Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney tumor and accounts for nearly 80% of all renal cancer
cases in adults (1). It is
described as one of the most lethal urological cancers worldwide
(2). Although various small renal
tumors can be detected at an early stage, one in 3 RCC tumors is
detected at a later metastatic stage (3). These late-stage tumors are associated
with a poor prognosis due to the fact that metastatic RCC is highly
resistant to many therapies including radiotherapy and chemotherapy
(4,5). Although recently developed targeted
therapies for advanced RCC have achieved certain improvement in the
treatment of selective patients, the majority of advanced RCC
patients remain refractory to treatment (6,7). Thus,
understanding the molecular mechanisms involved in RCC progression
is important in order to identify new targets for novel and more
effective therapy.
Emerging evidence shows that microRNAs (miRNAs), a
group of small non-coding RNAs of ~22 nucleotides in length,
negatively regulate gene expression, primarily by targeting the
3′-untranslated region (3′-UTR) of messenger RNAs (mRNAs) (8,9).
miRNAs are known to contribute to tumorigenesis in a wide spectrum
of human cancers, including RCC (10). Recent studies have demonstrated the
regulatory functions of miRNAs in RCC cell proliferation (11–13),
apoptosis (14,15), invasion and metastasis (16–18). A
predominant and systemic alteration in miRNA expression during
renal carcinogenesis has been indicated by studies of miRNA
expression profiling (16).
miR-204 is an miRNA which is enriched in germline
and mesoderm-derived tissues, such as the testis, uterus, kidney,
lung, heart and bladder (19).
Numerous studies have shown that miR-204 is downregulated and acts
as a tumor suppressor in various human malignancies, including head
and neck (20), gastric (21) and pancreatic cancer (22). Recent studies indicate that miR-204
is downregulated in RCC and is a critical regulator of tumor growth
via inhibition of autophagy (23),
implying the potential role of miR-204 in renal tumors. However,
the exact function of miR-204 in RCC remains elusive.
In the present study, we identified miR-204 as one
of the most significantly downregulated miRNAs in RCC tissues and
cells and a critical suppressor of RCC cell proliferation and
invasion both in vitro and in vivo. We further
demonstrated that attenuation of miR-204 expression in RCC is
associated with RAB22A as a potential candidate target gene. In
addition, our data based on clinical RCC tissues indicated that
miR-204 may be a potential biomarker for discriminating between RCC
and normal tissues.
Materials and methods
Human samples
Surgical specimens (paired normal and cancerous
tissues) were obtained from 65 patients with kidney tumors at the
Department of Urology, Chengdu Third People Hospital and were
freshly frozen in liquid nitrogen until use. Informed consent was
obtained from the patients, and the study was approved by the
Institutional Review Board of Sichuan University.
Cell culture and transfection
The human RCC cell lines, OSRC-2, 786-O, ACHN, A498
and 769P, and the primary renal tubular cell line HK2 were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The OSRC-2, 786-O, ACHN, A498, 769P and HK2
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS) in a humidified 5%
CO2 environment at 37°C.
The miRNA-204 expression plasmid was generated by
cloning the genomic pre-miR-204 gene, with a 200-bp sequence on
each flanking side, into retroviral transfer plasmid pWPI to
generate the plasmid pWPI-miR-204.
To generate RAB22A-overexpressing stable clones,
OSRC-2 and 786-O cells were transfected with lentiviral vectors,
pWPI-RAB22A/pWPI-Vec, with the PAX2 packaging plasmid and PMD2G
envelope plasmid. Then, these were transfected into OSRC-2 and
786-O cells for 48 h to obtain a lentivirus soup. The lentivirus
soup was collected and frozen at −80°C for use. Anti-miR-204 or
negative control inhibitors (RiboBio Co., Ltd.) were transfected
into confluent cells with Lipofectamine 3000 (Life Technologies,
Inc.). To obtain stably infected OSRC-2 and 786-O cells, the cells
were cultured to ~70% of the plates, and then added by a
concentration of 1.0×104 cells with miR-204 and mock or
anti-miR-204 and anti-miR-NC.
Real-time RT-PCR assays
The miRNA RT-PCR assays were performed using the
TaqMan miRNA reverse transcription kit and the 7500 Fast Real-Time
PCR system (both from Applied Biosystems) for quantitative miRNA
detection. Each miRNA TaqMan PCR probe was purchased from Applied
Biosystems. The real-time RT-PCR assays were performed using the
7500 Fast Real-Time PCR system for quantitative mRNA detection and
with iTaq Fast SYBR-Green Supermix (Bio-Rad). The primers used for
real-time PCR and vector construction are listed in Table I.
 | Table IPrimers used for real-time PCR and
vector construction. |
Table I
Primers used for real-time PCR and
vector construction.
| Primer names | Sequences |
|---|
| miR-204 |
5′-UUCCCUUUGUCAUCCUAUGCCU-3′ |
| RAB22A | F
5′-TTGTAGTTGCCATTGCAGGA-3′ |
| R
5′-AGGCTGTCTTCGGAGTTTGA-3′ |
| GAPDH | F
5′-GGAGCGAGATCCCTCCAAAAT-3′ |
| R
5′-GGCTGTTGTCATACTTCTCATGG-3′ |
| miR-204
inhibitor |
GGCUACAGUCUUUCUUCAUGUGACUCGUGGACUUCCCUUU |
| oe-miR-204 | F
5′-CCTTAATTAAGAAGGCAAAGGGACGTTCAA-3′ |
| R
5′-CCGGCGCGCCGTTTAAACTTCATCCAAGCAGCTCTGGA-3′ |
| oe-RAB22A | F
5′-CCTTAATTAAATGGCGCTGAGGGAGCTCAAAGTGTGTC-3′ |
| R
5′-CCGGCGCGCCGTTTAAACTCAGCAGCAGCTCCGCTTTGGCTCTGA-3′ |
Western blot analysis
Cells were lysed in RIPA buffer and proteins (20
µg) were separated on 8–10% SDS/PAGE gel and then
transferred onto PVDF membranes (Millipore, Billerica, MA, USA).
After blocking the membranes, they were incubated with appropriate
dilutions of specific primary antibodies. The blots were then
incubated with HRP-conjugated secondary antibodies and visualized
using an ECL system (Thermo Fisher Scientific, Rochester, NY, USA).
Immunoblots were performed with primary monoclonal antibodies
RAB22A (Abcam, Cambridge, UK) and GAPDH (Sigma, USA).
RCC cell invasion assay
The invasion capability of RCC cells was determined
by the Transwell assay. Before seeding the cells, 10 ml of Matrigel
(BD, Inc.) was dissolved in 50 ml serum-free DMEM, applied to the
upper chamber of 8-mm pore-size polycarbonate membrane filters
(Corning, Inc., Corning, NY, USA), and put into an incubator for 5
h. RCC cells were then harvested and seeded with serum-free DMEM
into the upper chamber at 1×105 cells/well, and the
bottom chamber of the apparatus contained DMEM with 10% FBS. The
Transwells were then incubated for 48 h at 37°C. Following
incubation, the invaded cells that had attached to the lower
surface of the membrane were fixed with 4% paraformaldehyde and
stained with 1% toluidine blue. Cell numbers were counted in five
randomly chosen microscopic fields (magnification, ×100) per
membrane.
RCC cell proliferation assay
RCC cells were seeded in 24-well plates (3,000
cells/well) and cultured for 24, 48, 72 and 96 h. Cells were
harvested, and the cell number was calculated using MTT agent. DMSO
was used as the control.
Luciferase assay
Cells were cultured in 24-well plates and
transfected with 0.2 mg of either wild-type or mutant 3′-UTR of
RAB22A. The plasmid contained firefly luciferase. After 48 h of
transfection, the cells were lysed with 1X reporter lysis buffer
and firefly and Renilla luciferase activities were measured
using the Dual Luciferase Reporter Assay (Promega) according to the
manufacturer's instructions. Firefly luciferase activity was
standardized to the Renilla activity as a control. All
transfection experiments were conducted in triplicate and repeated
3 times independently.
In vivo metastasis studies
Male 6- to 8-week-old nude mice were purchased from
NCI. Sixteen mice were divided into 2 groups (n=8). The first group
of mice was injected into the renal capsule with 1×106
oe-miR-204 OSRC-2 cells (mixture with Matrigel, 1:1); the second
group of mice was injected with 1×106 miR-NC OSRC-2
cells. Metastasis in mice was assessed using a fluorescent imager
(IVIS Spectrum; Caliper Life Sciences, Hopkinton, MA, USA) at 4
different time points (1, 4, 5 and 6 weeks after injection). After
monitoring with the imager, the mice were sacrificed and the
metastatic sites were further examined.
Statistical analysis
Data are expressed as the mean ± SEM from at least 3
independent experiments. Statistical analyses were carried out with
a paired t-test using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). In
the in vivo study, measurements of tumor metastasis among
the 3 groups were analyzed by one-way ANOVA coupled with the
Newman-Keuls test. P<0.05 was considered to indicate a
statistically significant result.
Results
miR-204 is downregulated in RCC tissues
and cell lines
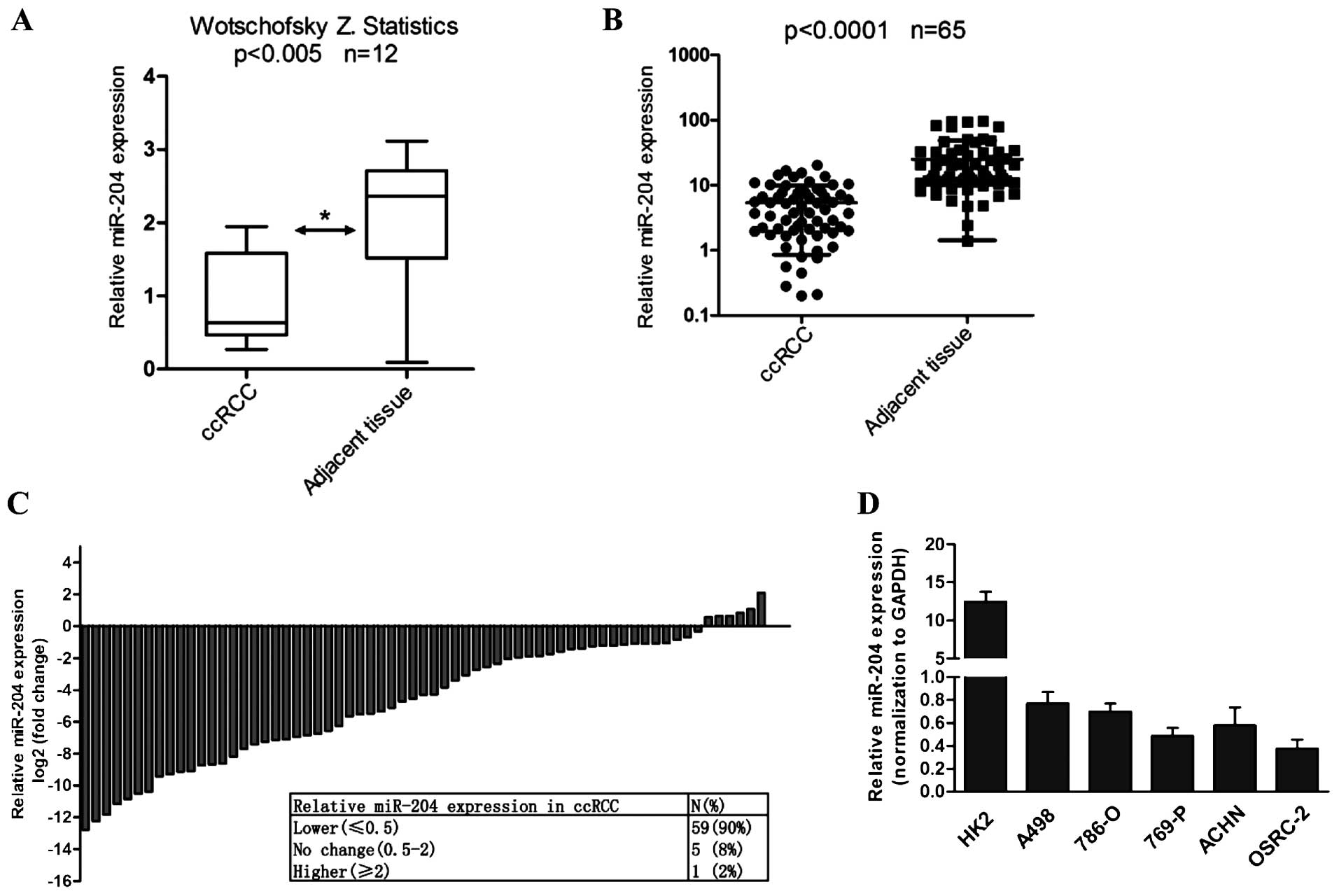
We firstly analyzed the expression of miR-204 in 65
human RCC tissues with paired adjacent normal tissues by real-time
PCR. Consistent with the statistical result of Wotschofsky et
al from GSE37989 in GEO database (24) (Fig.
1A), real-time PCR indicated that miR-204 expression was
markedly downregulated in 90% (59/65) of the RCC tissues
(P<0.0001; Fig. 1B and C).
Although the miR-204 mRNA level was not associated with tumor side,
its expression was found to be gradually decreased during tumor
progression (Table II). Notably,
miR-204 was lowly expressed in RCC cell lines, including A498,
OSRC-2, 769P, ACHN and OSRC-2 cells when compared with its
expression level in the HK2 cell line (Fig. 1D). The above results revealed that
miR-204 is downregulated in RCC, in keeping with the previous study
that the miR-204 level was downregulated in RCC compared with
matched normal renal tissues (23).
 | Table IIThe relative miR-204 expression
levels in the RCC samples. |
Table II
The relative miR-204 expression
levels in the RCC samples.
| Parameters | miR-204 expression
|
|---|
| N | High | Low | P-value |
|---|
| Gender | | | | |
| Male | 40 | 18 | 22 | 0.128 |
| Female | 25 | 12 | 13 | |
| Age (years) | | | | |
| <60 | 35 | 15 | 20 | 0.212 |
| ≥60 | 30 | 12 | 18 | |
| Tumor size
(cm) | | | | |
| ≤4 | 16 | 10 | 6 | 0.191 |
| >4 and ≤7 | 28 | 13 | 15 | |
| >7 | 21 | 10 | 11 | |
| pT stage | | | | |
| T1–2 | 42 | 18 | 24 | 0.036a |
| T3–4 | 23 | 15 | 8 | |
| Fuhrman | | | | |
| I–II | 41 | 25 | 16 | 0.042a |
| III–IV | 24 | 16 | 8 | |
| Tumor side | | | | |
| Left | 30 | 15 | 15 | 0.345 |
| Right | 35 | 19 | 16 | |
| Metastasis | | | | |
| Positive | 48 | 14 | 34 | 0.023a |
| Negative | 17 | 5 | 12 | |
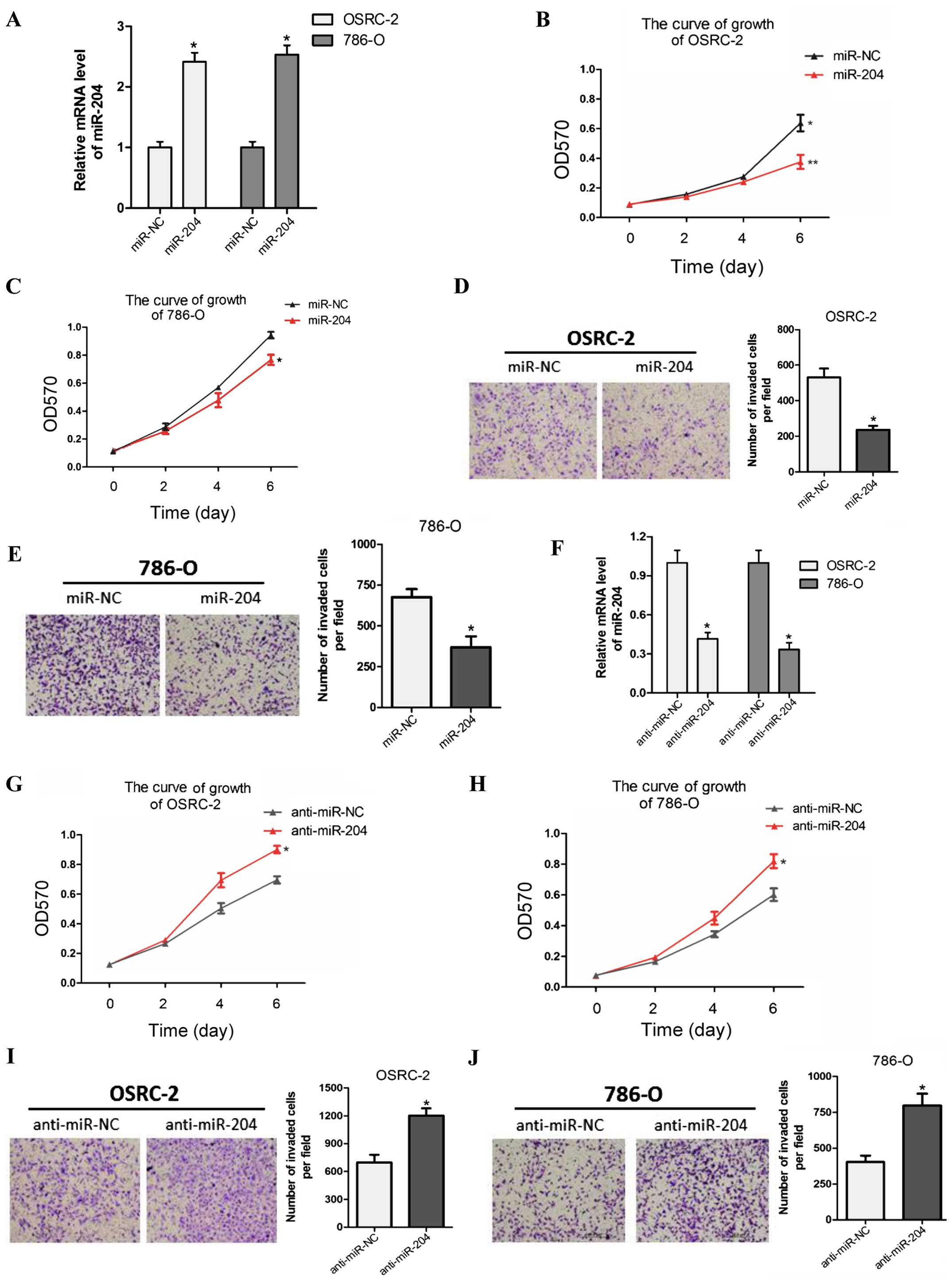
miR-204 attenuates RCC cell proliferation
and invasion in vitro
Previous research found that miR-204 suppressed RCC
proliferation via inhibition of LC3B-mediated autophagy (23). To explore the biological
significance of miR-204, we stably overexpressed miR-204 in two RCC
cell lines OSRC-2 and 786-O with lentiviruses carrying miR-204 and
its control (miR-NC). The efficacy of infection was tested by
qRT-PCR (Fig. 2A). Overexpression
of miR-204 markedly reduced RCC cell proliferation compared with
miR-NC in the OSRC-2 and 786-O cell lines (Fig. 2B and C). Similarly, miR-204
overexpression suppressed cell invasion in the two tested RCC cell
lines (Fig. 2D and E).
Conversely, we examine the phenotype in both cell
lines with anti-miR-204. RT-PCR indicated that anti-miR-204 was
effective when we treated the cells with miR-204 inhibitor
(Fig. 2F). MTT proliferation and
Transwell invasion assays showed that anti-miR-204 significantly
increased the growth and invasive capability of both RCC cell lines
compared with the anti-miR-NC group (Fig. 2G–J).
Taken together, these findings indicate that miR-204
acts as a tumor suppressor in RCC by inhibiting cell proliferation
and invasion.
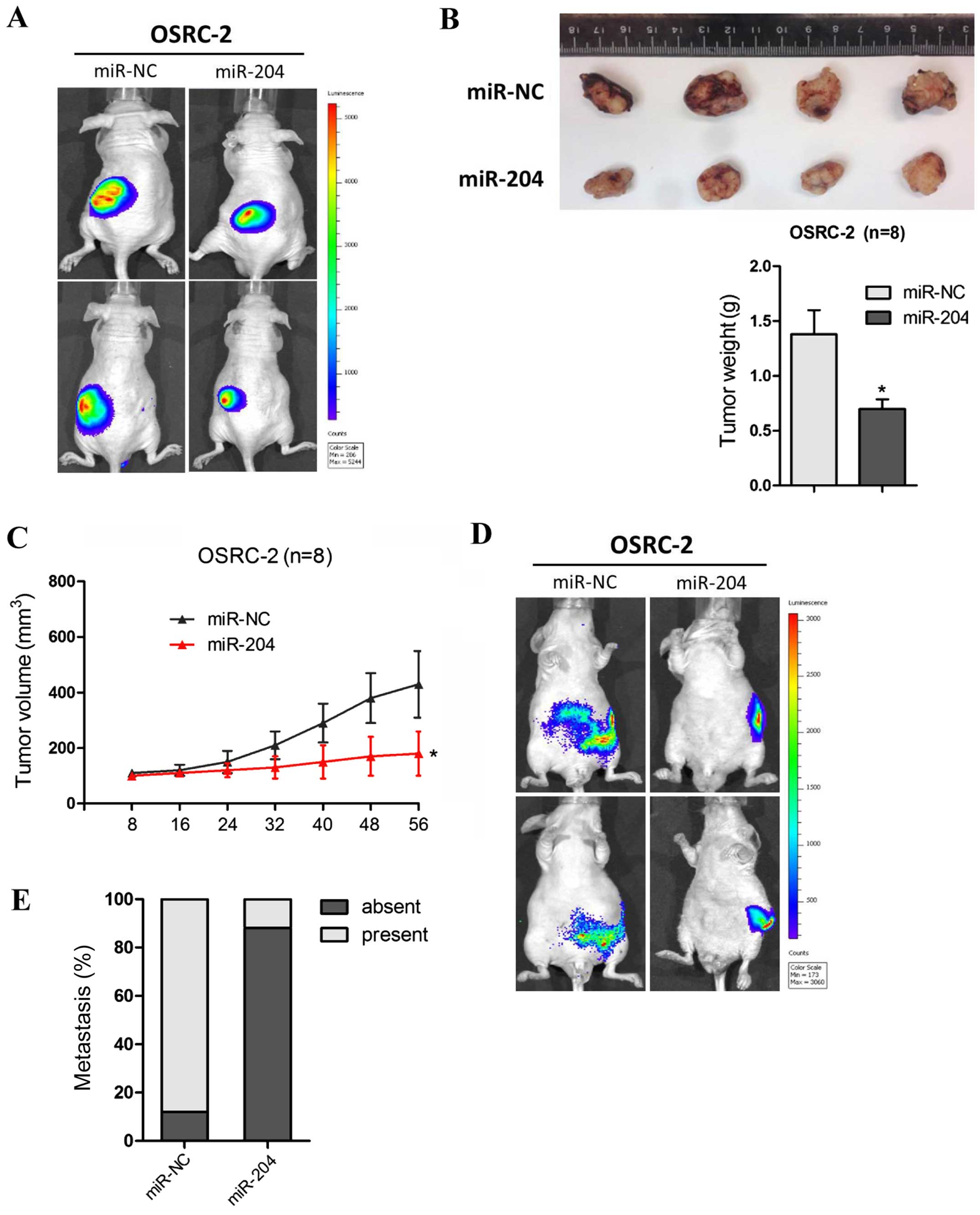
miR-204 suppresses RCC cell proliferation
and invasion in vivo
To identify the antitumorigenic roles of miR-204 in
RCC in vivo, OSRC-2 cells overexpressing miR-204 or cells
transfected with miR-NC were injected into the left kidney of nude
mice. Tumor growth and metastasis were then assessed by detecting
GFP expression using an automated fluorescence imaging system. As
shown in Fig. 3A, fluorescence
imaging showed a marked reduction in tumor growth in the
miR-204-overexpressing cells as early as week 3, as revealed by the
determination of tumor mass (Fig.
3B). Consistently, the tumor volume measurement also confirmed
the conclusion that overexpression of miR-204 inhibited
tumorigenicity of the OSRC-2 cells (Fig. 3C).
In addition, no macroscopic metastases were found in
the tumors with miR-204 overexpression while the miR-NC tumors were
prone to local invasion and metastasis through the automated
fluorescence imaging system (Fig.
3D). As shown in Fig. 3E, the
miR-204 tumors derived from the OSRC-2 cells metastasized to the
lung less frequently than the miR-NC tumors. These data above
further demonstrated that miR-204 functions as a critical tumor
suppressor in RCCs by suppressing tumorigenesis and metastatic
colonization.
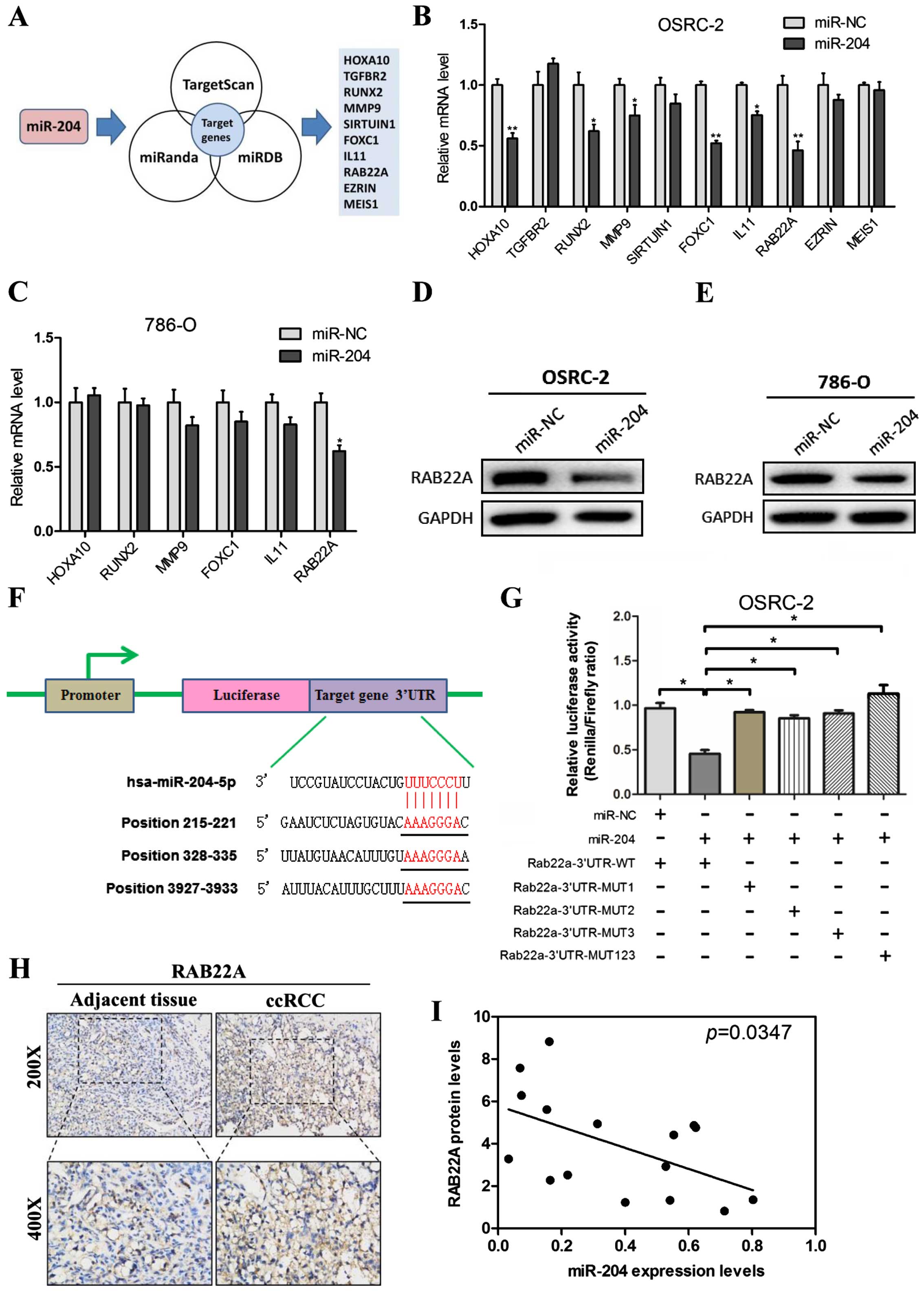
miR-204 directly targets RAB22A
To dissect the potential downstream gene which is
targeted by miR-204 in RCC cells, we predicted the potential
miR-204 targets using three bioinformatic analysis softwares
(TargetScan, miRanda and miRDB) (Fig.
4A), and explored the common 10 downregulated transcripts and
potential targets of miR-204 (HOXA10, TGFBR2, RUNX2, MMP9,
SIRTUIN1, FOXC1, IL11, RAB22A, EZRIN and MEIS1). To validate these
10 candidates, we firstly introduced miR-204 overexpression
lentiviruses into the OSRC-2 cell line. We next applied the RT-PCR
assay to validate the 11 predicted miRNA target genes in the OSRC-2
cells with overexpression of miR-204. We found 6 candidate genes
(HOXA10, RUNX2, MMP9, FOXC1, IL11 and RAB22A) that were
downregulated by overexpression of miR-204 in the OSRC-2
miR-204-transfected cells (Fig.
4B). We then used another RCC 786-O cell line to further
identify the candidate genes. Furthermore, RT-PCR analysis showed
that miR-204 led to a decrease in RAB22A expression in the 786-O
cell line (Fig. 4C).
To further examine RAB22A as a direct target gene of
miR-204, OSRC-2 and 786-O cells were transfected with miR-204
overexpression lentiviruses, respectively. The protein level of
RAB22A was substantially decreased after ectopic miR-204
transfection in both cell lines (Fig.
4D and E).
Based on TargetScan prediction analysis, 3
miR-204-targeting sites were identified in the 3′-UTR of RAB22A
mRNA (Fig. 4F), and mutant vectors
of RAB22A 3′-UTR containing four mutated bases on the predicted
binding sites were constructed (MUT-1, MUT-2, MUT-3 and MUT-1-3).
Transient transfection of OSRC-2 cells with the above wild-type or
mutant vectors and miR-204 or mock control resulted in partial
rescue of the inhibition (Fig.
4G).
Immunohistochemical staining was carried out to
assess RAB22A expression in the ccRCC tissues. Higher expression
was noted in the tissues than the level in the adjacent normal
tissues (Fig. 4H). miR-204
expression was negatively related to RAB22A protein expression in
the ccRCC samples (Fig. 4I).
Together, results from Fig. 4
demonstrated that RAB22A is a direct target of miR-204 and was
inversely correlated with the endogenous miR-204 expression in
primary ccRCC tissues.
miR-204-suppressed tumor proliferation
and invasion are modulated by RAB22A
The findings above indicate that RAB22A is a
candidate effector to mediate the biological functions of miR-204.
We then identified whether overexpression of RAB22A (oe-RAB22A)
could recapitulate the inhibitory effects of miR-204 on RCC cell
proliferation and invasion. OSRC-2 and 786-O cells were transfected
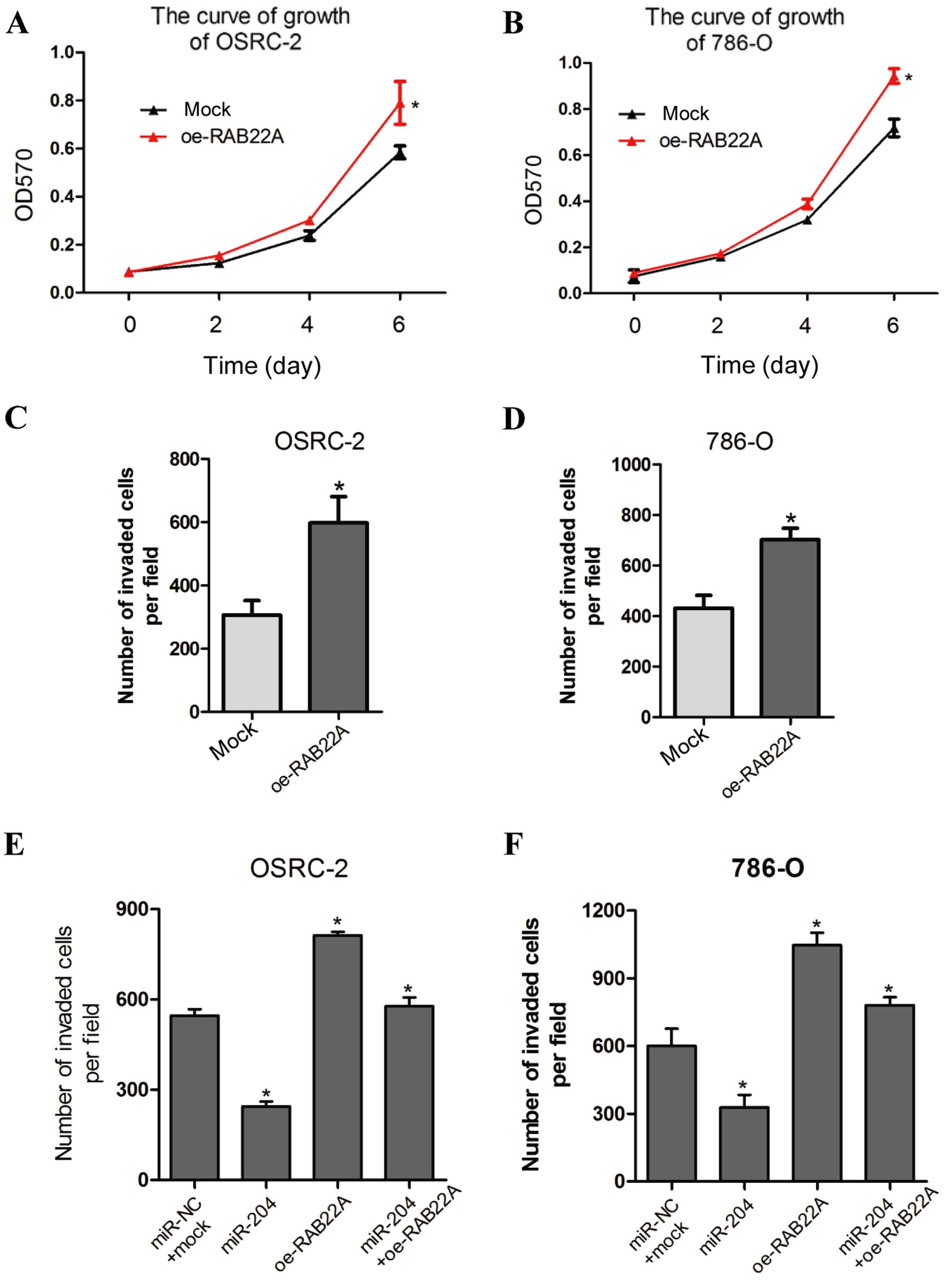
with oe-RAB22A vs. oe-mock. Significantly, oe-RAB22A in RCC cells
induced cell growth (Fig. 5A and
B), similarly to the phenotypic alterations upon miR-204
overexpression. In addition, oe-RAB22A led to enhance cell invasion
in the OSRC-2 and 786-O cells (Fig. 5C
and D).
To further determine whether RAB22A is a direct and
functional mediator of miR-204-suppressed cell invasion, we
performed a rescue experiment by co-transfection of oe-RAB22A (vs.
mock) and oe-miR-204 (vs. miR-NC) into the OSRC-2 and 786-O cell
lines. The enhancement in RCC cell invasion reduced by miR-204
overexpression was effectively reversed by oe-RAB22A (Fig. 5E and F). Collectively, these
findings indicate that RAB22A is an essential functional effector
of miR-204 in RCC.
Discussion
Recent studies have found that miRNAs play a
fundamental role in the proliferation, invasion and metastasis of
malignant human cancers (25),
involving renal cell carcinoma (RCC) (26). Previous studies indicate that
miR-204 is downregulated in various human tumors and acts as a
tumor suppressor negatively correlated with tumorigenesis and
progression, including endometrial and gastric cancer, and
nasopharyngeal carcinoma (27–29).
In the present study, we found that the expression of miR-204 in
RCC tissues and cell lines was downregulated compared to that in
surrounding normal tissues and normal HK2 cell lines, similar to a
study by Mikhaylova et al (23).
Next, results from in vitro and in
vivo experiments demonstrated that miR-204 markedly suppressed
the growth and invasion of RCC cell lines OSRC-2 and 786-O.
Furthermore, we noted that the lower miR-204 levels were associated
with promotion of RCC proliferation and invasion. Taken together,
all these data strongly support the conclusion that miR-204 can be
a potential biomarker in RCC progression by inhibition of cell
proliferation and invasion.
A previous study demonstrated that RAB22A is a
direct target of several miRNAs. Zhang et al found that
miR-373 targeting of the Rab22a oncogene suppressed tumor invasion
and metastasis in ovarian cancer (30). miR-203 has been reported as a
tumor-suppressor gene in osteosarcoma by regulating RAB22A
(31). The present study identified
that miR-204 inhibited RAB22A expression by directly targeting
3′UTR of RAB22A in RCC. Overexpression of miR-204 in RCC cell lines
led to decreased expression of RAB22A, and then suppressed RCC
proliferation and invasion. The interruption approach using RAB22A
overexpression reversed the oe-miR-204 effects on RCC OSRC-2 and
786-O cell proliferation and invasion. In contrast, knockdown of
miR-204 resulted in increased RAB22A expression and enhanced cell
growth and invasive capability in RCC. More importantly, the
expression of miR-204 was negatively correlated with the RAB22A
expression level in RCC tissues.
RAB22A, acting as a small GTPase, serves as a member
of the RAB family endocytic pathway (32). Several studies have shown that
RAB22A plays an oncogenic role in liver cancer and ovarian cancer
(30,33). The explored mechanism indicates that
RAB22A is mainly associated with EMT signaling to modulate cancer
cell progression. In addition, hypoxia-induced HIF-dependent RAB22A
expression is associated with breast cancer patient mortality
(34). However, the role of RAB22A
in RCC was not well established. Based on our findings,
overexpression of RAB22A promoted cell proliferation and invasion
in OSRC-2 and 786-O RCC cell lines. In conclusion, data presented
in the present study provided a firm foundation that RAB22A holds
great promise as a therapeutic target in RCC.
In summary, we demonstrated the marked alteration of
miR-204 expression in human RCC tissues and cell lines, and we
found that ectopic expression of miR-204 could efficiently suppress
cell proliferation and invasion of RCC cells in vitro and
in vivo by directly targeting RAB22A 3′-UTR. These specific
functional mediators may provide novel mechanisms in tumor
progression and additional diagnostic and/or therapeutic targets.
The present study extends our knowledge that the specification of
the miR-204-RAB22A pathway also has fundamental importance in RCC
progression.
Acknowledgments
The authors thank the patients and clinicians for
their contributions to the present study.
References
|
1
|
Compérat E and Camparo P: Histological
classification of malignant renal tumours at a time of major
diagnostic and therapeutic changes. Diagn Interv Imaging.
93:221–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramana J: RCDB: Renal Cancer Gene
Database. BMC Res Notes. 5:2462012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stadler WM: Targeted agents for the
treatment of advanced renal cell carcinoma. Cancer. 104:2323–2333.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Polyzos A: Activity of SU11248, a
multitargeted inhibitor of vascular endothelial growth factor
receptor and platelet-derived growth factor receptor, in patients
with metastatic renal cell carcinoma and various other solid
tumors. J Steroid Biochem Mol Biol. 108:261–266. 2008. View Article : Google Scholar
|
|
6
|
Atkins MB, Ernstoff MS, Figlin RA,
Flaherty KT, George DJ, Kaelin WG Jr, Kwon ED, Libermann TA,
Linehan WM, McDermott DF, et al: Innovations and challenges in
renal cell carcinoma: Summary statement from the Second Cambridge
Conference. Clin Cancer Res. 13:667s–670s. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Longo R, D'Andrea MR, Sarmiento R, Salerno
F and Gasparini G: Integrated therapy of kidney cancer. Ann Oncol.
18(Suppl 6): vi141–vi148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bagga S, Bracht J, Hunter S, Massirer K,
Holtz J, Eachus R and Pasquinelli AE: Regulation by let-7 and lin-4
miRNAs results in target mRNA degradation. Cell. 122:553–563. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stallings RL: MicroRNA involvement in the
pathogenesis of neuroblastoma: Potential for microRNA mediated
therapeutics. Curr Pharm Des. 15:456–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jung M, Mollenkopf HJ, Grimm C, Wagner I,
Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach
H, et al: MicroRNA profiling of clear cell renal cell cancer
identifies a robust signature to define renal malignancy. J Cell
Mol Med. 13:3918–3928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Majid S, Saini S, Dar AA, Hirata H,
Shahryari V, Tanaka Y, Yamamura S, Ueno K, Zaman MS, Singh K, et
al: MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal
cancer. Cancer Res. 71:2611–2621. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirata H, Hinoda Y, Ueno K, Nakajima K,
Ishii N and Dahiya R: MicroRNA-1826 directly targets beta-catenin
(CTNNB1) and MEK1 (MAP2K1) in VHL-inactivated renal cancer.
Carcinogenesis. 33:501–508. 2012. View Article : Google Scholar :
|
|
13
|
Doberstein K, Steinmeyer N, Hartmetz AK,
Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R,
Pfeilschifter J and Gutwein P: MicroRNA-145 targets the
metalloprotease ADAM17 and is suppressed in renal cell carcinoma
patients. Neoplasia. 15:218–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schickel R, Park SM, Murmann AE and Peter
ME: miR-200c regulates induction of apoptosis through CD95 by
targeting FAP-1. Mol Cell. 38:908–915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saini S, Yamamura S, Majid S, Shahryari V,
Hirata H, Tanaka Y and Dahiya R: MicroRNA-708 induces apoptosis and
suppresses tumorigenicity in renal cancer cells. Cancer Res.
71:6208–6219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koo S, Martin GS, Schulz KJ, Ronck M and
Toussaint LG: Serial selection for invasiveness increases
expression of miR-143/miR-145 in glioblastoma cell lines. BMC
Cancer. 12:1432012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar
|
|
18
|
Lu L, Xue X, Lan J, Gao Y, Xiong Z, Zhang
H, Jiang W, Song W and Zhi Q: MicroRNA-29a upregulates MMP2 in oral
squamous cell carcinoma to promote cancer invasion and
anti-apoptosis. Biomed Pharmacother. 68:13–19. 2014. View Article : Google Scholar
|
|
19
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu
Y, Li J, Hasina R, Cheng C, Lingen MW, et al: Network modeling
identifies molecular functions targeted by miR-204 to suppress head
and neck tumor metastasis. PLOS Comput Biol. 6:e10007302010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sacconi A, Biagioni F, Canu V, Mori F, Di
Benedetto A, Lorenzon L, Ercolani C, Di Agostino S, Cambria AM,
Germoni S, et al: miR-204 targets Bcl-2 expression and enhances
responsiveness of gastric cancer. Cell Death Dis. 3:e4232012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Z, Sangwan V, Banerjee S, Mackenzie
T, Dudeja V, Li X, Wang H, Vickers SM and Saluja AK: miR-204
mediated loss of Myeloid cell leukemia-1 results in pancreatic
cancer cell death. Mol Cancer. 12:1052013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et
al: VHL-regulated miR-204 suppresses tumor growth through
inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wotschofsky Z, Liep J, Meyer HA, Jung M,
Wagner I, Disch AC, Schaser KD, Melcher I, Kilic E, Busch J, et al:
Identification of metastamirs as metastasis-associated microRNAs in
clear cell renal cell carcinomas. Int J Biol Sci. 8:1363–1374.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.
|
|
26
|
Petillo D, Kort EJ, Anema J, Furge KA,
Yang XJ and Teh BT: MicroRNA profiling of human kidney cancer
subtypes. Int J Oncol. 35:109–114. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chung TK, Lau TS, Cheung TH, Yim SF, Lo
KW, Siu NS, Chan LK, Yu MY, Kwong J, Doran G, et al: Dysregulation
of microRNA-204 mediates migration and invasion of endometrial
cancer by regulating FOXC1. Int J Cancer. 130:1036–1045. 2012.
View Article : Google Scholar
|
|
28
|
Zhou X, Li L, Su J and Zhang G: Decreased
miR-204 in H. pylori-associated gastric cancer promotes cancer cell
proliferation and invasion by targeting SOX4. PLoS One.
9:e1014572014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma L, Deng X, Wu M, Zhang G and Huang J:
Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and
facilitates invasion of EBV-associated nasopharyngeal carcinoma
cells. FEBS Lett. 588:1562–1570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang D, Liu G and Wang K: miR-203 acts as
a tumor suppressor gene in osteosarcoma by regulating RAB22A. PLoS
One. 10:e01322252015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mesa R, Salomón C, Roggero M, Stahl PD and
Mayorga LS: Rab22a affects the morphology and function of the
endocytic pathway. J Cell Sci. 114:4041–4049. 2001.PubMed/NCBI
|
|
33
|
He H, Dai F, Yu L, She X, Zhao Y, Jiang J,
Chen X and Zhao S: Identification and characterization of nine
novel human small GTPases showing variable expressions in liver
cancer tissues. Gene Expr. 10:231–242. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang T, Gilkes DM, Takano N, Xiang L, Luo
W, Bishop CJ, Chaturvedi P, Green JJ and Semenza GL:
Hypoxia-inducible factors and RAB22A mediate formation of
microvesicles that stimulate breast cancer invasion and metastasis.
Proc Natl Acad Sci USA. 111:E3234–E3242. 2014. View Article : Google Scholar : PubMed/NCBI
|