|
1
|
Zauber AG, Winawer SJ, O'Brien MJ,
Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH,
Schapiro M, Panish JF, et al: Colonoscopic polypectomy and
long-term prevention of colorectal-cancer deaths. N Engl J Med.
366:687–696. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Irving AA, Yoshimi K, Hart ML, Parker T,
Clipson L, Ford MR, Kuramoto T, Dove WF and Amos-Landgraf JM: The
utility of Apc-mutant rats in modeling human colon cancer. Dis
Model Mech. 7:1215–1225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thompson BA, Goldgar DE, Paterson C,
Clendenning M, Walters R, Arnold S, Parsons MT, Michael DW,
Gallinger S, Haile RW, et al Colon Cancer Family Registry: A
multifactorial likelihood model for MMR gene variant classification
incorporating probabilities based on sequence bioinformatics and
tumor characteristics: A report from the Colon Cancer Family
Registry. Hum Mutat. 34:200–209. 2013. View Article : Google Scholar :
|
|
4
|
Schneikert J, Ruppert JG, Behrens J and
Wenzel EM: Different roles for the axin interactions with the SAMP
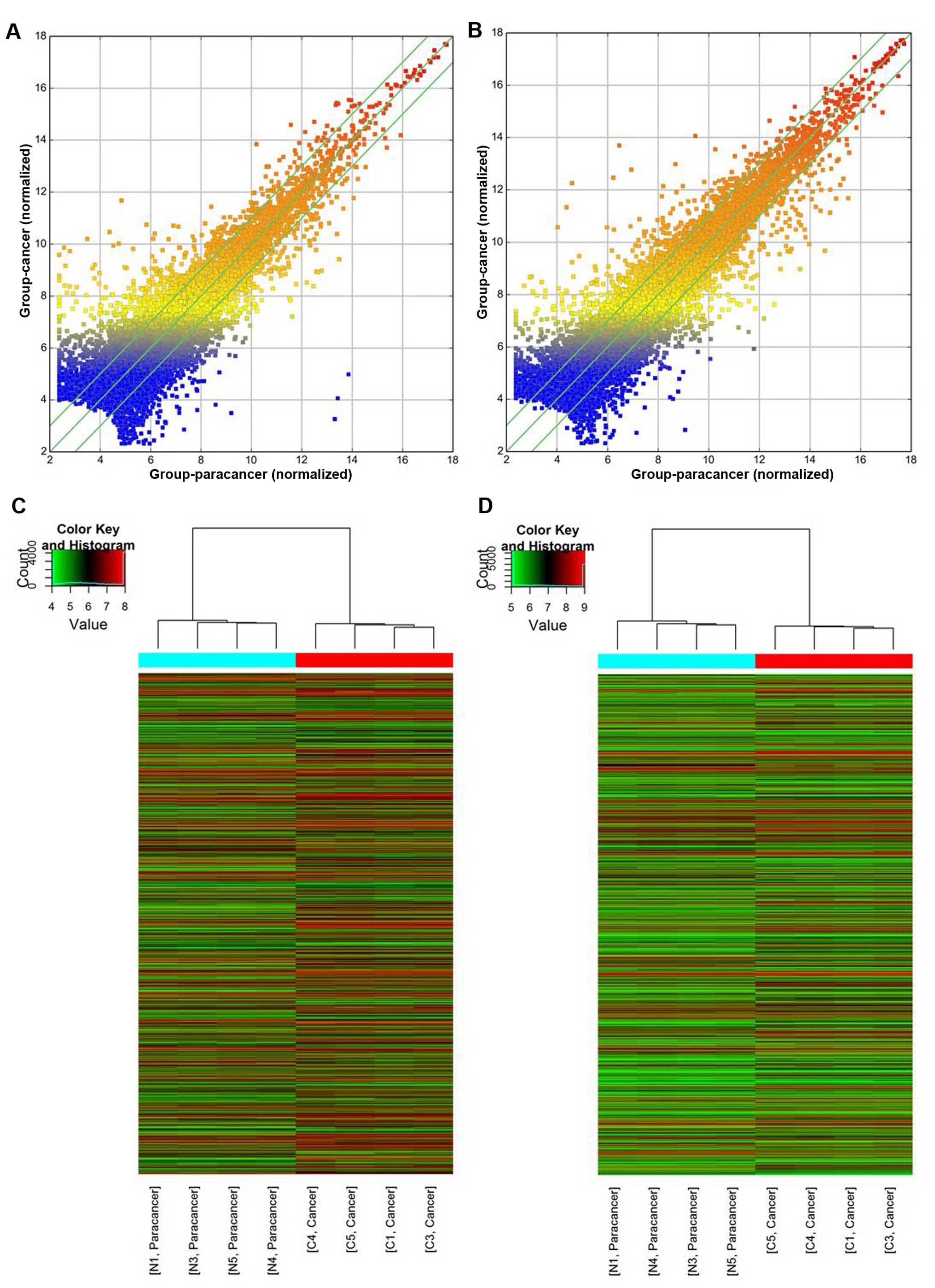
versus the second twenty amino acid repeat of adenomatous polyposis
coli. PLoS One. 9:e944132014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. Biochem Mol Biol Rep. 45:604–611.
2012.
|
|
6
|
Kim ED and Sung S: Long noncoding RNA:
Unveiling hidden layer of gene regulatory networks. Trends Plant
Sci. 17:16–21. 2012. View Article : Google Scholar
|
|
7
|
Clark MB and Mattick JS: Long noncoding
RNAs in cell biology. Semin Cell Dev Biol. 22:366–376. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu ZH, Wang XL, Tang HM, Jiang T, Chen J,
Lu S, Qiu GQ, Peng ZH and Yan DW: Long non-coding RNA HOTAIR is a
powerful predictor of metastasis and poor prognosis and is
associated with epithelial-mesenchymal transition in colon cancer.
Oncol Rep. 32:395–402. 2014.PubMed/NCBI
|
|
11
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
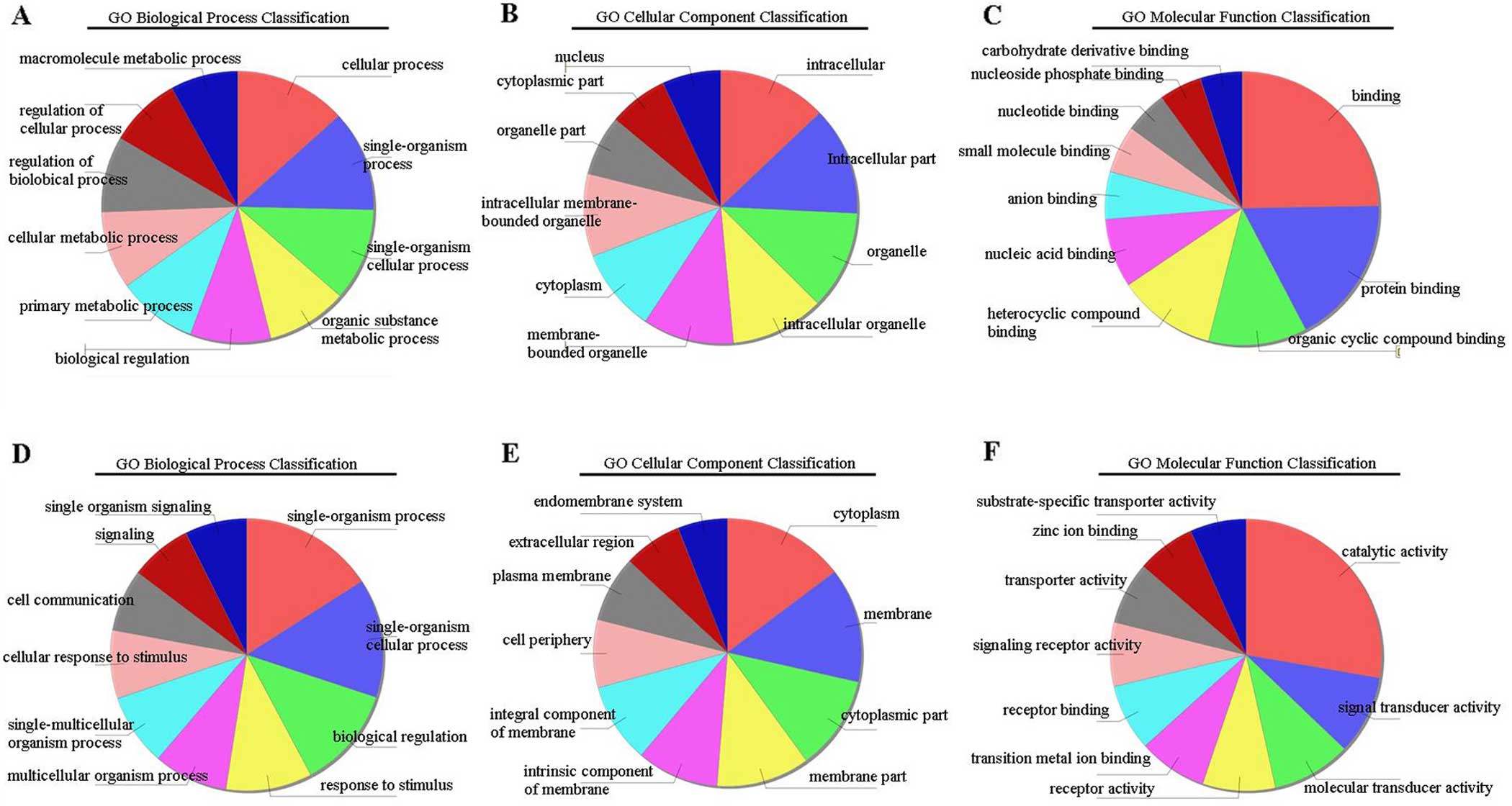
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium Nat Genet. 25:25–29. 2000.
|
|
13
|
Watanabe K, Biesinger J, Salmans ML,
Roberts BS, Arthur WT, Cleary M, Andersen B, Xie X and Dai X:
Integrative ChIP-seq/microarray analysis identifies a CTNNB1 target
signature enriched in intestinal stem cells and colon cancer. PLoS
One. 9:e923172014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de Sousa E, Melo F, Colak S, Buikhuisen J,
Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR,
Fessler E, van den Bergh SP, et al: Methylation of
cancer-stem-cell-associated Wnt target genes predicts poor
prognosis in colorectal cancer patients. Cell Stem Cell. 9:476–485.
2011. View Article : Google Scholar
|
|
15
|
Naba A, Clauser KR, Whittaker CA, Carr SA,
Tanabe KK and Hynes RO: Extracellular matrix signatures of human
primary metastatic colon cancers and their metastases to liver. BMC
Cancer. 14:5182014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang F, Ruan XJ and Zhang HY: BDE-99
(2,2′,4,4′,5-pentabro-modiphenyl ether) triggers
epithelial-mesenchymal transition in colorectal cancer cells via
PI3K/Akt/Snail signaling pathway. Tumori. 101:238–245. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Simpson ER: Sources of estrogen and their
importance. J Steroid Biochem Mol Biol. 86:225–230. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maglietta R, Liuzzi VC, Cattaneo E, Laczko
E, Piepoli A, Panza A, Carella M, Palumbo O, Staiano T, Buffoli F,
et al: Molecular pathways undergoing dramatic transcriptomic
changes during tumor development in the human colon. BMC Cancer.
12:6082012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brighenti E, Calabrese C, Liguori G,
Giannone FA, Trerè D, Montanaro L and Derenzini M: Interleukin 6
downregulates p53 expression and activity by stimulating ribosome
biogenesis: a new pathway connecting inflammation to cancer.
Oncogene. 33:4396–4406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bernatsky S, Ramsey-Goldman R, Labrecque
J, Joseph L, Boivin JF, Petri M, Zoma A, Manzi S, Urowitz MB,
Gladman D, et al: Cancer risk in systemic lupus: An updated
international multi-centre cohort study. J Autoimmun. 42:130–135.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
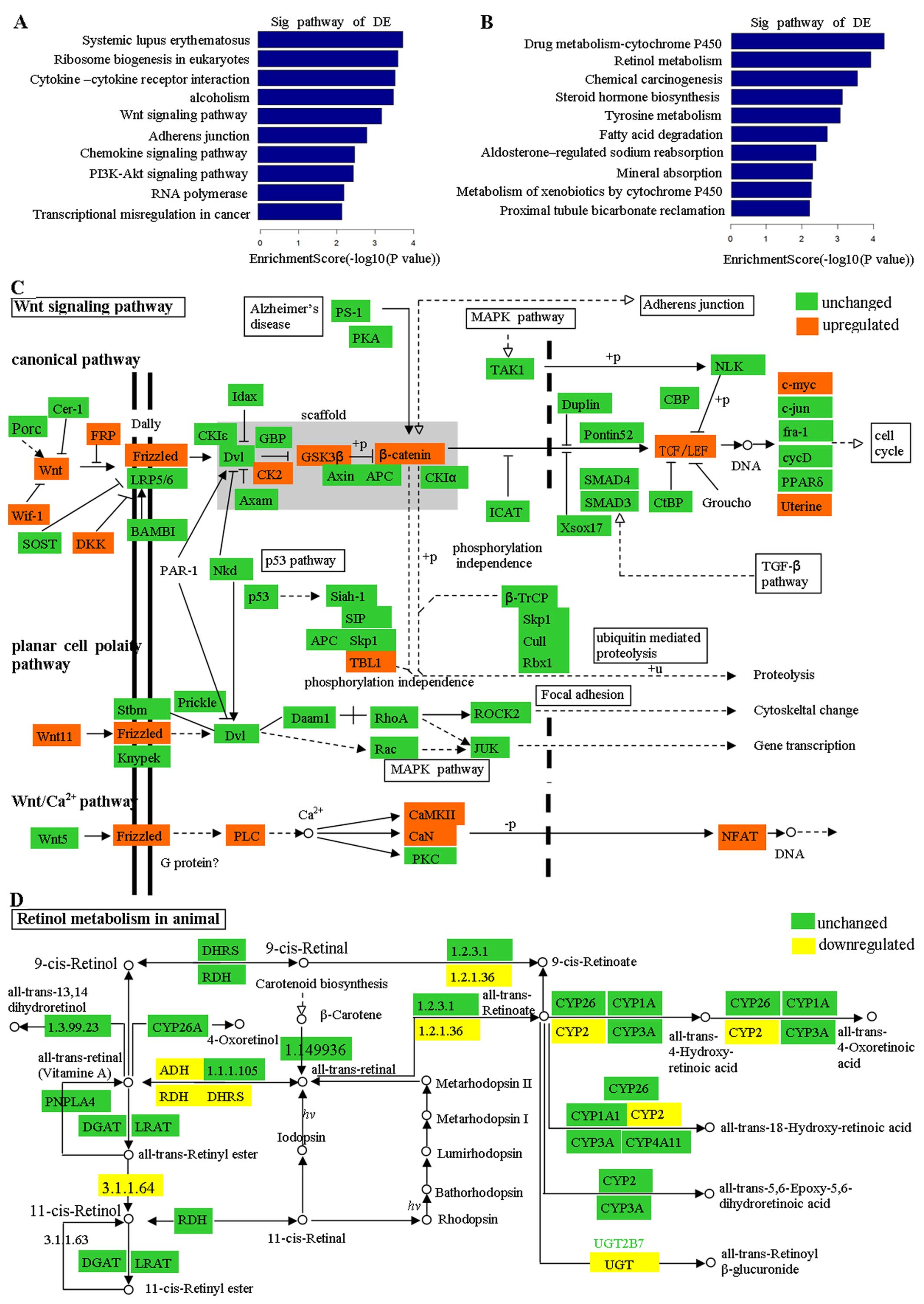
Bienz M and Clevers H: Linking colorectal
cancer to Wnt signaling. Cell. 103:311–320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Polakis P: Wnt signaling in cancer. Cold
Spring Harb Perspect Biol. 4:a0080522012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.PubMed/NCBI
|
|
24
|
Siamakpour-Reihani S, Caster J, Bandhu
Nepal D, Courtwright A, Hilliard E, Usary J, Ketelsen D, Darr D,
Shen XJ, Patterson C, et al: The role of calcineurin/NFAT in SFRP2
induced angiogenesis - a rationale for breast cancer treatment with
the calcineurin inhibitor tacrolimus. PLoS One. 6:e204122011.
View Article : Google Scholar
|
|
25
|
Ebert MP, Tänzer M, Balluff B,
Burgermeister E, Kretzschmar AK, Hughes DJ, Tetzner R, Lofton-Day
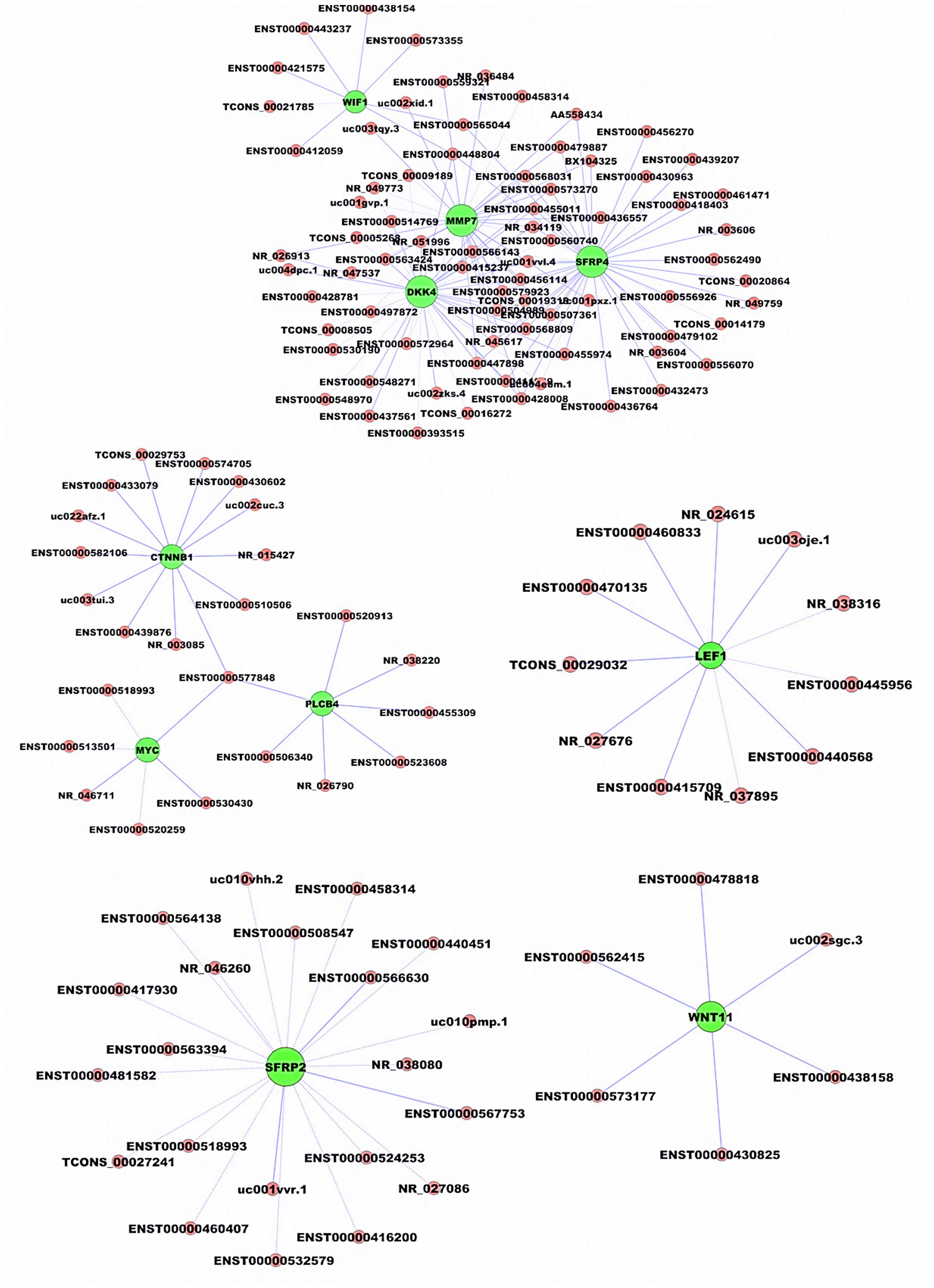
C, Rosenberg R, Reinacher-Schick AC, et al: TFAP2E-DKK4 and
chemoresistance in colorectal cancer. N Engl J Med. 366:44–53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ford CE, Jary E, Ma SS, Nixdorf S,
Heinzelmann-Schwarz VA and Ward RL: The Wnt gatekeeper SFRP4
modulates EMT, cell migration and downstream Wnt signalling in
serous ovarian cancer cells. PLoS One. 8:e543622013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Warrier S, Bhuvanalakshmi G, Arfuso F,
Rajan G, Millward M and Dharmarajan A: Cancer stem-like cells from
head and neck cancers are chemosensitized by the Wnt antagonist,
sFRP4, by inducing apoptosis, decreasing stemness, drug resistance
and epithelial to mesenchymal transition. Cancer Gene Ther.
21:381–388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu Y, Bai J, Li Z, Wang F, Cao L, Liu C,
Yu S, Yu G and Zhang Y: Low expression of secreted frizzled-related
protein 4 in aggressive pituitary adenoma. Pituitary. 18:335–342.
2015. View Article : Google Scholar
|
|
29
|
Huang D, Yu B, Deng Y, Sheng W, Peng Z,
Qin W and Du X: SFRP4 was overexpressed in colorectal carcinoma. J
Cancer Res Clin Oncol. 136:395–401. 2010. View Article : Google Scholar
|
|
30
|
Hartman J, Edvardsson K, Lindberg K, Zhao
C, Williams C, Ström A and Gustafsson JA: Tumor repressive
functions of estrogen receptor beta in SW480 colon cancer cells.
Cancer Res. 69:6100–6106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rawłuszko AA, Antoniucci M, Horbacka K,
Lianeri M, Krokowicz P and Jagodziński PP: Reduced expression of
steroid sulfatase in primary colorectal cancer. Biomed
Pharmacother. 67:577–582. 2013. View Article : Google Scholar
|
|
32
|
Shimodaira M, Nakayama T, Sato I, Sato N,
Izawa N, Mizutani Y, Furuya K and Yamamoto T: Estrogen synthesis
genes CYP19A1, HSD3B1, and HSD3B2 in hypertensive disorders of
pregnancy. Endocrine. 42:700–707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee HK, Hsu AK, Sajdak J, Qin J and
Pavlidis P: Coexpression analysis of human genes across many
microarray data sets. Genome Res. 14:1085–1094. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Villegas VE and Zaphiropoulos PG:
Neighboring gene regulation by antisense long non-coding RNAs. Int
J Mol Sci. 16:3251–3266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang D, Chen N, Peng M, Xu Q and Zhou J:
Construction of a eukaryotic expression vector of TM4SF1 and its
effect on migration and invasion of colorectal cancer cells. Nan
Fang Yi Ke Da Xue Xue Bao. 34:847–851. 2014.In Chinese. PubMed/NCBI
|
|
38
|
Tacha D, Zhou D and Cheng L: Expression of
PAX8 in normal and neoplastic tissues: A comprehensive
immunohistochemical study. Appl Immunohistochem Mol Morphol.
19:293–299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nomura M1, Shiiba K, Katagiri C, Kasugai
I, Masuda K, Sato I, Sato M, Kakugawa Y, Nomura E, Hayashi K, et
al: Novel function of MKP-5/DUSP10, a phosphatase of
stress-activated kinases, on ERK-dependent gene expression, and
upregulation of its gene expression in colon carcinomas. Oncol Rep.
28:931–936. 2012.PubMed/NCBI
|
|
40
|
He G, Zhang L, Li Q and Yang L:
miR-92a/DUSP10/JNK signaling axis promotes human pancreatic cancer
cells proliferation. Biomed Pharmacother. 68:25–30. 2014.
View Article : Google Scholar
|
|
41
|
Zhang D, Jiang P, Xu Q and Zhang X:
Arginine and glutamate-rich 1 (ARGLU1) interacts with mediator
subunit 1 (MED1) and is required for estrogen receptor-mediated
gene transcription and breast cancer cell growth. J Biol Chem.
286:17746–17754. 2011. View Article : Google Scholar : PubMed/NCBI
|