene.
p53 acts as a tumor suppressor by initiating cell
cycle arrest, apoptosis, and senescence in response to cellular
stress to maintain the integrity of the genome (83). In response to the anticancer drug
adriamycin, RUNX3 was induced to accumulate in the cell nucleus and
co-localized with p53. COOH-terminal portion of p53 is required for
the interaction with RUNX3. RUNX3 induced the phosphorylation of
p53 at Ser-15, thereby promoting p53-dependent apoptosis (84). Murine double minute 2 (MDM2) is a
well known E3 ubiquitin ligase that binds p53 at its
transactivation domain blocking p53-mediated transcriptional
regulation, while simultaneously promoting its polyubiquitination
and proteasome-dependent degradation (83). Intriguingly, RUNX3 directly binds
MDM2 through its runt-related DNA-binding domain. MDM2 blocks RUNX3
transcriptional activity by interacting with RUNX3 through an
acidic domain adjacent to the p53-binding domain of MDM2 and
ubiquitinates RUNX3 on key lysine residues to mediate nuclear
export and proteasomal degradation (52). Consequently, as a cofactor of p53,
RUNX3 and the ubiquitous p53 protein are both principal responders
of the p14 (ARF)-MDM2 cell surveillance pathway that prevents
pathologic consequences of abnormal oncogene activation.
In addition, metastatic cells must develop a
mechanism to evade cell death resulting from recognition and
destruction by cytotoxic lymphocytes such as natural killer (NK)
cells. Natural cytotoxicity receptor 1 (NCR1) is an NK
lymphocyte-activating receptor. The key cis-regulatory
element in the immediate vicinity upstream of the gene contains a
RUNX recognition motif that preferentially binds RUNX3. Higher
level of RUNX3 expression in NK cells may result in preferred
binding of RUNX3 at the NCR1 promoter and lead to NCR1 expression.
Thus, the role of RUNX3 in the control of an important
NK-activating receptor may promote cell death resulting from
recognition NK cells (85).
Moreover, RUNX3 impaired interleukin-15 (IL-15)-dependent
accumulation of mature NK cells. RUNX3 cooperates with ETS and
T-box transcription factors to drive the IL-15-mediated
transcription program during activation of these cells (86).
Resistance to chemotherapy is a significant barrier
for effective cancer treatment. miRNAs promote chemoresistance of
cancer cells, and suppress drug-induced apoptosis by target RUNX3
in human cancers. MiR-106a in highly expressed in gastric cancer
and promotes chemoresistance of gastric cancer cells, accelerates
ADR efflux, and suppresses drug-induced apoptosis. miR-106a may
modulate drug resistance of gastric cancer cells partially by
targeting RUNX3 (87). In
additional, Li et al identified miR-185 to be a contributor
to chemosensitivity in gastric cancer. Knockdown of endogenous
miR-185 prevented high-dose chemotherapy-induced apoptosis via a
direct target apoptosis repressor with caspase recruitment domain
is of miR-185. RUNX3 was involved in the activation of miR-185 at
the transcriptional level (88).
RUNX3 and autophagy
Autophagy is an evolutionarily conserved catabolic
process in which intracellular membrane structures package protein
complexes and organelles to degrade and renew these cytoplasmic
components (89). The role of
autophagy in cancer metastasis is complex, as reports have
indicated both pro-metastatic and antimetastatic roles of
autophagy. Stage-specificity may affect the cellular response to
autophagy during cancer metastasis (90). During the early stage of cancer
metastasis, autophagy may act as an inhibitor of metastasis by
restricting tumor necrosis and inflammatory cell infiltration and
by alleviating oncogene-induced senescence. These processes may
help to suppress the invasion and dissemination of cancer cells
from the primary site. During the advanced stages of metastasis,
autophagy tends to act as a promoter of metastasis by promoting ECM
detached metastatic cell survival and colonization in a distant
site and by inducing metastatic cells that fail to establish
contact with the ECM in the new environment to enter dormancy.
RUNX3 has been reported to be involved in regulating
autophagy mediated by microRNA in dysfunction of endothelial
progenitor cells (EPC). miR-130a is important for maintaining
normal autophagy levels and promoting the survival of EPC.
Overexpression of miR-130a decreased the number of autophagosomes,
cell death and Beclin 1 expression, but promoted Bcl2 expression;
these effects were mediated by RUNX3 (91). RUNX3 may be an autophagy regulator
linking apoptosis and the autophagy of EPC. However, the role of
RUNX3 in regulating autophagy in human cancers has not been
explored. Thus, further investigations needs to be carried out to
focus on the role of RUNX3 in autophagy in human cancers.
RUNX3 in migration and invasion
Tumor cell migration and invasion are essential
steps in the process of metastasis. The deregulation of cell
migration during cancer progression determines the capacity of
tumor cells to escape from the primary tumors. Then, tumor cells
invade adjacent tissues to finally form metastases. As cell
migration and invasion a prerequisite for metastasis, strategies
targeting cell migration and invasion are a major focus of current
efforts to develop cancer treatments.
Cancer cell migration and invasion are regulated by
numerous, interconnected molecular networks. In fact, there are
many intracellular molecules belonging to the Wnt, Notch, sonic
Hedgehog, NF-κB, Ras/Raf/MEK/MAPK, as well as the AKT/ERK signaling
pathways, which control every aspect of each of the stages of
cancer cell migration and invasion (4,5,10). In
contrast, extracellular molecules also contribute in a critical way
to the progress of cancer cell invasion. These molecules can be: i)
part of the ECM; ii) secreted in the ECM; iii) secreted but also
attached on the cell surface; and iv) cell membrane proteins such
as receptors. The role of RUNX3 in regulating these molecules that
related cancer cell migration and invasion may elucidate how RUNX3
function in cancer metastasis from another points.
OPN
Osteopontin (OPN) is a secreted multifunctional
glycophosphoprotein that is expressed in various tissues and plays
important roles in a wide range of biological processes, such as
inflammation, angiogenesis, bone remodeling, cell adhesion and
migration (92). OPN binds to
various receptors, including integrins and CD44, which potentially
allow it to stimulate different signaling pathways and ultimately
leading to tumor progression (93).
Studies have indicated that OPN induces the PI3K-Akt signaling
pathway through the αvβ3 integrin-mediated pathway and promotes
cell migration (94,95). The constitutive expression of OPN is
involved in the invasion, progression and metastasis in different
cancers (96,97). Elevated OPN and its receptor CD44v9,
are correlated with the degree of lymphatic vessel invasion and
lymph node metastasis in gastric cancer (98). Cheng et al identified one
RUNX3 binding site in the OPN promoter. The binding of RUNX3 to the
OPN promoter significantly decreased OPN promoter activity. The
knockdown of OPN or overexpression of RUNX3 inhibited cell
migration in gastric cancer cells; however, the coexpression of
RUNX3 and OPN reversed the RUNX3-reduced migration ability in these
cells. Obviously, RUNX3 is important to control cell migration
through OPN, which promotes migration in gastric cancer cells
(99).
MMPs
Invasion of basement membrane (BM) and extracellular
matrix (ECM) is an essential event in tumor invasion and
metastasis. ECM provides both structural support and extracellular
cues that regulate invasive tumor growth, and tumor-associated
changes in ECM contribute to cancer progression. MMPs are a family
of zinc-binding endopeptidases that participate in the ECM
degradation molecular machinery during tumor invasion (100,101). In order for tumor
neo-vascularization and cell invasion processes to occur,
degradation of the basement membrane as well as matrix remodeling
is essential. Amongst the many known MMPs, MMP-2 and MMP-9 degrade
gelatin as well as type IV collagen, the central component of the
basement membrane. These MMPs are secreted in an inactive form and
acquire their active form extracellularly (102,103). MMP-2 and MMP-9 are enriched in the
invadopodia where they contribute to ECM degradation in
vitro and in vivo (104). In our previous studies, we
provided evidence that RUNX3 can suppress MMP-2 activities in
prostate and breast cancers, but MMP-9 in renal cell carcinoma
(18,24,105).
We suppose that RUNX3 regulate cell invasion through different MMPs
depending on the cancer tissue.
MMPs activity is controlled by specific, endogenous
tissue inhibitors of metalloproteinases (TIMPs). TIMPs bind
non-covalently with 1:1 stoichiometry to the active form of MMPs to
inhibit their proteolytic activity (106). We found that the suppression of
RUNX3 on MMP-2 may be due to upregulation of TIMP-2 in prostate
cancer. However, in gastric cancer, the inhibition of MMP-9 induced
by RUNX3 overexpression attributed to TIMP-1. Two RUNX3 binding
sites were identified in the TIMP-1 promoter and direct interaction
of RUNX3 with the TIMP-1 promoter was confirmed in vitro and
in vivo.
5. Conclusion
In the present review, we highlighted important
findings that shape our current understanding of RUNX3 as a tumor
suppressor in the process of cancer metastasis. RUNX3 is deeply
involved in multiple cancer metastasis processes, such as
apoptosis, angiogenesis, EMT, migration and invasion in various
types of tissues. Identification of key tissue- and stage-specific
target genes of RUNX3 functioning as a tumor suppressor or an
oncogene is an important step forward, and combination of these
genes would improve the potential of usage of RUNX3 as a biomarker
for cancer diagnosis. Aberrant methylation is the leading reason
that results in RUNX3 inactivation in human cancers. Reversal of
DNA methylation by demethylating agents and inhibitors of
DNA-methyltransferases restores RUNX3 activation. Therefore,
development of drugs targeting DNA methylation of RUNX3 may have
vital clinical implications for the treatment or prevention of
cancers. An understanding of the upstream and downstream signaling
pathways of RUNX3 in the context of physiological and pathological
processes is necessary if we are to harness our growing knowledge
of RUNX3 for successful applications in cancer therapeutics.
Acknowledgments
The present review was supported by the National
Natural Science Foundation of China (no. 81302207).
References
|
1
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al-Mehdi AB, Tozawa K, Fisher AB, Shientag
L, Lee A and Muschel RJ: Intravascular origin of metastasis from
the proliferation of endothelium-attached tumor cells: A new model
for metastasis. Nat Med. 6:100–102. 2000. View Article : Google Scholar
|
|
4
|
Fidler IJ: The pathogenesis of cancer
metastasis: The 'seed and soil' hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fukushima-Nakase Y, Naoe Y, Taniuchi I,
Hosoi H, Sugimoto T and Okuda T: Shared and distinct roles mediated
through C-terminal subdomains of acute myeloid
leukemia/Runt-related transcription factor molecules in murine
development. Blood. 105:4298–4307. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wotton S, Terry A, Kilbey A, Jenkins A,
Herzyk P, Cameron E and Neil JC: Gene array analysis reveals a
common Runx transcriptional programme controlling cell adhesion and
survival. Oncogene. 27:5856–5866. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brady G and Farrell PJ: RUNX3-mediated
repression of RUNX1 in B cells. J Cell Physiol. 221:283–287. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukamachi H and Ito K: Growth regulation
of gastric epithelial cells by Runx3. Oncogene. 23:4330–4335. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Levanon D, Bettoun D, Harris-Cerruti C,
Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C,
Fliegauf M, et al: The Runx3 transcription factor regulates
development and survival of TrkC dorsal root ganglia neurons. EMBO
J. 21:3454–3463. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Woolf E, Brenner O, Goldenberg D, Levanon
D and Groner Y: Runx3 regulates dendritic epidermal T cell
development. Dev Biol. 303:703–714. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bagchi A and Mills AA: The quest for the
1p36 tumor suppressor. Cancer Res. 68:2551–2556. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Subramaniam MM, Chan JY, Yeoh KG, Quek T,
Ito K and Salto-Tellez M: Molecular pathology of RUNX3 in human
carcinogenesis. Biochim Biophys Acta. 1796:315–331. 2009.PubMed/NCBI
|
|
13
|
Oshimo Y, Oue N, Mitani Y, Nakayama H,
Kitadai Y, Yoshida K, Ito Y, Chayama K and Yasui W: Frequent loss
of RUNX3 expression by promoter hypermethylation in gastric
carcinoma. Pathobiology. 71:137–143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito
K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al: RUNX3 is
frequently inactivated by dual mechanisms of protein
mislocalization and promoter hypermethylation in breast cancer.
Cancer Res. 66:6512–6520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahlquist T, Lind GE, Costa VL, Meling GI,
Vatn M, Hoff GS, Rognum TO, Skotheim RI, Thiis-Evensen E and Lothe
RA: Gene methylation profiles of normal mucosa, and benign and
malignant colorectal tumors identify early onset markers. Mol
Cancer. 7:942008. View Article : Google Scholar
|
|
16
|
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada
K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al: RUNX3, a
novel tumor suppressor, is frequently inactivated in gastric cancer
by protein mislocalization. Cancer Res. 65:7743–7750.
2005.PubMed/NCBI
|
|
17
|
Voon DC, Wang H, Koo JK, Nguyen TA, Hor
YT, Chu YS, Ito K, Fukamachi H, Chan SL, Thiery JP, et al: Runx3
protects gastric epithelial cells against epithelial-mesenchymal
transition-induced cellular plasticity and tumorigenicity. Stem
Cells. 30:2088–2099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen F, Bai J, Li W, Mei P, Liu H, Li L,
Pan Z, Wu Y and Zheng J: RUNX3 suppresses migration, invasion and
angiogenesis of human renal cell carcinoma. PLoS One. 8:e562412013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sakakura C, Hasegawa K, Miyagawa K,
Nakashima S, Yoshikawa T, Kin S, Nakase Y, Yazumi S, Yamagishi H,
Okanoue T, et al: Possible involvement of RUNX3 silencing in the
peritoneal metastases of gastric cancers. Clin Cancer Res.
11:6479–6488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yano T, Ito K, Fukamachi H, Chi XZ, Wee
HJ, Inoue K, Ida H, Bouillet P, Strasser A, Bae SC, et al: The
RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells
undergoing transforming growth factor beta-induced apoptosis. Mol
Cell Biol. 26:4474–4488. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peng Z, Wei D, Wang L, Tang H, Zhang J, Le
X, Jia Z, Li Q and Xie K: RUNX3 inhibits the expression of vascular
endothelial growth factor and reduces the angiogenesis, growth, and
metastasis of human gastric cancer. Clin Cancer Res. 12:6386–6394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mei PJ, Bai J, Liu H, Li C, Wu YP, Yu ZQ
and Zheng JN: RUNX3 expression is lost in glioma and its
restoration causes drastic suppression of tumor invasion and
migration. J Cancer Res Clin Oncol. 137:1823–1830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Z, Chen G, Cheng Y, Martinka M and
Li G: Prognostic significance of RUNX3 expression in human
melanoma. Cancer. 117:2719–2727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen F, Wang M, Bai J, Liu Q, Xi Y, Li W
and Zheng J: Role of RUNX3 in suppressing metastasis and
angiogenesis of human prostate cancer. PLoS One. 9:e869172014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen W, Salto-Tellez M, Palanisamy N,
Ganesan K, Hou Q, Tan LK, Sii LH, Ito K, Tan B, Wu J, et al:
Targets of genome copy number reduction in primary breast cancers
identified by integrative genomics. Genes Chromosomes Cancer.
46:288–301. 2007. View Article : Google Scholar
|
|
26
|
Finak G, Bertos N, Pepin F, Sadekova S,
Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu
A, et al: Stromal gene expression predicts clinical outcome in
breast cancer. Nat Med. 14:518–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sakakura C, Miyagawa K, Fukuda KI,
Nakashima S, Yoshikawa T, Kin S, Nakase Y, Ida H, Yazumi S,
Yamagishi H, et al: Frequent silencing of RUNX3 in esophageal
squamous cell carcinomas is associated with radioresistance and
poor prognosis. Oncogene. 26:5927–5938. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chuang LS and Ito Y: RUNX3 is
multifunctional in carcinogenesis of multiple solid tumors.
Oncogene. 29:2605–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu YY, Chen C, Kong FF and Zhang W:
Clinicopathological significance and potential drug target of RUNX3
in breast cancer. Drug Des Devel Ther. 8:2423–2430. 2014.PubMed/NCBI
|
|
30
|
Wang D, Cui W, Wu X, Qu Y, Wang N, Shi B
and Hou P: RUNX3 site-specific hypermethylation predicts papillary
thyroid cancer recurrence. Am J Cancer Res. 4:725–737.
2014.PubMed/NCBI
|
|
31
|
Katayama Y, Takahashi M and Kuwayama H:
Helicobacter pylori causes runx3 gene methylation and its loss of
expression in gastric epithelial cells, which is mediated by nitric
oxide produced by macrophages. Biochem Biophys Res Commun.
388:496–500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng AS, Culhane AC, Chan MW, Venkataramu
CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J,
et al: Epithelial progeny of estrogen-exposed breast progenitor
cells display a cancer-like methylome. Cancer Res. 68:1786–1796.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fujii S, Ito K, Ito Y and Ochiai A:
Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by
increasing histone H3 methylation. J Biol Chem. 283:17324–17332.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee SH, Kim J, Kim WH and Lee YM: Hypoxic
silencing of tumor suppressor RUNX3 by histone modification in
gastric cancer cells. Oncogene. 28:184–194. 2009. View Article : Google Scholar
|
|
35
|
Varambally S, Cao Q, Mani RS, Shankar S,
Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al:
Genomic loss of microRNA-101 leads to overexpression of histone
methyl-transferase EZH2 in cancer. Science. 322:1695–1699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et
al Singapore Gastric Cancer Consortium: MicroRNA-130b regulates the
tumour suppressor RUNX3 in gastric cancer. Eur J Cancer.
46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kitago M, Martinez SR, Nakamura T, Sim MS
and Hoon DS: Regulation of RUNX3 tumor suppressor gene expression
in cutaneous melanoma. Clin Cancer Res. 15:2988–2994. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH,
Jang JW, Li YH, Chi XZ, Lee KS, Wee H, et al: Src kinase
phosphorylates RUNX3 at tyrosine residues and localizes the protein
in the cytoplasm. J Biol Chem. 285:10122–10129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim HR, Oh BC, Choi JK and Bae SC: Pim-1
kinase phosphorylates and stabilizes RUNX3 and alters its
subcellular localization. J Cell Biochem. 105:1048–1058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim JH, Choi JK, Cinghu S, Jang JW, Lee
YS, Li YH, Goh YM, Chi XZ, Lee KS, Wee H, et al: Jab1/CSN5 induces
the cytoplasmic localization and degradation of RUNX3. J Cell
Biochem. 107:557–565. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sheen YY, Kim MJ, Park SA, Park SY and Nam
JS: Targeting the transforming growth factor-β signaling in cancer
therapy. Biomol Ther. 21:323–331. 2013. View Article : Google Scholar
|
|
42
|
Ito Y and Miyazono K: RUNX transcription
factors as key targets of TGF-beta superfamily signaling. Curr Opin
Genet Dev. 13:43–47. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chi XZ, Yang JO, Lee KY, Ito K, Sakakura
C, Li QL, Kim HR, Cha EJ, Lee YH, Kaneda A, et al: RUNX3 suppresses
gastric epithelial cell growth by inducing p21WAF1/Cip1
expression in cooperation with transforming growth factor
β-activated SMAD. Mol Cell Biol. 25:8097–8107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamamura Y, Lee WL, Inoue K, Ida H and Ito
Y: RUNX3 cooperates with FoxO3a to induce apoptosis in gastric
cancer cells. J Biol Chem. 281:5267–5276. 2006. View Article : Google Scholar
|
|
45
|
Yao H, Ashihara E and Maekawa T: Targeting
the Wnt/β-catenin signaling pathway in human cancers. Expert Opin
Ther Targets. 15:873–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tanaka S, Shiraha H, Nakanishi Y, Nishina
S, Matsubara M, Horiguchi S, Takaoka N, Iwamuro M, Kataoka J,
Kuwaki K, et al: Runt-related transcription factor 3 reverses
epithelial-mesenchymal transition in hepatocellular carcinoma. Int
J Cancer. 131:2537–2546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ito K, Lim AC, Salto-Tellez M, Motoda L,
Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H, et al: RUNX3
attenuates beta-catenin/T cell factors in intestinal tumorigenesis.
Cancer Cell. 14:226–237. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ju X, Ishikawa TO, Naka K, Ito K, Ito Y
and Oshima M: Context-dependent activation of Wnt signaling by
tumor suppressor RUNX3 in gastric cancer cells. Cancer Sci.
105:418–424. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Palmero I, Pantoja C and Serrano M:
p19ARF links the tumour suppressor p53 to Ras. Nature.
395:125–126. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lee KS, Lee YS, Lee JM, Ito K, Cinghu S,
Kim JH, Jang JW, Li YH, Goh YM, Chi XZ, et al: Runx3 is required
for the differentiation of lung epithelial cells and suppression of
lung cancer. Oncogene. 29:3349–3361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kruse JP and Gu W: Modes of p53
regulation. Cell. 137:609–622. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chi XZ, Kim J, Lee YH, Lee JW, Lee KS, Wee
H, Kim WJ, Park WY, Oh BC, Stein GS, et al: Runt-related
transcription factor RUNX3 is a target of MDM2-mediated
ubiquitination. Cancer Res. 69:8111–8119. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Efeyan A and Serrano M: p53: Guardian of
the genome and policeman of the oncogenes. Cell Cycle. 6:1006–1010.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH,
Li YH, Kim MK, Kim DM, Choi BS, Kim EG, et al: Runx3 inactivation
is a crucial early event in the development of lung adenocarcinoma.
Cancer Cell. 24:603–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hnilicová J, Hozeifi S, Stejskalová E,
Dušková E, Poser I, Humpolíčková J, Hof M and Staněk D: The
C-terminal domain of Brd2 is important for chromatin interaction
and regulation of transcription and alternative splicing. Mol Biol
Cell. 24:3557–3568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK,
Kim WJ, Lee KY and Bae SC: Transforming growth factor-beta
stimulates p300-dependent RUNX3 acetylation, which inhibits
ubiquitination-mediated degradation. J Biol Chem. 279:29409–29417.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Padua D and Massagué J: Roles of TGFbeta
in metastasis. Cell Res. 19:89–102. 2009. View Article : Google Scholar
|
|
59
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lee JM, Shin JO, Cho KW, Hosoya A, Cho SW,
Lee YS, Ryoo HM, Bae SC and Jung HS: Runx3 is a crucial regulator
of alveolar differentiation and lung tumorigenesis in mice.
Differentiation. 81:261–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chang CJ, Chao CH, Xia W, Yang JY, Xiong
Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al: p53 regulates
epithelial-mesenchymal transition and stem cell properties through
modulating miRNAs. Nat Cell Biol. 13:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kumarswamy R, Mudduluru G, Ceppi P,
Muppala S, Kozlowski M, Niklinski J, Papotti M and Allgayer H:
MicroRNA-30a inhibits epithelial-to-mesenchymal transition by
targeting Snai1 and is downregulated in non-small cell lung cancer.
Int J Cancer. 130:2044–2053. 2012. View Article : Google Scholar
|
|
63
|
Yamasaki T, Seki N, Yamada Y, Yoshino H,
Hidaka H, Chiyomaru T, Nohata N, Kinoshita T, Nakagawa M and
Enokida H: Tumor suppressive microRNA-138 contributes to cell
migration and invasion through its targeting of vimentin in renal
cell carcinoma. Int J Oncol. 41:805–817. 2012.PubMed/NCBI
|
|
64
|
Cheng CW, Wang HW, Chang CW, Chu HW, Chen
CY, Yu JC, Chao JI, Liu HF, Ding SL and Shen CY: MicroRNA-30a
inhibits cell migration and invasion by downregulating vimentin
expression and is a potential prognostic marker in breast cancer.
Breast Cancer Res Treat. 134:1081–1093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu Z, Chen L, Zhang X, Xu X, Xing H,
Zhang Y, Li W, Yu H, Zeng J and Jia J: RUNX3 regulates vimentin
expression via miR-30a during epithelial-mesenchymal transition in
gastric cancer cells. J Cell Mol Med. 18:610–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Folkman J: Tumor angiogenesis: Therapeutic
implications. N Engl J Med. 285:1182–1186. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Nishida N, Yano H, Nishida T, Kamura T and
Kojiro M: Angiogenesis in cancer. Vasc Health Risk Manag.
2:213–219. 2006. View Article : Google Scholar
|
|
68
|
Hanahan D and Folkman J: Patterns and
emerging mechanisms of the angiogenic switch during tumorigenesis.
Cell. 86:353–364. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Jain RK: Antiangiogenesis strategies
revisited: From starving tumors to alleviating hypoxia. Cancer
Cell. 26:605–622. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang M, Kuang X, Pan Y, Tan M, Lu B, Lu J,
Cheng Q and Li J: Clinicopathological characteristics of vascular
endothelial growth factor expression in uveal melanoma: A
meta-analysis. Mol Clin Oncol. 2:363–368. 2014.PubMed/NCBI
|
|
71
|
Lee JM, Lee DJ, Bae SC and Jung HS:
Abnormal liver differentiation and excessive angiogenesis in mice
lacking Runx3. Histochem Cell Biol. 139:751–758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lee JM, Kwon HJ, Lai WF and Jung HS:
Requirement of Runx3 in pulmonary vasculogenesis. Cell Tissue Res.
356:445–449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Semenza GL: Angiogenesis in ischemic and
neoplastic disorders. Annu Rev Med. 54:17–28. 2003. View Article : Google Scholar
|
|
74
|
Harris AL: Hypoxia - a key regulatory
factor in tumour growth. Nat Rev Cancer. 2:38–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lee SH, Bae SC, Kim KW and Lee YM: RUNX3
inhibits hypoxia-inducible factor-1α protein stability by
interacting with prolyl hydroxylases in gastric cancer cells.
Oncogene. 33:1458–1467. 2014. View Article : Google Scholar
|
|
76
|
Meng S, Cao J, Zhang X, Fan Y, Fang L,
Wang C, Lv Z, Fu D and Li Y: Downregulation of microRNA-130a
contributes to endothelial progenitor cell dysfunction in diabetic
patients via its target Runx3. PLoS One. 8:e686112013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Luzzi KJ, MacDonald IC, Schmidt EE,
Kerkvliet N, Morris VL, Chambers AF and Groom AC: Multistep nature
of metastatic inefficiency: Dormancy of solitary cells after
successful extravasation and limited survival of early
micrometastases. Am J Pathol. 153:865–873. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Nagasaka A, Kawane K, Yoshida H and Nagata
S: Apaf-1-independent programmed cell death in mouse development.
Cell Death Differ. 17:931–941. 2010. View Article : Google Scholar
|
|
79
|
Burgess DJ: Apoptosis: Refined and lethal.
Nat Rev Cancer. 13:792013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nakanishi Y, Shiraha H, Nishina S, Tanaka
S, Matsubara M, Horiguchi S, Iwamuro M, Takaoka N, Uemura M, Kuwaki
K, et al: Loss of runt-related transcription factor 3 expression
leads hepatocellular carcinoma cells to escape apoptosis. BMC
Cancer. 11:32011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhai FX, Liu XF, Fan RF, Long ZJ, Fang ZG,
Lu Y, Zheng YJ and Lin DJ: RUNX3 is involved in caspase-3-dependent
apoptosis induced by a combination of 5-aza-CdR and TSA in
leukaemia cell lines. J Cancer Res Clin Oncol. 138:439–449. 2012.
View Article : Google Scholar
|
|
82
|
Liu Z, Zhang X, Xu X, Chen L, Li W, Yu H,
Sun Y, Zeng J and Jia J: RUNX3 inhibits survivin expression and
induces cell apoptosis in gastric cancer. Eur J Cell Biol.
93:118–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Meng X, Franklin DA, Dong J and Zhang Y:
MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res.
74:7161–7167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yamada C, Ozaki T, Ando K, Suenaga Y,
Inoue K, Ito Y, Okoshi R, Kageyama H, Kimura H, Miyazaki M, et al:
RUNX3 modulates DNA damage-mediated phosphorylation of tumor
suppressor p53 at Ser-15 and acts as a co-activator for p53. J Biol
Chem. 285:16693–16703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lai CB and Mager DL: Role of runt-related
transcription factor 3 (RUNX3) in transcription regulation of
natural cytotoxicity receptor 1 (NCR1/NKp46), an activating natural
killer (NK) cell receptor. J Biol Chem. 287:7324–7334. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Levanon D, Negreanu V, Lotem J, Bone KR,
Brenner O, Leshkowitz D and Groner Y: Transcription factor Runx3
regulates interleukin-15-dependent natural killer cell activation.
Mol Cell Biol. 34:1158–1169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang Y, Lu Q and Cai X: MicroRNA-106a
induces multidrug resistance in gastric cancer by targeting RUNX3.
FEBS Lett. 587:3069–3075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li Q, Wang JX, He YQ, Feng C, Zhang XJ,
Sheng JQ and Li PF: MicroRNA-185 regulates chemotherapeutic
sensitivity in gastric cancer by targeting apoptosis repressor with
caspase recruitment domain. Cell Death Dis. 5:e11972014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Boya P, Reggiori F and Codogno P: Emerging
regulation and functions of autophagy. Nat Cell Biol. 15:713–720.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kenific CM, Thorburn A and Debnath J:
Autophagy and metastasis: Another double-edged sword. Curr Opin
Cell Biol. 22:241–245. 2010. View Article : Google Scholar :
|
|
91
|
Xu Q, Meng S, Liu B, Li MQ, Li Y, Fang L
and Li YG: MicroRNA-130a regulates autophagy of endothelial
progenitor cells through Runx3. Clin Exp Pharmacol Physiol.
41:351–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Denhardt DT, Lopez CA, Rollo EE, Hwang SM,
An XR and Walther SE: Osteopontin-induced modifications of cellular
functions. Ann NY Acad Sci. 760:127–142. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Anborgh PH, Mutrie JC, Tuck AB and
Chambers AF: Role of the metastasis-promoting protein osteopontin
in the tumour microenvironment. J Cell Mol Med. 14:2037–2044. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Fong YC, Liu SC, Huang CY, Li TM, Hsu SF,
Kao ST, Tsai FJ, Chen WC, Chen CY and Tang CH: Osteopontin
increases lung cancer cells migration via activation of the
alphavbeta3 integrin/FAK/Akt and NF-kappaB-dependent pathway. Lung
Cancer. 64:263–270. 2009. View Article : Google Scholar
|
|
95
|
Wang Y, Yan W, Lu X, Qian C, Zhang J, Li
P, Shi L, Zhao P, Fu Z, Pu P, et al: Overexpression of osteopontin
induces angiogenesis of endothelial progenitor cells via the
avβ3/PI3K/AKT/eNOS/NO signaling pathway in glioma cells. Eur J Cell
Biol. 90:642–648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Singhal H, Bautista DS, Tonkin KS,
O'Malley FP, Tuck AB, Chambers AF and Harris JF: Elevated plasma
osteopontin in metastatic breast cancer associated with increased
tumor burden and decreased survival. Clin Cancer Res. 3:605–611.
1997.PubMed/NCBI
|
|
97
|
Schneider S, Yochim J, Brabender J, Uchida
K, Danenberg KD, Metzger R, Schneider PM, Salonga D, Hölscher AH
and Danenberg PV: Osteopontin but not osteonectin messenger RNA
expression is a prognostic marker in curatively resected non-small
cell lung cancer. Clin Cancer Res. 10:1588–1596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ue T, Yokozaki H, Kitadai Y, Yamamoto S,
Yasui W, Ishikawa T and Tahara E: Co-expression of osteopontin and
CD44v9 in gastric cancer. Int J Cancer. 79:127–132. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cheng HC, Liu YP, Shan YS, Huang CY, Lin
FC, Lin LC, Lee L, Tsai CH, Hsiao M and Lu PJ: Loss of RUNX3
increases osteopontin expression and promotes cell migration in
gastric cancer. Carcinogenesis. 34:2452–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Min KW, Kim DH, Do SI, Kim K, Lee HJ, Chae
SW, Sohn JH, Pyo JS, Oh YH, Kim WS, et al: Expression patterns of
stromal MMP-2 and tumoural MMP-2 and -9 are significant prognostic
factors in invasive ductal carcinoma of the breast. APMIS.
122:1196–1206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Stellas D and Patsavoudi E: Inhibiting
matrix metalloproteinases, an old story with new potentials for
cancer treatment. Anticancer Agents Med Chem. 12:707–717. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Galis ZS and Khatri JJ: Matrix
metalloproteinases in vascular remodeling and atherogenesis: The
good, the bad, and the ugly. Circ Res. 90:251–262. 2002.PubMed/NCBI
|
|
103
|
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng
GQ, Wan XX, He QY, Li JH, Qu JQ, et al: Activation of EGFR promotes
squamous carcinoma SCC10A cell migration and invasion via inducing
EMT-like phenotype change and MMP-9-mediated degradation of
E-cadherin. J Cell Biochem. 112:2508–2517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Murphy DA and Courtneidge SA: The 'ins'
and 'outs' of podosomes and invadopodia: Characteristics, formation
and function. Nat Rev Mol Cell Biol. 12:413–426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bai ZK, Guo B, Tian XC, Li DD, Wang ST,
Cao H, Wang QY and Yue ZP: Expression and regulation of Runx3 in
mouse uterus during the peri-implantation period. J Mol Histol.
44:519–526. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jinga DC, Blidaru A, Condrea I, Ardeleanu
C, Dragomir C, Szegli G, Stefanescu M and Matache C: MMP-9 and
MMP-2 gelatinases and TIMP-1 and TIMP-2 inhibitors in breast
cancer: Correlations with prognostic factors. J Cell Mol Med.
10:499–510. 2006. View Article : Google Scholar : PubMed/NCBI
|