Currently, the development of powerful and smart
high-throughput tools in translational medicine along with
nanotechnological applications enabling information-based medicine
have revived the hope for the broader application of
pharmacogenomics (PGx) for most, if not all, individual patients
(see Table I for term definition)
(1–10). By extending such a notion from PGx
to routine healthcare and drug prescription (Rx), it means that
this transition is better served by achieving pharmacotyping (PTx)
in drug delivery (6,8,9).
Further support is gained through expanding the clinical
application of innovative approaches and technicalities happening
in the era of: i) genomic profiling; ii) biomedical imaging; iii)
synthetic, biology-based, cell engineering to advance drug delivery
entities; iv) population-based modeling as predictive tools of
pharmacokinetic (PK) parameters of drugs (absorption, distribution,
metabolism and excretion; ADME) and v) innovative approaches and
tools on patient electronic data management, clinical support and
routine healthcare. These scientific breakthroughs empower the
speed and the productivity in developing innovative molecularly
targeted drugs and nanotheranostics with improved clinical safety
and efficacy profiles (11–20). The latter coincides with the
movement from an Rx process mainly based on the physician's own
experience into a more highly digitized and integrated workflow
platform to aid the administration of individualized drug dosage
schemes for personalized medicine.
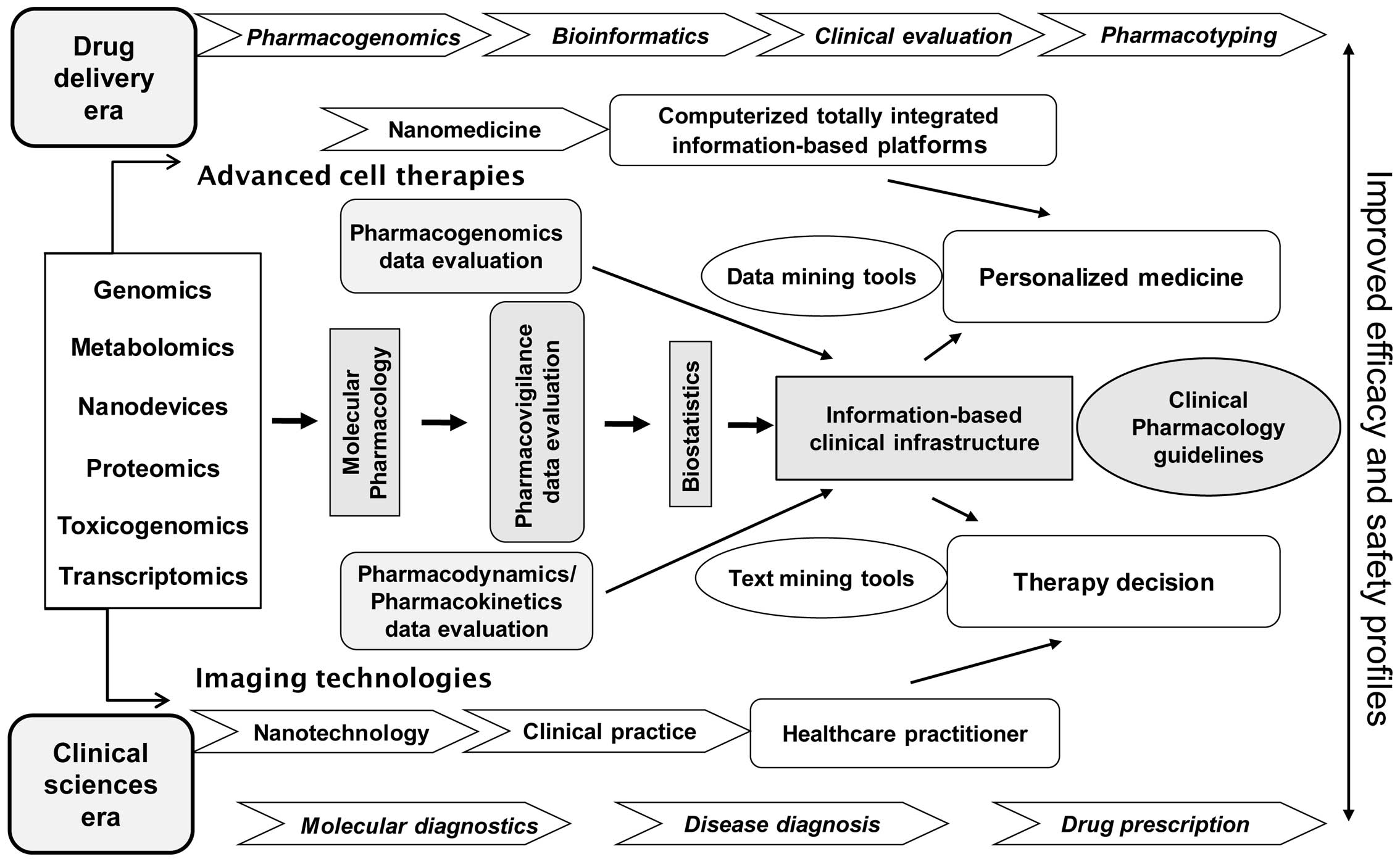
As a matter of fact, the interdisciplinary
infrastructure and methodologies needed to broadly enable PTx in
the clinical setting is depicted in Fig. 1 (for a more extensive overview see
ref. 7). In particular, such a
diagrammatic presentation exemplifies the environment to maximize
outcomes and achieve improved efficacy and safety profiles of
clinical practical utility for personalized medicine decisions.
That means that the proper translational capacity for all the
involved disciplines and technologies has to be clinically
validated in terms of precise clinical diagnosis and drug
prescription dosage scheme selection by healthcare practitioners.
The latter refers to advancements such as: i) the ʻomicsʼ-related
for molecular diagnostics and PGx; ii) the
computerized/information-based platforms in data and text mining
for bioinformatics and biostatistics; and iii) the
PK/pharmacodynamic (PK/PD) guidelines for pharmacovigilance,
molecular and clinical pharmacology. By enabling this direction,
the establishment of PTx in drug prescription is served. Last, but
not least, the integration of nanotechnology, imaging technologies
and advanced cell therapies impacting nanomedicine applications are
also better facilitated and implemented in the clinical setting, as
shown in Fig. 1.
Following such a roadmap in Rx it must also be
considered that the inter-individual response heterogeneity
(patient phenotype) noted in clinical pharmacology and the
incidence of adverse drug reactions (ADRs) are related to various
factors and parameters affecting the risk/benefit therapeutic ratio
that refer to: i) the molecular pathophysiology of diseases as well
as their severity; ii) the co-administration of medicines that
might result in drug interactions; iii) any existed co-morbidities;
iv) the functional status of vital organs, i.e. liver and kidneys
contributing to PK behavior of drugs; v) the age of the individual
patient; vi) the person's lifestyle, i.e. smoking, caffeine intake;
vii) the patient's compliance (adherence) to the physician's
guidance to prescribed therapy; and also viii) the personal genetic
make-up, or alternatively, the existing human genome variability in
genes involved in PK/PD processes ensuring drug effects in the
body; i.e. the genetic polymorphism of various gene alleles
referring to drug metabolizing enzymes, transporters and receptors
(21–32). Simultaneously, the successful
address of such PK/PD issues represents the gold-standard target
upon developing drug nanodelivery vehicles.
It has been estimated that drug response
heterogeneity for marketed drugs significantly contributes to the
increase of healthcare costs, the rate of in-patient
hospitalization and mortality index (33–37).
Although there is a widespread interest in personalized medicine,
the broad application of PGx testing implies that the validation of
the clinical improvement outcome must be clearly demonstrated in a
cost-effective manner. Undoubtedly, however, only a limited number
of studies have addressed cost-effectiveness issues of PGx
applications in the clinical setting (38–41).
To this end, recent studies assessing the pharmacoeconomic benefits
of thiopurine methyltransferase (TPMT) PGx testing have shown a
favorable cost-effectiveness ratio, by analyzing either the outcome
achieved in children suffering from acute lymphoblastic leukemia
(ALL), or relevant pharmacovigilance data referring to the
emergence of ADRs (39,41). On the contrary, the lack of
standardized PGx economic models has undoubtedly emerged in the
case of CYP genetic variation, by trying to evaluate the economic
burden vs. the clinical benefits of PGx testing upon prescribing
antipsychotics in routine healthcare (38). In addition, the complexity in
undertaking a trial-based evaluation of the cost-effectiveness of
PGx tests such as the application of TPMT for prescribing
azathioprine in patients suffering autoimmune diseases has recently
been shown (40). Moreover, the
economic evaluation of PGx testing is now considered a main barrier
hindering the implementation of clinical practice with PGx
knowledge. Importantly, before moving toward the routine
application of PGx concepts in the healthcare system and
establishing PTx, the clear demonstration of the economic benefits
gained must be addressed to accompany the already assured clinical
benefits of any PGx testing. Such a direction will allow
cost-effectiveness analysis to document the relative cost/benefit
ratios of PGx interventions compared to current clinical practices
and create the framework for healthcare providers to make
reimbursement decisions. Importantly, the limited evidence
accumulated thus far in analyzing the economic value of
personalized medicine tests also restricts any proper informed
decision-making and assessment of genomic priorities (42). Complementary, in order to advance
practical clinical utility of personalized medicine decisions, the
cost-effectiveness in addition to the clinical effectiveness of PGx
tests for decision makers should simultaneously be undertaken in a
robust and timely manner (43).
The PK and PD behavior of drugs is known to be
affected by either drug interactions or genetic polymorphism of
genes involved in drug actions, thus leading to altered plasma
therapeutic concentrations (inefficiency or toxicity) and also to
modulated receptor affinity (sensitivity) (8,9). The
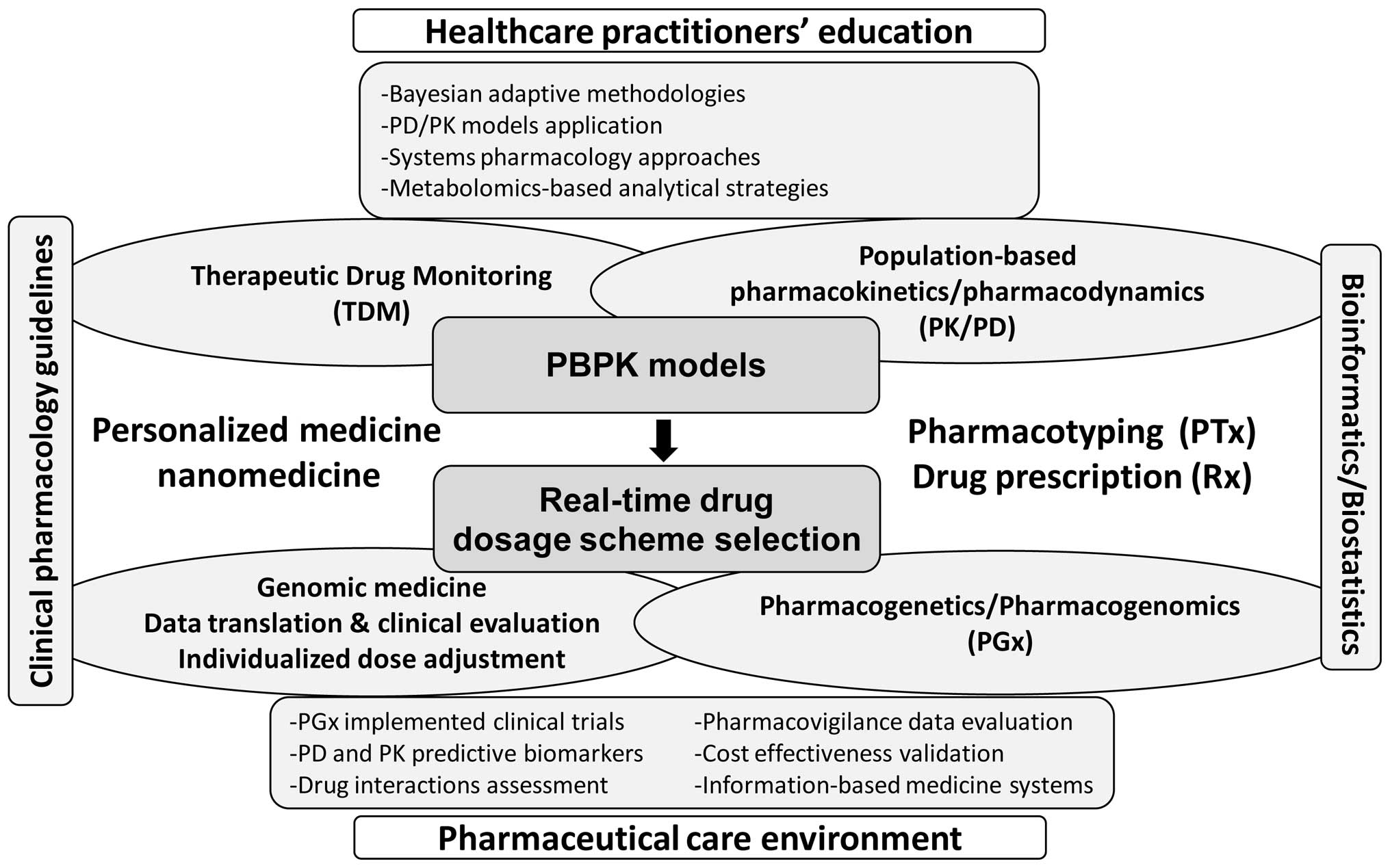
future application of physiologically based pharmacokinetic (PBPK)
models to shape the practical utility of personalized/stratified
medicine and ensure PTx is shown in Fig. 2. Indeed, the accuracy and
predictability potential of the applied methodologies and model
approaches for assessing the drug plasma concentration-time profile
and clearance in humans has been recently verified (44,45).
This, however, implies the development of algorithms to improve the
statistical power and robustness upon mathematical integration and
PBPK modeling of drug effects by facilitating the clinical
exploitation and inference of genome-wide association studies and
PGx tests (46–48). In contrast, PGx knowledge for drugs
is now considered an integral part of therapeutics and drug
development, since a number of predictive PGx biomarkers to assess
the safety and clinical efficacy profiles of individual marketed
drugs have been validated by drug regulatory agencies (e.g. the FDA
and EMA) (32,49). To this end, the development of PBPK
models, implemented together with system pharmacology approaches
(assessing predictive PGx biomarkers), represents a promising
platform where in real-time the assessment of both patient- and
drug-related factors can be inter-correlated for individual patient
populations. Alternatively, the latter means the elaboration of a
multidisciplinary environment in order to assess both drug
interactions and PGx data to be effectively incorporated to guide
Rx, thus achieving PTx.
By taking into consideration the deficit in
scientific knowledge, the approach to accelerate the development of
more powerful innovative anticancer therapeutics is by discovering
methods by which to improve early clinical anticancer drug
evaluation through structured and rational trial designs that
incorporate predictive PK, PD, PGx and intermediate end-point
biomarkers (51). In addition, it
is also crucial to discover ways to enrich methodologies
facilitating the integration of PGx into know-how strategy of the
mainstay drug development pipeline (55). The latter implies that the suitable
organization and application of population-based PK/PD ex
vivo pharmacology modeling is also vital to support the
integration of genomic knowledge in routine clinical practice and
Rx by hastening the movement toward personalized medicine (56). Moreover, it is evident from the
previously gained in vivo pharmacology experience in drug
development that the efficient clinical translation of genomic
knowledge coincides with successfully addressing the issues related
to the adjustment of clinical pharmacology guidelines toward
personalized medicine concepts. Alternatively, the implementation
of clinical pharmacology guidelines with PGx knowledge through the
development of advanced PBPK models represents one of the most
crucial elements for enabling the practical utility of personalized
medicine and applying PTx concepts for drug delivery worldwide
(8,9). This direction also necessitates
overcoming barriers hindering efficient clinical translation and
implementation of PGx knowledge from bench to bedside. To this end,
it is notable that PGx guidelines for dose recommendation schemes
of specific pharmacological drug classes have already been
initiated and proposed (57–62).
To foster the applicability and practical utility of these
methodologies through the use of PBPK/PGx models, the assessment of
any ethical, social and cost-benefit impacts should also be
addressed before the transfer of techniques used in genomic-related
research laboratories to diagnostic laboratories (8,9,32,54).
Subsequently, the modeling of the tumor microenvironment will be
outlined to further stress the usefulness of the application of
PBPKs in the era of nanomedicine and personalized medicine.
One of the most challenging issues in pharmacology
and therapeutics relates to the capacity of predicting the PK
profile of any administered drug to individual patients and thus to
estimate the cellular concentrations reached at the site of action
referring either to the diseased organ, or tissue in order to be
within the therapeutic ratio range. This prediction of the behavior
of drugs plays fundamental and crucial roles in the development of
stratified and personalized treatment decisions. Indeed, the latter
offers opportunities for pharmacologists and clinicians to generate
models and test different scenarios of the effects of drugs on the
lesion (organ or tissue) by taking into consideration all the
variants, including different genomic backgrounds that eventually
might contribute to variation in the pharmacological response and
clinical phenotype. Particularly for cancer therapy, tumor
microenvironment heterogeneity represents a main obstacle hindering
the development of innovative anticancer therapeutics and also
limiting the clinical outcomes achieved thus far (19,63,64).
Particularly for solid tumors, the role of the lesion
microenvironment, blood flow, vessel leakage, oxygen supply, size,
specific gene expression and growth rate have been found to play an
important role in therapeutic outcomes since they are related with
drug delivery (i.e. PK) and consequently with pharmacological
action (i.e. PD) (65–68). Despite the fact that chemotherapy
dosage schemes in oncology always take into consideration various
patient characteristics (i.e. body weight), the applied therapeutic
protocols and scaling of the dose are based on nomograms and the
optimum fitting of the patient to them instead of considering
additional tumor characteristics. Although limited attempts to
apply PBPK models for nanoformulations have to date been made (see
Table II), the dynamics of such a
methodology and the way by which specific factors crucial for the
PK profile of nanomedicines could be quantified in models have
recently been presented (69,70).
Alternatively, the recent advancements in translation medicine,
nanomedicine and personal genome sequencing permit the
implementation of cancer genome testing into the clinical setting,
a fact with major opportunities for personalized therapeutic
decisions in oncology; i.e. precision cancer medicine to overcome
therapy obstacles and ameliorate the negative outcome that
oncologists face in their practice (71–74).
Moving forward, the genuine application of PBPK modeling in such an
innovative cancer therapy era could help to elucidate tumor
heterogeneity properties and predict drug behavior through
simulation of individual patient tumor characteristics and drug
properties. A better understanding of the microenvironment
complexity, phenotypic diversity and genetic heterogeneity of
tumors through PBPK modeling may lead to the development of early
approaches in the drug discovery process that will minimize the
attrition rate and increase the productivity outcome for novel
anticancer therapeutics. An example of personalized cancer therapy
is application of tamoxifen for estrogen receptor-positive breast
cancer patients (75,76). Tamoxifen is a pro-drug that
undergoes extensive hepatic and gut wall metabolism thus forming
metabolites with different response effects (77). Moreover, CYP2D6 function and genetic
polymorphism has been proven to be the rate-limiting step upon the
formation of its main active metabolite
4-hydroxy-N-desmethyltamoxifen (endoxifen). As a consequence,
CYP2D6 PGx knowledge allows the prediction of the individual
patient pharmacological response and hence the clinical outcome of
tamoxifen-treated breast cancer women (75,77,78).
In parallel, the clinical impact of drug-drug interactions with
concomitant use of selective serotonin reuptake inhibitors such as
paroxetine, a strong CYP2D6 inhibitor, has also been described
(79,80). Such clinical PGx knowledge has been
applied in tamoxifen-based PBPK approaches in order to provide a
more precise clinical understanding of any required in
vitro-in vivo extrapolation regarding a drug's PK
profile evaluation (81).
Consequently, tamoxifen-based PBPK modeling provides the
opportunity for infrastructure creation for assessing whether
potential co-administration of endoxifen with tamoxifen could
successfully address genetic variability chemotherapy issues and
achieve a sustained pharmacodynamic profile in treated breast
cancer patients (82). At the same
time, this knowledge appears to stimulate the design of multiscale
mechanistic PK/PD model-based approaches for individualizing the
therapeutic dosage schemes for tamoxifen and other anticancer
drugs, such as temozolamide for glioblastoma, thus introducing a
new era for modern PTx in cancer therapeutics (83,84).
The latter obviously may also help to apply the predictive capacity
of cancer chemotherapy-based PBPK models in nanomedicine
development to achieve optimum nanoformulation concentration
profiles needed in tumor-targeted cells. In contrast, scientific
interdisciplinary efforts in the era of nanomedicine hold promise
to seize the opportunity to apply novel model-based approaches and
enhance the capability for designing and analyzing the properties
and the pharmacological effects of cancer therapeutic
nanoformulations (82,85). Indicatively, there is on-going
research on nanoparticles loaded with tamoxifen for the development
of nanoformulations with improved PK profiles and tumor-targeted
delivery, thus attempting to overcome the PK/PD and PGx issues
negatively impacting the clinical outcome, as mentioned above
(86,87). Furthermore, in order for the broad
exploitation of such innovative approaches, nanoformulations of
anticancer drugs such as paclitaxel, docetaxel, cisplatin and
doxorubicin are in development and under clinical evaluation
(88–91).
In addition to drug-improved profiles and clinical
research, recently published model-based PK studies describe the
impact of the physicochemical properties of nanoformulations and
theranostics on their in vivo kinetic profile as delivery
systems and/or therapeutic molecules in tumors (69,92).
The combination of the above observations already positively
affects the development of novel nanomedicines where their
construction, size and physical properties contribute to their
targeted delivery to the lesion site. Moreover, it is indicative of
the future perspectives presented for the era of nanotechnology and
the adoption of all possible data and parameters (model-based,
biological, pharmacological and technological) in order to develop
nanoformulations with desired PK/PD properties to ensure maximum
clinical outcome for most, if not all, patients. To this respect,
PBPK modeling represents for complex and multifactorial diseases,
such as cancer, an essential and fruitful tool with which to
advance future research regarding the disposition of
nanoformulations at the site of their action in the body (70,93,94).
This needed collaboration from various scientific disciplines of
pharmacology, in silico modeling and pharmaceutical
nanotechnology shows a tremendous potential for designing
sophisticated and clinically effective and safe nanomedicines with
either diagnostic and/or therapeutic modality (95–98).
Importantly, the usefulness of such nanomedicine-focused PBPK
models could directly be exploited for specific cancer (or other
disease-related) patient groups such as the pediatric population to
help solve issues of pediatric drug development and administration
dosage scheme protocols (99). In
addition, moving forward, a similar beneficial approach could
eventually occur to impact the area of protein cancer therapeutics
and relevant formulations, since the peculiar characteristics of
this type of drug and their behavior in the body, clearly requires
a suitable simulation design and the predictive capacity of more
complex PBPK models (i.e. including immune system) to enhance
productivity, drug behavior profiles and clinical outcomes
(100–102).
Undoubtedly, the early applicability of PBPK models
for the development of small-molecule drug entities has advanced
the predictive capability of their subsequent PK/PD effect profiles
which has eventually enhanced the productivity outcomes within the
new drug development era (103).
Currently, however, PBPK modeling extends toward covering the
administration of therapeutic proteins based on the continuously
increasing number of biopharmaceuticals reaching the market
(102,104,105). From the mechanistic perspective,
modeling and simulation of PBPK are implemented by the central idea
that the drug molecules of interest (small chemicals or proteins)
are subject to PK processes within the body described by
distribution, metabolism and elimination routes in addition to the
absorption mechanisms applied for small chemicals. By considering
that these PK processes could be described mathematically, a number
of differential equations have been applied to mechanistically
express the underlying biological phenomena in a way that all body
organs are linked through blood circulation. The suitable clinical
validation of this approach then creates a framework where
knowledge of clinical utility is generated from data covering in
silico, in vitro and/or in vivo methodologies and
experimentation upon new drug development. The main challenge still
refers to the need for mutual understanding of both mathematics and
biology from the users in order to avoid developing models which
could be unrealistic or provide false results (106–108). Importantly in the case of
nanomedicines, PBPK approaches are considered more complex based on
the characteristics of the nanoparticles that make them different
from small-molecule drugs and therapeutic proteins, since their PK
profile is controlled by more multifaceted and interrelated
relationships between the nanosystem and body physiology (109,110). Nanostructured vehicles are
designed either as solid nanoparticles consisting of polymers or
inorganic materials or as liquid-based formulations that could be
described as nanoemulsions. Such nanosystems are then loaded with
small chemical drug molecules, therapeutic proteins, imaging probes
or other relevant compounds (small RNA molecules, e.g.
siRNAs/miRNAs) to cover a wide-range of medical uses from imaging
to therapeutic applications (70,111–115). One of the driving forces for
developing nanosystems for drug targeted delivery relies on their
drug-loaded capacity for improved in vivo PK/PD profiles due
to advanced metabolic stability and membrane permeability that
could lead to improved bioavailability (for per os
administration) and also to a prolonged pharmacological effect at
the site of action with limited toxicity (116–118). Although these pharmacological
advantages have intrigued the research on developing nanomedicines
they also obstruct the straightforward application of PBPK
approaches as they are applied today. This is due to several
reasons which are also attributed to the characteristics of
nanomedicines in order to obtain the above features of improved PK
profiles. For example, it is easily conceivable that for the
various systems of nanomedicines (i.e. dendrimers, nanocrystals,
emulsions, liposomes, solid lipid nanoparticles, micelles and
polymeric nanoparticles) different PBPK considerations should be
implemented, since they represent different drug-release systems.
The latter also adds more complexity to the models, since for each
one a different mechanism of drug release exists depending on the
administration route - i.e. absorption through gut for per
os delivery, release in the blood stream for intravenous
infusion, or even release on the site of action for targeted
administration. For example, in the typical setting of the PBPK
approach, the controlled or sustained drug release systems are
related with mechanisms of gut absorption (transporters, enzymes)
or of other routes (i.e. skin, lungs) in order to predict the
concentration of the drug that reaches the blood circulation which
thereafter is used to further estimate the tissue concentrations,
the pD profile on the site of action, and the elimination profiles
(119–121). In contrast to nanomedicines, which
act as drug nano-carriers, the controlled release often appears to
be in the blood stream or in the site of action (i.e. tumor lesion,
microenvironment) (122–126). These mechanisms add more
complexity to the systems since additional parameters are involved,
such as the mechanism of the release from the nanocarrier and the
permeability into the site of action for each specific diseased
tissue targeted, but also the drug's characteristic distribution
such as solubility in plasma, plasma to tissue ratio and fraction
of binding into tissue. In addition, the nanoparticle
concentrations in systemic circulation and/or in the organ locally
should be linked with drug release profiles related to either
top-down or bottom-up PBPK models. The latter presents added
complexity toward validation due to the difficulty to collect in
vivo and clinical data for nanomedicines that act as carriers
for targeted drug release. Taking into consideration the recent
advancements in PGx approaches, then the use of PBPK modeling for
nanomedicines also requires suitable adjustment to address
drug-specific genetic PK/PD information. For example, issues that
need to be addressed include the impact of polymorphisms of phase I
and II drug metabolizing enzymes on the controlled/sustained
release over time of the drug from its nano-carrier into the
systemic concentration (implying changes in the rate of the
enzymatic reaction), since it is unclear whether it remains the
same or even similar as for cases with classical sustained release
systems for prolonged absorption (120,127).
Based on the molecular knowledge accumulated thus
far for epithelial-mesenchymal transition (EMT) and the known
histological architecture of the tumor cells within the tissue, one
can consider to build such a core PBPK model by suitably developing
algorithms that incorporate parameters referring also to the
metastatic behavior of malignant cells such as: i) cell adhesion
molecule levels (e.g. for adherent junctions E-cadherin, for focal
adhesion junctions integrin β1, and for desmosomal junctions
desmoglein-3); ii) the neovascularization extent (angiogenesis
level) and the computationally depicted histological structure in
the tumor area that could also contain measurable additional
molecular markers involved in these processes [e.g. expression
levels of vascular endothelial growth factor (VEGF), αvβ3
integrin]. The clinical validation of any relevant adhesion
molecule specification as metastatic biomarker, however, is needed
for each tumor type, as recently shown in the case of oral squamous
cell carcinoma (128). Finally,
taking into consideration the known histological, genomic and
cellular heterogeneity that exists within the tumor in various
organs, one can further add information to specific components
depending on the desired target diseased tissue (63). All the above parameters could
contribute to the design of additional compartments (representing
primary and metastatic tumor sites) although, to date, there are no
published data available combining PBPK and metastatic tumors
despite the fact that clinical trials are already executed for
metastatic tumors and nanoparticles based on the preclinical data
available (129).
Furthermore, much work exists for tumor growth
modeling (usually for xerographs) and how they can possibly be
coupled with PBPK approaches regarding the estimation of
chemotherapy concentrations in tumor compartments and the improved
adjustment of dosing regimen (83,130–132). Moreover, in addition to
xenograft-based models, there is much effort toward the development
of multi-scale in silico models aimed at the improved
comprehension of tumor growth and the underlying mechanisms that
lead to diverse outcomes of tumor lesions (133–141). Although both approaches sometimes
require a tedious and time-consuming process due to the required
individual extrapolation of data and validation of various factors
contributing to cancer pathophysiology, diversity and modeling,
they show encouraging results towards improved, personalized or
stratified approaches to cancer treatment particularly for novel
chemotherapeutic drug delivery schemes. The application of
nanomedicines to prevent cancer metastasis provides new
opportunities toward the development of improved therapies
(142–146). It must also be emphasized that
these approaches could be implemented at two levels of personalized
approaches. The first level could be the stratification of patients
based on primary and metastatic tumor regions and the second one
the individual dosage regiments as proposed by in silico
clinical trial models based on PBPK approaches. In addition, by
successfully generating a metastatic behavior-based PBPK model, its
application to assess the PK/PD profile of novel therapeutics very
early in the drug developmental process may lead to improved
productivity of drug candidate products with enhanced efficacy and
clinical safety profiles, particularly in the more complex cases of
metastatic tumors.
From the above mentioned, more focused efforts are
needed toward establishing the optimum PK/PD correlations regarding
the nanosystem physicochemical properties, the biological
environment and the underlying molecular markers for balancing
benefit-to-risk ratio in the preclinical/clinical development
process. Drug-release kinetics either in systemic circulation or in
the local diseased-tissue microenvironment (e.g. pulsatile vs.
continuous release) is a crucial factor affecting the efficacy of
nanomedicines and their safety behavior. It is thus urgent that a
more thorough understanding of human physiology and pathophysiology
and the interactions of biological systems with therapeutics be
achieved. The latter implies a direction where the enhancement of
pharmacological translational efficiency is facilitated through
exploitation of the underlying developmental biology dynamics, the
cellular diversity and the genomic heterogeneity for individual
patients and for most illnesses. To successfully move on, the
nanosystem formulation platforms, the pharmacological and
preclinical assessment methodologies as well as the clinical
development design must comply with such interdisciplinary research
scheme requirements to readily overcome various obstructing
parameters from bench to the clinic. Recently published work for
cancer nanosystem platform methodologies and PBPK modeling add new
insights to such a direction. In one study, the polymeric
nanoparticle design platform was based on a combinatorial nature of
simultaneously assessing their physicochemical diversity (size,
surface hydrophilicity, targeting ligand density, drug loading and
drug release) with the best clinical performance profiling for PK,
biodistribution and efficacy upon encapsulation with
chemotherapeutic agents (129).
Importantly, the designed docetaxel-loaded polymeric nanoparticles
targeted to the prostate-specific surface antigen exhibited similar
PK, biodistribution, and safety profiles in three animal species
(mice, rats and monkeys). In particular, their PK characteristics
achieved much higher plasma concentrations of docetaxel for an
extended duration that coincided with prolonged circulation in the
vasculature and controlled release pattern in the body. To this
end, docetaxel-loaded polymeric nanoparticles have been tested in
humans with promising results confirming the previous data observed
in animal species, whereas a phase II clinical trial is now running
to evaluate their PK/PD behavior in patients with advanced
metastatic prostate cancer (ClinicalTrials.gov Identifier: NCT01812746). In
another similar study using the bisphosphonate ligand alendronate
to target bone cancer lesions, the successful formulation of
targeted polymeric nanoparticles for the controlled delivery of
bortezomib to diminish off-target effects and increase tumor cell
concentration was achieved (147).
Bortezomib, a proteasome inhibitor, is clinically used for the
therapy of multiple myeloma, a cancer primarily originating in bone
marrow resulting in osteolytic lesions. Again, these bone
microenvironment-targeted polymeric nanoparticles loaded with
bortezomib have shown a favored PK/PD profile in terms of high
retention, accumulation and bone homing capacity as well as
enhanced survival and decreased tumor burden in mouse models of
multiple myeloma. In a third study, the development of a PK model
to quantify the effect of vascular physiology and permeability in
individual patients for cancer nanotherapeutics was conducted
(69). notably, the specific
particle size range of cancer nanosystems and the timed drug
release profile were found to be crucial predictive rate-limiting
formulation factors in order for the benefit-to-risk ratio to be
maximized and clinically validated. The latter was correlated with
the enhanced permeability and retention effect, attributed to the
tumor microenvironment vascular pore size along with that exhibited
for normal tissues, meaning that both normal physiology and
pathophysiology of the vasculature contributed to the PK/PD cancer
nanotherapeutic behavior. Unanimously, these results present
evidence that the exploitation of PBPK modeling early in the
development of cancer therapy nanomedicine represents the required
solid ground and suitable framework for increasing the productivity
later on in clinical phases and ensuring better efficacy and safety
profiles for the respective marketed nanotherapeutics.
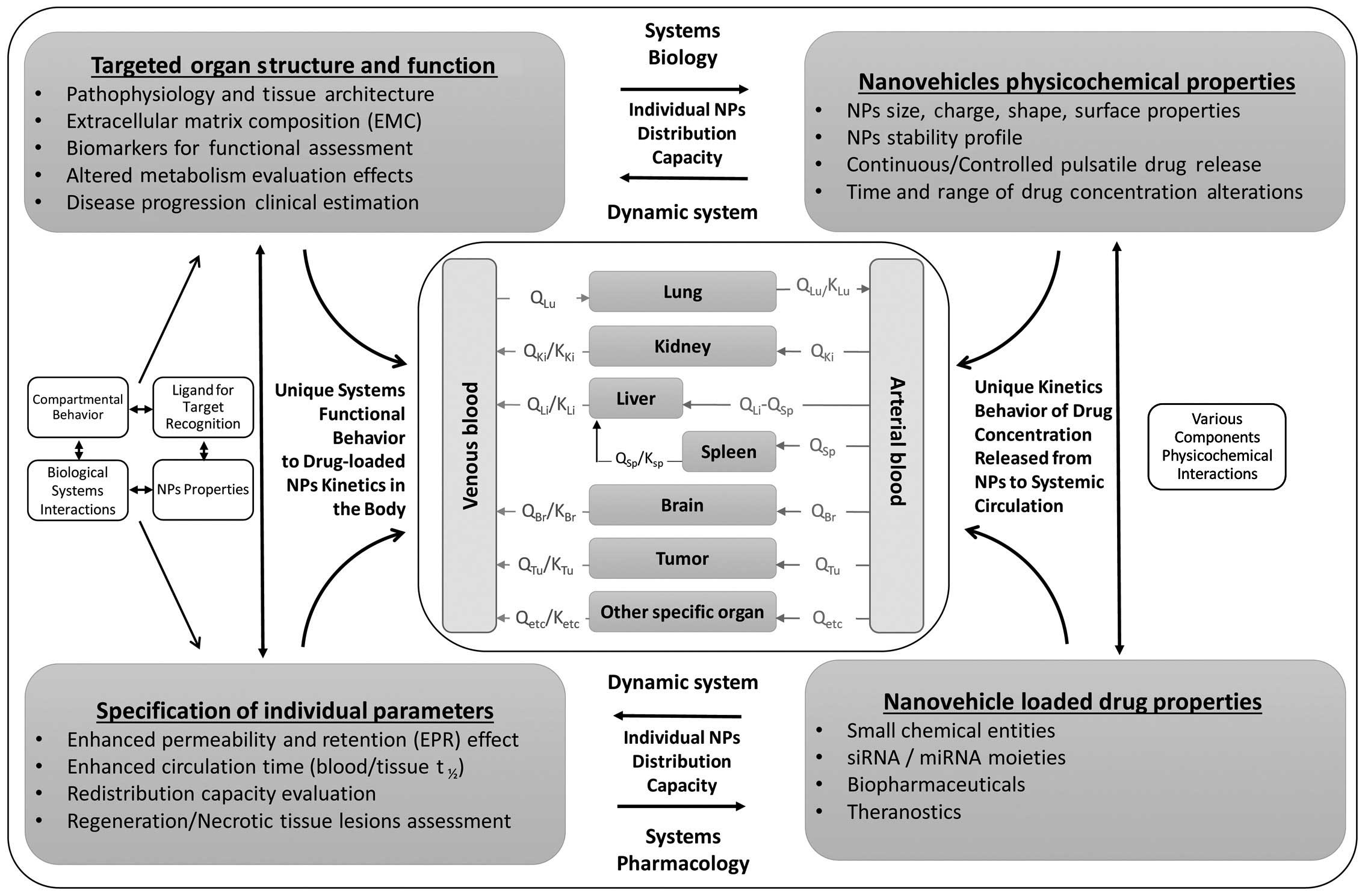
Based on the above, a proposed tumor-focused PBPK
model for nanomedicines (PBPK-NM) is presented in Fig. 3. This vital need for successfully
addressing the different PK behaviors of nanoparticles and thus
nanomedicines in the body compared to that of their small-molecule
drug counterparts, has already led the FDA to initiate projects
such as ʻPBPK Modeling of nanomedicineʼ in order to apply
computational modeling approaches to assess the safety of
nano-scale materials. In particular, liposomal doxorubicin (Doxil),
the first nanomedicine that entered the market and is used in the
clinic for various solid tumors (ovarian cancer and myeloma) has
been selected. The project launched in November 2014 is currently
at the half of the road stage and it is expected to be concluded at
the end of 2015 aimed at the development of computational
approaches to be utilized to monitor Doxil in the blood and in
different sites of the body (http://www.accessdata.fda.gov/FDATrack/track-proj?program=nctr&id=NCTR-OSC-PM-PBPK-Modeling-of-Nanomedicine).
The predictive capacity of PBPK models allows them
to also be successfully applied in assessing impacts on dose and
particle size as well as on the in vivo performance of
nano-formulations, either for medicines under development or for
marketed drugs (148). However,
fundamental issues exist that are hampering the advancement of
personalized medicine and that highlight the lack of emerging
predictive tools that could serve as a decision support mechanism
for physicians to personalize treatment (149,150). Particularly, this seems even more
difficult for drug delivery by attempting to unify knowledge from
heterogeneous data derived e.g. from personal genome sequencing,
structural, functional and chemical genomics applications, as well
as the PK/PD drug profiles including the clinical setting. In
contrast, PBPK models represent platforms where organ- and
tissue-specific parameters, genomic variation data and
physicochemical drug properties can be efficiently inter-correlated
to allow the prediction of PK/PD drug behavior in the body for
specific populations or individual patients. Importantly, the
enrichment of the predictive capacity of PBPK models towards
empowering individualized drug response phenotype decisions could
be achieved through incorporation of: i) genome-wide drug-target
molecular interactions; ii) temporal and spatial scale
macromolecular conformation state behavior; iii) drug-driven
molecular circuit pathways; and iv) in real-time monitoring the
impact of environmental and clinical parameters. To this end, the
structure-enabled integrative modeling to effectively predict QT
interval prolongation and minimize the drug-induced arrhythmia
profile of delivered medicines in the clinical setting has been
recently exemplified (150). The
latter implements crucial clinical information related to
molecular, genomics, pharmacological and chemical drug-related
aspects including those of pharmacovigilance that have been
previously established in regulatory legislation for the
arrhythmogenic behavior of drugs (8).
As far as drug delivery is concerned, new tools are
tilting health-care control from physicians to patients with the
paradigm of the delivery of insulin using telemedicine (151). Three dimensional (3D) printing
technologies emerge as an innovative approach to design
pharmaceutical dosage forms for personalized medicine (152). Recently, 3D printing was used for
the production of tablets capable of satisfying regulatory tests
and matching the release of standard commercial tablets (153). Remote controlled capsules (RCCs)
have been extensively used in the field of site-specific drug
delivery using micro-electronic mechanical system (MEMS) technology
154). MEMS combined with medical devices (e.g. contact lenses) can
be used for ocular diagnostics (155). Finally dosing devices and solid
dosage forms might allow oral individual drug therapy (156).
The availability of information science technologies
aimed to develop advanced tools and databases for electronic health
records for patients raise the question of whether virtual
population profiles gained from epidemiologic recorded data with
data generated from the patients and/or hospital driven databases
and relative software could match. The latter, if occurred, would
permit the fitting of modeled data of virtual profiles with
profiles of real life patients and thus initiate personalized or
stratified medicine approaches. Moreover, it will also advance the
next step for the development of unique decision-making tools or
for novel approaches during R&D but also for PTx procedures.
However, the absence of a roadmap on how the regulatory environment
in drug development and healthcare is gaining major clinical
benefit and economic value from personalized medicine applications
has clearly restricted the required evidence to informed
decision-making and assessment of genomic priorities (42). It is therefore of important priority
to work and present evidence by establishing infrastructure and
methodologies capable of confirming cost-effectiveness in addition
to clinical effectiveness in a robust and timely manner for
decision makers (43); and by doing
so, the parallel advancement of PBPK models will empower the
translation capacity and practical clinical utility of personalized
medicine decisions, thus benefitting healthcare and stabilizing PTx
worldwide (157). These new
technologies are expected to revolutionize existing healthcare and
drug delivery, in a way to enrich personalized medicine
capabilities and precision for broader clinical utility of
translational medicine therapeutic interventions worldwide.
|
1
|
Braeckmans K, De Smedt SC, Leblans M,
Pauwels R and Demeester J: Encoding microcarriers: Present and
future technologies. Nat Rev Drug Discov. 1:447–456. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ginsburg GS and Willard HF: Genomic and
personalized medicine: Foundations and applications. Transl Res.
154:277–287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hertz DL and McLeod HL: Use of
pharmacogenetics for predicting cancer prognosis and treatment
exposure, response and toxicity. J Hum Genet. 58:346–352. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaddurah-Daouk R and Weinshilboum RM;
Pharmacometabolomics Research Network: Pharmacometabolomics:
Implications for clinical pharmacology and systems pharmacology.
Clin Pharmacol Ther. 95:154–167. 2014. View Article : Google Scholar
|
|
5
|
Roses AD, Saunders AM, Lutz MW, Zhang N,
Hariri AR, Asin KE, Crenshaw DG, Budur K, Burns DK and Brannan SK:
New applications of disease genetics and pharmacogenetics to drug
development. Curr Opin Pharmacol. 14:81–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vizirianakis IS: Challenges in current
drug delivery from the potential application of pharmacogenomics
and personalized medicine in clinical practice. Curr Drug Deliv.
1:73–80. 2004. View Article : Google Scholar
|
|
7
|
Vizirianakis IS: Improving pharmacotherapy
outcomes by pharmacogenomics: From expectation to reality?
Pharmacogenomics. 6:701–711. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vizirianakis IS: Clinical translation of
genotyping and haplotyping data: Implementation of in vivo
pharmacology experience leading drug prescription to
pharmacotyping. Clin Pharmacokinet. 46:807–824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vizirianakis IS: Advancement of
pharmacogenomics toward pharmacotyping in drug prescription:
Concepts, challenges, and perspectives for personalized medicine.
Handbook of Personalized Medicine: Advances in Nanotechnology, Drug
Delivery and Therapy. Vizirianakis IS: Pan Stanford Publishing;
Singapore: pp. 893–952. 2014, View Article : Google Scholar
|
|
10
|
Pirmohamed M: Personalized
pharmacogenomics: Predicting efficacy and adverse drug reactions.
Annu Rev Genomics Hum Genet. 15:349–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahn K, Luo J, Berg A, Keefe D and Wu R:
Functional mapping of drug response with
pharmacodynamic-pharmacokinetic principles. Trends Pharmacol Sci.
31:306–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Daka A and Peer D: RNAi-based
nanomedicines for targeted personalized therapy. Adv Drug Deliv
Rev. 64:1508–1521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Debbage P: Targeted drugs and
nanomedicine: Present and future. Curr Pharm Des. 15:153–172. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huttenhower C and Hofmann O: A quick guide
to large-scale genomic data mining. PLOS Comput Biol.
6:e10007792010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Janowski M, Bulte JW and Walczak P:
Personalized nano-medicine advancements for stem cell tracking. Adv
Drug Deliv Rev. 64:1488–1507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mura S and Couvreur P: Nanotheranostics
for personalized medicine. Adv Drug Deliv Rev. 64:1394–1416. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Petersen AL, Hansen AE, Gabizon A and
Andresen TL: Liposome imaging agents in personalized medicine. Adv
Drug Deliv Rev. 64:1417–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ryu JH, Koo H, Sun IC, Yuk SH, Choi K, Kim
K and Kwon IC: Tumor-targeting multi-functional nanoparticles for
theragnosis: New paradigm for cancer therapy. Adv Drug Deliv Rev.
64:1447–1458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vizirianakis IS, Chatzopoulou M,
Bonovolias ID, Nicolaou I, Demopoulos VJ and Tsiftsoglou AS: Toward
the development of innovative bifunctional agents to induce
differentiation and to promote apoptosis in leukemia: Clinical
candidates and perspectives. J Med Chem. 53:6779–6810. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wieland M and Fussenegger M: Reprogrammed
cell delivery for personalized medicine. Adv Drug Deliv Rev.
64:1477–1487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cook D, Brown D, Alexander R, March R,
Morgan P, Satterthwaite G and Pangalos MN: Lessons learned from the
fate of Astrazeneca's drug pipeline: A five-dimensional framework.
Nat Rev Drug Discov. 13:419–431. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cree IA: Designing personalised cancer
treatments. J Control Release. 172:405–409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hillgren KM, Keppler D, Zur AA, Giacomini
KM, Stieger B, Cass CE and Zhang L; International Transporter
Consortium: Emerging transporters of clinical importance: An update
from the International Transporter Consortium. Clin Pharmacol Ther.
94:52–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang M, Shen A, Ding J and Geng M:
Molecularly targeted cancer therapy: Some lessons from the past
decade. Trends Pharmacol Sci. 35:41–50. 2014. View Article : Google Scholar
|
|
25
|
Ingelman-Sundberg M: Pharmacogenetics of
cytochrome P450 and its applications in drug therapy: The past,
present and future. Trends Pharmacol Sci. 25:193–200. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jain KK: Innovative diagnostic
technologies and their significance for personalized medicine. Mol
Diagn Ther. 14:141–147. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee JW, Aminkeng F, Bhavsar AP, Shaw K,
Carleton BC, Hayden MR and Ross CJ: The emerging era of
pharmacogenomics: Current successes, future potential, and
challenges. Clin Genet. 86:21–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ntziachristos V and Razansky D: Molecular
imaging by means of multispectral optoacoustic tomography (MSOT).
Chem Rev. 110:2783–2794. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ntziachristos V, Schellenberger EA, Ripoll
J, Yessayan D, Graves E, Bogdanov A Jr, Josephson L and Weissleder
R: Visualization of antitumor treatment by means of fluorescence
molecular tomography with an annexin V-Cy5.5 conjugate. Proc Natl
Acad Sci USA. 101:12294–12299. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pirmohamed M: Pharmacogenetics: Past,
present and future. Drug Discov Today. 16:852–861. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sadee W: Pharmacogenomic biomarkers:
Validation needed for both the molecular genetic mechanism and
clinical effect. Pharmacogenomics. 12:675–680. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vizirianakis IS: Nanomedicine and
personalized medicine toward the application of pharmacotyping in
clinical practice to improve drug-delivery outcomes. Nanomedicine.
7:11–17. 2011. View Article : Google Scholar
|
|
33
|
Flowers CR and Veenstra D: The role of
cost-effectiveness analysis in the era of pharmacogenomics.
Pharmacoeconomics. 22:481–493. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Payne K and Shabaruddin FH:
Cost-effectiveness analysis in pharmacogenomics. Pharmacogenomics.
11:643–646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sorich MJ, Wiese MD and Pekarsky B:
Cost-effectiveness of geno-typing to guide treatment.
Pharmacogenomics. 15:727–729. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wong WB, Carlson JJ, Thariani R and
Veenstra DL: Cost effectiveness of pharmacogenomics: A critical and
systematic review. Pharmacoeconomics. 28:1001–1013. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu AHB, Babic N and Yeo KT: Implementation
of pharmacogenomics into the clinical practice of therapeutics:
Issues for the clinician and the laboratorian. Per Med. 6:315–327.
2009. View Article : Google Scholar
|
|
38
|
Fleeman N, McLeod C, Bagust A, Beale S,
Boland A, Dundar Y, Jorgensen A, Payne K, Pirmohamed M, Pushpakom
S, et al: The clinical effectiveness and cost-effectiveness of
testing for cytochrome P450 polymorphisms in patients with
schizophrenia treated with antipsychotics: A systematic review and
economic evaluation. Health Technol Assess. 14:1–157. iii2010.
View Article : Google Scholar
|
|
39
|
Gurwitz D, Rodríguez-Antona C, Payne K,
Newman W, Gisbert JP, de Mesa EG and Ibarreta D: Improving
pharmacovigilance in Europe: TPMT genotyping and phenotyping in the
uk and spain. Eur J Hum Genet. 17:991–998. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Thompson AJ, Newman WG, Elliott RA,
Roberts SA, Tricker K and Payne K: The cost-effectiveness of a
pharmacogenetic test: A trial-based evaluation of TPMT genotyping
for azathioprine. Value Health. 17:22–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
van den Akker-van Marle ME, Gurwitz D,
Detmar SB, Enzing CM, Hopkins MM, Gutierrez de Mesa E and Ibarreta
D: Cost-effectiveness of pharmacogenomics in clinical practice: A
case study of thiopurine methyltransferase genotyping in acute
lymphoblastic leukemia in Europe. Pharmacogenomics. 7:783–792.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Phillips KA, Ann Sakowski J, Trosman J,
Douglas MP, Liang SY and Neumann P: The economic value of
personalized medicine tests. Genet Med. 16:251–257. 2014.
View Article : Google Scholar :
|
|
43
|
Shabaruddin FH and Payne K: Evaluating the
cost-effectiveness of pharmacogenomics in clinical practice.
Handbook of personalized Medicine: Advances in nanotechnology, Drug
Delivery and Therapy. Vizirianakis IS: Pan stanford publishing;
Singapore: pp. 779–812. 2014, View Article : Google Scholar
|
|
44
|
Poulin P, Jones RD, Jones HM, Gibson CR,
Rowland M, Chien JY, Ring BJ, Adkison KK, Ku MS, He H, et al: PHRMA
CPCDC initiative on predictive models of human pharmacokinetics,
part 5: Prediction of plasma concentration-time profiles in human
using the physiologically-based pharmacokinetic modeling approach.
J Pharm Sci. 100:4127–4157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ring BJ, Chien JY, Adkison KK, Jones HM,
Rowland M, Jones RD, Yates JW, Ku MS, Gibson CR, He H, et al: PHRMA
CPCDC initiative on predictive models of human pharmacokinetics,
part 3: Comparative assessment of prediction methods of human
clearance. J Pharm Sci. 100:4090–4110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bates S: Progress towards personalized
medicine. Drug Discov Today. 15:115–120. 2010. View Article : Google Scholar
|
|
47
|
Fu G, Liu J, Luo J, Zhong W, Wang Y, Wang
N and Wu R: Systems mapping: A computational tool for personalized
medicine. Handbook of personalized Medicine: Advances in
nanotechnology, Drug Delivery and Therapy. Vizirianakis IS: Pan
Stanford Publishing; Singapore: pp. 321–340. 2014, View Article : Google Scholar
|
|
48
|
Wu R, Tong C, Wang Z, Mauger D, Tantisira
K, Szefler SJ, Chinchilli VM and Israel E: A conceptual framework
for pharmacodynamic genome-wide association studies in
pharmacogenomics. Drug Discov Today. 16:884–890. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hong H, Perkins R, Shi L, Fang H, Mendrick
DL and Tong W: Molecular biomarkers for personalized medicine.
Handbook of personalized Medicine: Advances in nanotechnology, Drug
Delivery and Therspy. Vizirianakis IS: Pan stanford publishing;
Singapore: pp. 607–644. 2014, View Article : Google Scholar
|
|
50
|
Gonzalez de Castro D, Clarke PA,
Al-Lazikani B and Workman P: Personalized cancer medicine:
Molecular diagnostics, predictive biomarkers, and drug resistance.
Clin Pharmacol Ther. 93:252–259. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yap TA, Sandhu SK, Workman P and de Bono
JS: Envisioning the future of early anticancer drug development.
Nat Rev Cancer. 10:514–523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rubin EH and Gilliland DG: Drug
development and clinical trials - the path to an approved cancer
drug. Nat Rev Clin Oncol. 9:215–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Vizirianakis IS and Fatouros DG:
Personalized nanomedicine: Paving the way to the practical clinical
utility of genomics and nanotechnology advancements. Adv Drug Deliv
Rev. 64:1359–1362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Vizirianakis IS: Handbook of personalized
Medicine: Advances in Nanotechnology, Drug Delivery, and Therapy.
Pan Stanford Publishing; Singapore: 2014, View Article : Google Scholar
|
|
55
|
Swanson TW, Akkari PA, Arbuckle JB,
Grossman I, Sundseth SS and Roses AD: Methodology to enable
integration of genomic knowledge into drug development. Handbook of
Personalized Medicine: Advances in nanotechnology, Drug Delivery
and Therpy. Vizirianakis IS: Pan stanford publishing; Singapore:
pp. 645–684. 2014, View Article : Google Scholar
|
|
56
|
Jamei M, Rowland YK and Rostami-Hodjegan
A: Framework, organization, and applications of the Simcyp
population-based simulator to support new drug development.
Handbook of personalized Medicine: Advances in nanotechnology, Drug
Delivery and Therapy. Vizirianakis IS: Pan stanford publishing;
Singapore: pp. 685–726. Pan stanford publishing; Singapore: 2014,
View Article : Google Scholar
|
|
57
|
Amstutz U and Carleton BC: Pharmacogenetic
testing: Time for clinical practice guidelines. Clin Pharmacol
Ther. 89:924–927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Johnson JA, Gong L, Whirl-Carrillo M, Gage
BF, Scott SA, Stein CM, Anderson JL, Kimmel SE, Lee MT, Pirmohamed
M, et al Clinical pharmacogenetics Implementation Consortium:
Clinical Pharmacogenetics Implementation Consortium Guidelines for
CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol
Ther. 90:625–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Relling MV, Gardner EE, Sandborn WJ,
Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE and
Klein TE; Clinical Pharmacogenetics Implementation Consortium:
Clinical Pharmacogenetics Implementation Consortium guidelines for
thiopurine methyltransferase genotype and thiopurine dosing. Clin
Pharmacol Ther. 89:387–391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Relling MV and klein TE: CPIC: Clinical
pharmacogenetics Implementation Consortium of the Pharmacogenomics
Research network. Clin Pharmacol Ther. 89:464–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Scott SA, Sangkuhl K, Gardner EE, Stein
CM, Hulot JS, Johnson JA, Roden DM, Klein TE and Shuldiner AR;
Clinical Pharmacogenetics Implementation Consortium: Clinical
Pharmacogenetics Implementation Consortium guidelines for
cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy.
Clin Pharmacol Ther. 90:328–332. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Swen JJ, Nijenhuis M, de Boer A, Grandia
L, Maitland-van der zee AH, Mulder H, Rongen GA, van Schaik RH,
Schalekamp T, Touw DJ, et al: Pharmacogenetics: From bench to byte
- an update of guidelines. Clin Pharmacol Ther. 89:662–673. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Workman P: The opportunities and
challenges of personalized genome-based molecular therapies for
cancer: Targets, technologies, and molecular chaperones. Cancer
Chemother. 52(Suppl 1): S45–S56. 2003. View Article : Google Scholar
|
|
65
|
Jain RK: Normalizing tumor
microenvironment to treat cancer: Bench to bedside to biomarkers. J
Clin Oncol. 31:2205–2218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Khawar IA, Kim JH and Kuh HJ: Improving
drug delivery to solid tumors. J Control Release. 201:78–89. 2015.
View Article : Google Scholar
|
|
67
|
Scott JG, Hjelmeland AB, Chinnaiyan P,
Anderson AR and Basanta D: Microenvironmental variables must
influence intrinsic phenotypic parameters of cancer stem cells to
affect tumourigenicity. PLOS Comput Biol. 10:e10034332014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Watnick RS: The role of the tumor
microenvironment in regulating angiogenesis. Cold Spring Harb
Perspect Med. 2:a0066762012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kirtane AR, Siegel RA and Panyam J: A
pharmacokinetic model for quantifying the effect of vascular
permeability on the choice of drug carrier: A framework for
personalized nanomedicine. J Pharm Sci. 104:1174–1186. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Moss DM and siccardi M: Optimizing
nanomedicine phar-macokinetics using physiologically based
pharmacokinetics modelling. Br J Pharmacol. 171:3963–3979. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Andre F, Mardis E, Salm M, Soria JC, Siu
LL and Swanton C: Prioritizing targets for precision cancer
medicine. Ann Oncol. 25:2295–2303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Collins FS and Varmus H: A new initiative
on precision medicine. N Engl J Med. 372:793–795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Doudican NA, Kumar A, Singh NK, Nair PR,
Lala DA, Basu K, Talawdekar AA, Sultana Z, Tiwari KK, Tyagi A, et
al: Personalization of cancer treatment using predictive
simulation. J Transl Med. 13:432015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Roychowdhury S and Chinnaiyan AM:
Translating genomics for precision cancer medicine. Annu Rev
Genomics Hum Genet. 15:395–415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Binkhorst L, Mathijssen RH, Jager A and
van Gelder T: Individualization of tamoxifen therapy: Much more
than just CYP2D6 genotyping. Cancer Treat Rev. 41:289–299. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Smith GL: The long and short of tamoxifen
therapy: A review of the ATLAs trial. J Adv Pract Oncol. 5:57–60.
2014.PubMed/NCBI
|
|
77
|
Borges S, Desta Z, Li L, Skaar TC, Ward
BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, et al:
Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen
metabolism: Implication for optimization of breast cancer
treatment. Clin Pharmacol Ther. 80:61–74. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Flockhart D: CYP2D6 genotyping and the
pharmacogenetics of tamoxifen. Clin Adv Hematol Oncol. 6:493–494.
2008.PubMed/NCBI
|
|
79
|
Jin Y, Desta Z, Stearns V, Ward B, Ho H,
Lee KH, Skaar T, Storniolo AM, Li L, Araba A, et al: CYP2D6
genotype, antidepressant use, and tamoxifen metabolism during
adjuvant breast cancer treatment. J Natl Cancer Inst. 97:30–39.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Stearns V, Johnson MD, Rae JM, Morocho A,
Novielli A, Bhargava P, Hayes DF, Desta Z and Flockhart DA: Active
tamoxifen metabolite plasma concentrations after coadministration
of tamoxifen and the selective serotonin reuptake inhibitor
paroxetine. J Natl Cancer Inst. 95:1758–1764. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Dickschen K, Willmann S, Thelen K, Lippert
J, Hempel G and Eissing T: Physiologically based pharmacokinetic
modeling of tamoxifen and its metabolites in women of different
CYP2D6 phenotypes provides new insight into the tamoxifen mass
balance. Front Pharmacol. 3:922012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Drbohlavova J, Chomoucka J, Adam V,
Ryvolova M, Eckschlager T, Hubalek J and Kizek R: Nanocarriers for
anti-cancer drugs - new trends in nanomedicine. Curr Drug Metab.
14:547–564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ballesta A, Zhou Q, Zhang X, Lv H and
Gallo JM: Multiscale design of cell-type-specific
pharmacokinetic/pharmacodynamic models for personalized medicine:
Application to temozolomide in brain tumors. CPT Pharmacometrics
Syst Pharmacol. 3:e1122014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Block M: Physiologically based
pharmacokinetic and pharmacodynamic modeling in cancer drug
development: Status, potential and gaps. Expert Opin Drug Metab
Toxicol. 11:743–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Upreti M, Jyoti A and Sethi P: Tumor
microenvironment and nanotherapeutics. Transl Cancer Res.
2:309–319. 2013.
|
|
86
|
Maji R, Dey NS, Satapathy BS, Mukherjee B
and Mondal S: Preparation and characterization of Tamoxifen citrate
loaded nanoparticles for breast cancer therapy. Int J Nanomedicine.
9:3107–3118. 2014.PubMed/NCBI
|
|
87
|
Pandey SK, Ghosh S, Maiti P and Haldar C:
Therapeutic efficacy and toxicity of tamoxifen loaded PLA
nanoparticles for breast cancer. Int J Biol Macromol. 72:309–319.
2015. View Article : Google Scholar
|
|
88
|
Hersh EM, O'Day SJ, Ribas A, Samlowski WE,
Gordon MS, Shechter DE, Clawson AA and Gonzalez R: A phase 2
clinical trial of nab-paclitaxel in previously treated and
chemotherapy-naive patients with metastatic melanoma. Cancer.
116:155–163. 2010.
|
|
89
|
Jehn CF, Boulikas T, Kourvetaris A, Kofla
G, Possinger K and Lüftner D: First safety and response results of
a randomized phase III study with liposomal platin in the treatment
of advanced squamous cell carcinoma of the head and neck (SCCHN).
Anticancer Res. 28:3961–3964. 2008.
|
|
90
|
Mamot C, Ritschard R, Wicki A, Stehle G,
Dieterle T, Bubendorf L, Hilker C, Deuster S, Herrmann R and
Rochlitz C: Tolerability, safety, pharmacokinetics, and efficacy of
doxorubicin-loaded anti-EGFR immunoliposomes in advanced solid
tumours: A phase 1 dose-escalation study. Lancet oncol.
13:1234–1241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhao P and Astruc D: Docetaxel
nanotechnology in anticancer therapy. ChemMedChem. 7:952–972. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Baish JW, Stylianopoulos T, Lanning RM,
Kamoun WS, Fukumura D, Munn LL and Jain RK: Scaling rules for
diffusive drug delivery in tumor and normal tissues. Proc Natl Acad
Sci USA. 108:1799–1803. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Bachler G, von Goetz N and Hungerbühler K:
A physiologically based pharmacokinetic model for ionic silver and
silver nanoparticles. Int J Nanomedicine. 8:3365–3382.
2013.PubMed/NCBI
|
|
94
|
Bachler G, von Goetz N and Hungerbuhler K:
Using physiologically based pharmacokinetic (PBPK) modeling for
dietary risk assessment of titanium dioxide (TiO2)
nanoparticles. Nanotoxicology. 9:373–380. 2015. View Article : Google Scholar
|
|
95
|
Liu J, Zheng X, Yan L, Zhou L, Tian G, Yin
W, Wang L, Liu Y, Hu Z, Gu Z, et al: Bismuth sulfide nanorods as a
precision nano-medicine for in vivo multimodal imaging-guided
photothermal therapy of tumor. ACS Nano. 9:696–707. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Mouffouk F, Simão T, Dornelles DF, Lopes
AD, Sau P, Martins J, Abu-salah KM, Alrokayan SA, Rosa da Costa AM
and dos Santos NR: Self-assembled polymeric nanoparticles as new,
smart contrast agents for cancer early detection using magnetic
resonance imaging. Int J Nanomedicine. 10:63–76. 2015.PubMed/NCBI
|
|
97
|
Perera RH, Hernandez C, Zhou H, Kota P,
Burke A and Exner AA: Ultrasound imaging beyond the vasculature
with new generation contrast agents. Wiley Interdiscip Rev Nanomed
Nanobiotechnol. 7:593–608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Torchilin VP: Multifunctional,
stimuli-sensitive nanoparticulate systems for drug delivery. Nat
Rev Drug Discov. 13:813–827. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Barrett JS, Della Casa Alberighi O, Läer S
and Meibohm B: Physiologically based pharmacokinetic (PBPK)
modeling in children. Clin Pharmacol Ther. 92:40–49. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chetty M, Li L, Rose R, Machavaram K,
Jamei M, Rostami-Hodjegan A and Gardner I: Prediction of the
pharmacokinetics, pharmacodynamics, and efficacy of a monoclonal
antibody, using a physiologically based pharmacokinetic FcRn model.
Front Immunol. 5:6702014.
|
|
101
|
Diao L and Meibohm B: Pharmacokinetics and
pharmacokinetic-pharmacodynamic correlations of therapeutic
peptides. Clin Pharmacokinet. 52:855–868. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Dostalek M, Gardner I, Gurbaxani BM, Rose
RH and Chetty M: Pharmacokinetics, pharmacodynamics and
physiologically-based pharmacokinetic modelling of monoclonal
antibodies. Clin Pharmacokinet. 52:83–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Jones HM, Chen Y, Gibson C, Heimbach T,
Parrott N, Peters SA, Snoeys J, Upreti VV, Zheng M and Hall SD:
Physiologically based pharmacokinetic modeling in drug discovery
and development: A pharmaceutical industry perspective. Clin
pharmacol Ther. 97:247–262. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
104
|
Dranitsaris G, Amir E and Dorward K:
Biosimilars of biological drug therapies: Regulatory, clinical and
commercial considerations. Drugs. 71:1527–1536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang J, Iyer S, Fielder PJ, Davis JD and
Deng R: Projecting human pharmacokinetics of monoclonal antibodies
from nonclinical data: Comparative evaluation of prediction
approaches in early drug development. Biopharm Drug Dispos. Apr
13–2015.Epub ahead of print. View Article : Google Scholar
|
|
106
|
Bouzom F, Ball K, Perdaems N and Walther
B: Physiologically based pharmacokinetic (PBPK) modelling tools:
How to fit with our needs? Biopharm Drug Dispos. 33:55–71. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Nyberg J, Bazzoli C, Ogungbenro K, Aliev
A, Leonov S, Duffull S, Hooker AC and Mentré F: Methods and
software tools for design evaluation in population
pharmacokinetics-pharmacodynamics studies. Br J Clin Pharmacol.
79:6–17. 2015. View Article : Google Scholar :
|
|
108
|
Rowland M, Peck C and Tucker G:
Physiologically-based pharmacokinetics in drug development and
regulatory science. Annu Rev Pharmacol Toxicol. 51:45–73. 2011.
View Article : Google Scholar
|
|
109
|
Moghimi SM, Hunter AC and Andresen TL:
Factors controlling nanoparticle pharmacokinetics: An integrated
analysis and perspective. Annu Rev Pharmacol Toxicol. 52:481–503.
2012. View Article : Google Scholar
|
|
110
|
Zhang XQ, Xu X, Bertrand N, Pridgen E,
Swami A and Farokhzad OC: Interactions of nanomaterials and
biological systems: Implications to personalized nanomedicine. Adv
Drug Deliv Rev. 64:1363–1384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Davis ME, Zuckerman JE, Choi CH, Seligson
D, Tolcher A, Alabi CA, Yen Y, Heidel JD and Ribas A: Evidence of
RNAi in humans from systemically administered siRNA via targeted
nanoparticles. Nature. 464:1067–1070. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Gao W, Xiao Z, Radovic-Moreno A, Shi J,
Langer R and Farokhzad OC: Progress in siRNA delivery using
multifunctional nanoparticles. Methods Mol Biol. 629:53–67. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhou J, Shum KT, Burnett JC and Rossi JJ:
Nanoparticle-based delivery of RNAi therapeutics: Progress and
challenges. Pharmaceuticals. 6:85–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Abakumov MA, Nukolova NV, Sokolsky-Papkov
M, Shein SA, Sandalova TO, Vishwasrao HM, Grinenko NF, Gubsky IL,
Abakumov AM, Kabanov AV, et al: VEGF-targeted magnetic
nanoparticles for MRI visualization of brain tumor. Nanomedicine.
11:825–833. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Yohan D and Chithrani BD: Applications of
nanoparticles in nanomedicine. J Biomed nanotechnol. 10:2371–2392.
2014. View Article : Google Scholar
|
|
116
|
Pridgen EM, Alexis F and Farokhzad OC:
Polymeric nanoparticle technologies for oral drug delivery. Clin
Gastroenterol Hepatol. 12:1605–1610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wright J: Deliver on a promise. Sci Am.
311:S12–S13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhang L, Gu FX, Chan JM, Wang AZ, Langer
RS and Farokhzad OC: Nanoparticles in medicine: Therapeutic
applications and developments. Clin Pharmacol Ther. 83:761–769.
2008. View Article : Google Scholar
|
|
119
|
Aoyama T, Omori T, Watabe S, Shioya A,
Ueno T, Fukuda N and Matsumoto Y: Pharmacokinetic/pharmacodynamic
modeling and simulation of rosuvastatin using an extension of the
indirect response model by incorporating a circadian rhythm. Biol
Pharm Bull. 33:1082–1087. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Chetty M, Rose RH, Abduljalil K, Patel N,
Lu G, Cain T, Jamei M and Rostami-Hodjegan A: Applications of
linking PBPK and PD models to predict the impact of genotypic
variability, formulation differences, differences in target binding
capacity and target site drug concentrations on drug responses and
variability. Front Pharmacol. 5:2582014. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Rose RH, Neuhoff S, Abduljalil K, Chetty
M, Rostami-Hodjegan A and Jamei M: Application of a physiologically
based pharmacokinetic model to predict OATP1B1-related variability
in pharmacodynamics of rosuvastatin. CPT Pharmacometrics Syst
Pharmacol. 3:e1242014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Abbad S, Wang C, Waddad AY, LV H and Zhou
J: Preparation, in vitro and in vivo evaluation of polymeric
nanoparticles based on hyaluronic acid-poly(butyl cyanoacrylate)
and D-alphatocopheryl polyethylene glycol 1000 succinate for
tumor-targeted delivery of morin hydrate. Int J Nanomedicine.
10:305–320. 2015.
|
|
123
|
Abouzeid AH, Patel NR, Sarisozen C and
Torchilin VP: Transferrin-targeted polymeric micelles co-loaded
with curcumin and paclitaxel: Efficient killing of
paclitaxel-resistant cancer cells. Pharm Res. 31:1938–1945. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Acharya S and Sahoo SK: PLGA nanoparticles
containing various anticancer agents and tumour delivery by EPR
effect. Adv Drug Deliv Rev. 63:170–183. 2011. View Article : Google Scholar
|
|
125
|
Chapman VP, Stylianopoulos T, Martin JD,
Popović Z, Chen O, Kamoun WS, Bawendi MG, Fukumura D and Jain RK:
Normalization of tumour blood vessels improves the delivery of
nanomedicines in a size-dependent manner. Nat Nanotechnol.
7:383–388. 2012. View Article : Google Scholar
|
|
126
|
Jain RK and Stylianopoulos T: Delivering
nanomedicine to solid tumors. Nat Rev Clin Oncol. 7:653–664. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Olivares-Morales A, Kamiyama Y, Darwich
AS, Aarons L and Rostami-Hodjegan A: Analysis of the impact of
controlled release formulations on oral drug absorption, gut wall
metabolism and relative bioavailability of CYP3A substrates using a
physiologically-based pharmacokinetic model. Eur J Pharm Sci.
67:32–44. 2015. View Article : Google Scholar
|
|
128
|
Kyrodimou M, Andreadis D, Drougou A,
Amanatiadou EP, Angelis L, Barbatis C, Epivatianos A and
Vizirianakis IS: Desmoglein-3/γ-catenin and E-cadherin/β-catenin
differential expression in oral leukoplakia and squamous cell
carcinoma. Clin Oral Investig. 18:199–210. 2014. View Article : Google Scholar
|
|
129
|
Hrkach J, Von Hoff D, Mukkaram Ali M,
Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M,
Horhota A, et al: Preclinical development and clinical translation
of a PSMA-targeted docetaxel nanoparticle with a differentiated
pharmacological profile. Sci Transl Med. 4:128ra392012. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hudachek SF and Gustafson DL:
Physiologically based pharmacokinetic model of lapatinib developed
in mice and scaled to humans. J pharmacokinet pharmacodyn.
40:157–176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Poulin P, Hop CE, Salphati L and Liederer
BM: Correlation of tissue-plasma partition coefficients between
normal tissues and subcutaneous xenografts of human tumor cell
lines in mouse as a prediction tool of drug penetration in tumors.
J Pharm Sci. 102:1355–1369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Zhou Q, Guo P, Kruh GD, Vicini P, Wang X
and Gallo JM: Predicting human tumor drug concentrations from a
preclinical pharmacokinetic model of temozolomide brain
disposition. Clin Cancer Res. 13:4271–4279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Castorina P, Carcò D, Guiot C and
Deisboeck TS: Tumor growth instability and its implications for
chemotherapy. Cancer Res. 69:8507–8515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Ganguly R and Puri IK: Mathematical model
for chemotherapeutic drug efficacy in arresting tumour growth based
on the cancer stem cell hypothesis. Cell Prolif. 40:338–354. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Hubbard ME and Byrne HM: Multiphase
modelling of vascular tumour growth in two spatial dimensions. J
Theor Biol. 316:70–89. 2013. View Article : Google Scholar
|
|
136
|
Johnson D, McKeever S, Stamatakos G,
Dionysiou D, Graf N, Sakkalis V, Marias K, Wang Z, Deisboeck TS,
Johnson D, et al: Dealing with diversity in computational cancer
modeling. Cancer Inform. 12:115–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Loessner D, Flegg JA, Byrne HM, Clements
JA and Hutmacher DW: Growth of confined cancer spheroids: A
combined experimental and mathematical modelling approach. Integr
Biol. 5:597–605. 2013. View Article : Google Scholar
|
|
138
|
Molina-Peña R and Álvarez MM: A simple
mathematical model based on the cancer stem cell hypothesis
suggests kinetic commonalities in solid tumor growth. PLoS One.
7:e262332012. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Sakkalis V, Sfakianakis S, Tzamali E,
Marias K, Stamatakos G, Misichroni F, Ouzounoglou E, Kolokotroni E,
Dionysiou D, Johnson D, et al: Web-based workflow planning platform
supporting the design and execution of complex multiscale cancer
models. IEEE J Biomed Health Inform. 18:824–831. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Sturrock M, Hao W, schwartzbaum J and
rempala GA: A mathematical model of pre-diagnostic glioma growth. J
Theor Biol. 380:299–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Tzamali E, Grekas G, Marias K and Sakkalis
V: Exploring the competition between proliferative and invasive
cancer phenotypes in a continuous spatial model. PLoS One.
9:e1031912014. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
He Q, Guo S, Qian Z and Chen X:
Development of individualized anti-metastasis strategies by
engineering nanomedicines. Chem Soc Rev. 44:6258–6286. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Hu C, Niestroj M, Yuan D, Chang S and Chen
J: Treating cancer stem cells and cancer metastasis using
glucose-coated gold nanoparticles. Int J nanomedicine.
10:2065–2077. 2015.PubMed/NCBI
|
|
144
|
Landesman-Milo D, Ramishetti S and Peer D:
Nanomedicine as an emerging platform for metastatic lung cancer
therapy. Cancer Metastasis Rev. 34:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Patil R, Ljubimov AV, Gangalum PR, Ding H,
Portilla-Arias J, Wagner S, Inoue S, Konda B, Rekechenetskiy A,
Chesnokova A, et al: MRI virtual biopsy and treatment of brain
metastatic tumors with targeted nanobioconjugates: Nanoclinic in
the brain. ACS Nano. 9:5594–5608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Rychahou P, Shu Y, Haque F, Hu J, Guo P
and Evers BM: Methods and assays for specific targeting and
delivery of RNA nanoparticles to cancer metastases. Methods Mol
Biol. 1297:121–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Swami A, Reagan MR, Basto P, Mishima Y,
Kamaly N, Glavey S, Zhang S, Moschetta M, Seevaratnam D, Zhang Y,
et al: Engineered nanomedicine for myeloma and bone
microenvironment targeting. Proc Natl Acad Sci USA.
111:10287–10292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Sjögren E, Westergren J, Grant I, Hanisch
G, Lindfors L, Lennernäs H, Abrahamsson B and Tannergren C: In
silico predictions of gastrointestinal drug absorption in
pharmaceutical product development: Application of the mechanistic
absorption model GI-Sim. Eur J Pharm Sci. 49:679–698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Jayachandran D, Ramkrishna U, Skiles J,
Renbarger J and Ramkrishna D: Revitalizing personalized medicine:
Respecting biomolecular complexities beyond gene expression. CPT
Pharmacometrics Syst Pharmacol. 3:e1102014. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Xie L, Ge X, Tan H, Xie L, Zhang Y, Hart
T, Yang X and Bourne PE: Towards structural systems pharmacology to
study complex diseases and personalized medicine. PLOS Comput Biol.
10:e10035542014. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Gröning R, Remmerbach S and Jansen AC:
Telemedicine: Insulin pump controlled via the Global System for
Mobile Communications (GSM). Int J Pharm. 339:61–65. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Alomari M, Mohamed FH, Basit AW and
Gaisford S: Personalised dosing: Printing a dose of one's own
medicine. Int J pharm. 494:568–577. 2015. View Article : Google Scholar
|
|
153
|
Khaled SA, Burley JC, Alexander MR and
Roberts CJ: Desktop 3D printing of controlled release
pharmaceutical bilayer tablets. Int J Pharm. 461:105–111. 2014.
View Article : Google Scholar
|
|
154
|
Xitian P, Hongying L, Kang W, Yulin L,
Xiaolin Z and Zhiyu W: A novel remote controlled capsule for
site-specific drug delivery in human GI tract. Int J Pharm.
382:160–164. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Farandos NM, Yetisen AK, Monteiro MJ, Lowe
CR and Yun SH: Contact lens sensors in ocular diagnostics. Adv
Healthc Mater. 4:792–810. 2015. View Article : Google Scholar
|
|
156
|
Wening K and Breitkreutz J: Oral drug
delivery in personalized medicine: Unmet needs and novel
approaches. Int J pharm. 404:1–9. 2011. View Article : Google Scholar
|
|
157
|
Vizirianakis IS: Harnessing
pharmacological knowledge for personalized medicine and
pharmacotyping: Challenges and lessons learned. World J Pharmacol.
3:110–119. 2014. View Article : Google Scholar
|
|
158
|
Li M, Panagi Z, Avgoustakis K and Reineke
J: Physiologically based pharmacokinetic modeling of PLGA
nanoparticles with varied mPEG content. Int J nanomedicine.
7:1345–1356. 2012.PubMed/NCBI
|
|
159
|
Shelley ML, Wagner AJ, Hussain SM and
Bleckmann C: Modeling the in vivo case with in vitro nanotoxicity
data. Int J Toxicol. 27:359–367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Lin P, Chen JW, Chang LW, Wu JP, Redding
L, Chang H, Yeh TK, Yang CS, Tsai MH, Wang HJ, et al: Computational
and ultrastructural toxicology of a nanoparticle, Quantum Dot 705,
in mice. Environ Sci Technol. 42:6264–6270. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Péry AR, Brochot C, Hoet PH, Nemmar A and
Bois FY: Development of a physiologically based kinetic model for
99m-technetium-labelled carbon nanoparticles inhaled by humans.
Inhal Toxicol. 21:1099–1107. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Lankveld DP, Oomen AG, Krystek P, Neigh A,
Troost-de Jong A, Noorlander CW, Van Eijkeren JC, Geertsma RE and
De Jong WH: The kinetics of the tissue distribution of silver
nanoparticles of different sizes. Biomaterials. 31:8350–8361. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Mager DE, Mody V, Xu C, Forrest A, Lesniak
WG, Nigavekar SS, Kariapper MT, Minc L, Khan MK and Balogh LP:
Physiologically based pharmacokinetic model for composite
nanodevices: Effect of charge and size on in vivo disposition.
Pharm Res. 29:2534–2542. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Pascal J, Ashley CE, Wang Z, Brocato TA,
Butner JD, Carnes EC, Koay EJ, Brinker CJ and Cristini V:
Mechanistic modeling identifes drug-uptake history as predictor of
tumor drug resistance and nano-carrier-mediated response. ACS Nano.
7:11174–11182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Li D, Johanson G, Emond C, Carlander U,
Philbert M and Jolliet O: Physiologically based pharmacokinetic
modeling of polyethylene glycol-coated polyacrylamide nanoparticles
in rats. nanotoxicology. 8(suppl 1): S128–S137. 2014. View Article : Google Scholar
|
|
166
|
Sweeney LM, MacCalman L, Haber LT, Kuempel
ED and Tran CL: Bayesian evaluation of a physiologically-based
pharmacokinetic (PBPK) model of long-term kinetics of metal
nanoparticles in rats. Regul Toxicol Pharmacol. 73:151–163. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Bachler G, Losert S, Umehara Y, von Goetz
N, Rodriguez-Lorenzo L, Petri-Fink A, Rothen-Rutishauser B and
Hungerbuehler K: Translocation of gold nanoparticles across the
lung epithelial tissue barrier: Combining in vitro and in silico
methods to substitute in vivo experiments. Part Fibre Toxicol.
12:182015. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Lin Z, Monteiro-Riviere NA and Riviere JE:
A physiolo gically based pharmacokinetic model for polyethylene
glycol-coated gold nanoparticles of different sizes in adult mice.
Nanotoxicology. 11:1–11. 2015. View Article : Google Scholar
|