Introduction
Gastric cancer (GC) is one of the most universal
malignancies, however the advances in recent decades have improved
long-term survival only slightly (1). Surgery and chemotherapy are the main
protocols for GC therapy; however, in many cases, the efficacy of
even the leading chemotherapy treatment is poor, regardless of
whether it is natural or acquired (2). The term 'multidrug resistance (MDR)'
is used to define this phenomenon and could provide an insight into
the reasons for the low 5-year survival rate of GC patients
(3,4). Pervasively, the molecular mechanisms
underlying MDR are complex, covering intricate courses, including
drug conveyance and drug-induced apoptosis (5). Until now, the mechanisms responsible
for MDR in GC have not been adequately determined.
MicroRNAs (miRNAs) are a type of non-coding RNAs
that are approximately 19–24 nucleotides long and that downregulate
the expression of genes by targeting the 3′-untranslated regions
(3′-UTRs) of particular mRNAs that are situated in the genomic
areas that are exclusively related to carcinoma (6,7),
miRNAs are consequently considered to be connected with
chemotherapy failure, and emerging evidence shows that MDR may be
regulated by altering miRNA (8–10). For
example, miR-23b-3p sensitizes GC cells to chemical agents by
regulating ATG12 and HMGB2 (11);
overexpression of miR-181b and miR-497 was able to increase the
sensitivity of cells to chemotherapy by targeting Bcl2, known as an
anti-apoptotic gene (12,13). Patnaik et al (14) proved that the expression of miR-1284
is much lower in lung adenocarcinoma than in their controls. Recent
research also suggested that compared with primary GC, miR-1284 is
downregulated in GC with lymph node metastases (15); however, there is still lack of
evidence as to the precise role of miR-1284 in MDR in GC.
To define the effects of miR-1284 in GC MDR, we
determined miR-1284 expression in GC tissue specimens with
metastasis and the vincristine-resistant (VCR) GC cell line SGC7901
(SGC7901/VCR). In addition, we established the SGC-7901/VCR cells
with a stable overexpression of miR-1284 and probed alterations in
IC50, cell cycles, apoptosis, and migration and
invasiveness. We also investigated the influence of miR-1284
overexpression on tumor growth in vivo. To ensure that the
underlying mechanism was identified, we surveyed the impact of
miR-1284 on the expression of genes associated with MDR; apoptosis;
and migration, including EIF4A1.
Materials and methods
Ethics statement
The study was approved by the Ethics Board of The
First Affiliated Hospital of Guangxi Medical University and
complied with the Declaration of Helsinki. All the patients gave
their written informed consent. The animal procedures were
conducted following the provisions of the Ethics Committee of The
First Affiliated Hospital of Guangxi Medical University in the
research. All efforts were made to minimize suffering.
Human tissue samples and cell lines
Gastric cancer tissues and adjacent non-tumor
tissues (located 5 cm away from the tumor) were collected during
surgery in The First Affiliated Hospital of Guangxi Medical
University (during in 2013–2015) and preserved in liquid nitrogen.
Vincristine-resistant SGC7901 (SGC7901/VCR) cells were obtained
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). SGC7901/VCR cells were cultivated in RPMI-1640 (GE
Healthcare Life Sciences, South Logan, UT, USA), with 50 mg/ml
penicillin, 100 mg/ml streptomycin, 10% fetal bovine serum, and 0.8
µg/ml vincristine to maintain drug resistance. Cells were
cultured at 37.8°C with 5.0% CO2.
Antibodies
Primary antibodies to EIF4A1 (1:1,000), JUN
(1:1,000), MMP12 (1:1,000), MYC (1:1,000), and GAPDH (1:1,000) were
provided by Abcam (Cambridge, UK). Secondary antibodies (1:10,000)
were provided by LI-COR Biosciences (Lincoln, NE, USA).
Quantitative reverse transcription
real-time polymerase chain reaction
RNA was extracted from SGC7901/VCR cells using
TRIzol (Invitrogen Corporation, Carlsbad, CA, USA). cDNA was
reverse transcribed from 1,000 ng RNA using the PrimeScript™ RT
reagent kit (Takara Bio, Inc., Tokyo, Japan). The mRNA or miRNA
expression levels were calculated by reference GAPDH (mRNA) or U6
(miRNA). All primer sequences, including those for miR-1284, U6,
EIF4A1, JUN, MMP12, MYC, and GAPDH, are listed in Table I. Quantitative reverse transcription
real-time polymerase chain reaction (qRT-PCR) methods were
developed on a SYBR® Premix Ex Taq™ II (Tli RNaseH Plus)
and a ROX Plus reagent kit (Takara Bio, Inc.) according to the
manufacturer's instructions. The mRNA and miRNA expressions were
analyzed using the 2−ΔΔCT method.
 | Table IThe sequences of primers for
quantitative reverse-transcriptase real-time polymerase chain
reaction. |
Table I
The sequences of primers for
quantitative reverse-transcriptase real-time polymerase chain
reaction.
| Gene | Primer
sequences |
|---|
| miR-1284 | F:
5′-CGTCTATACAGACCCTGGCTTTTC-3′ |
| R:
5′-CTCAACTGGTGTCGTGGA-3′ |
| U6 | F:
5′-TTATGGGTCCTAGCCTGAC-3′ |
| R:
5′-CACTATTGCGGGTCTGC-3′ |
| EAF4A1 | F:
5′-ATCCCAGAGGCTCTCCTCAC-3′ |
| R:
5′-CTACCATTTTCTCTCCCCTGCTT-3′ |
| JUN | F:
5′-ACCAAGAACTGCATGGACCTAACA-3′ |
| R:
5′-GCTCAGCCTCGCTCTCACAA-3′ |
| MMP12 | F:
5′-ACGTGGCATTCAGTCCCTGT-3′ |
| R:
5′-AACACTGGTCTTTGGTCTCTCAGAA-3′ |
| MYC | F:
5′-GCAGCTGCTTAGACGCTGGA-3′ |
| R:
5′-CGCAGTAGAAATACGGCTGCAC-3′ |
| GAPDH | F:
5′-GCACCGTCAAGGCTGAGAAC-3′ |
| F:
5′-TGGTGAAGACGCCAGTGGA-3′ |
Transfection of cell lines
The miR-1284 overexpression vector LV-miR-1284-GFP
and the null vector LV-GFP were provided by GeneChem (Shanghai,
China). After seeded into 6-well plates for 36 h, cells were
infected with the lentiviral vector at 100 PFU/cell [multiplicity
of infection (MOI) = 100] without penicillin or streptomycin. To
acquire stably transfected SGC7901/VCR cells, the cells were
cultured in 600 mg/ml G418 (Invitrogen Corporation) for 14–21 days.
Three groups of cells were identified as follows: gastric cancer
SGC7901/VCR cells in the LV-miR-1284-GFP group were transfected
with the recombinant lentivirus vector LV-miR-1284-GFP; the cells
in the LV-GFP group were transfected with the negative control
lentiviral vector LV-GFP, and the cells in the control group were
without any treatment. qRT-PCR was used to detect the miR-1284 in
the transfected cells.
Cytotoxicity assay
Cells were implanted into 96-well plates at a
density of 2.0×103 cells/well. After 24 h, vincristine
was divided into six concentrations (0, 0.2, 0.4, 0.8, 1.6, and 3.2
mg/ml) and each was added to a common medium. After incubation for
48 h, 10 µl Cell Counting Kit-8 reagent (Dojindo, Tokyo,
Japan) were added and development was sustained for 1 h at 37.8°C
with 5.0% CO2. The absorbance was detected at 450 nm,
and IC50 was calculated using the relative survival
curve. Each survey was processed in quadruplicate.
Cell cycle analysis
Cells were washed twice with PBS and fastened with
70% ethanol for 12 h at 4°C. The cells were hatched in a solution
that contained 200 ng/ml RNase and 0.05 mg/ml propidium iodide that
was incubated at ambient temperature for 0.5 h. The results were
tested by flow cytometry (BD Biosciences, Mountain View, CA,
USA).
Apoptosis assay
Apoptosis was tested using the Apoptosis Detection
kit (BD Biosciences) and following the manufacturer's instructions.
The cells were hatched in a solution containing 5 µl/ml
Annexin V-PE and 5 µl/ml 7-amino-acti-nomycin D at 4°C in
the dark. The results were tested by flow cytometry (BD
Biosciences).
Wound-healing assay
To inhibit cell proliferation, cells were cultivated
in 6-well plates with mitomycin C. A straight line as a wound was
created using a 200-µl sterile pipette tip and washed twice
lightly with PBS to remove any floating cells. The wound conditions
were noted at 0, 48, and 96 h by microscope and the relative
motility was calculated using the following formula: Relative
motility = (initial distance - a time point distance)/initial
distance × 100%.
Cell invasion assay
RPMI-1640 medium (75 µl) without serum
containing 1 µg/ml Matrigel (BD Biosciences) was used in the
above chamber (6.5 mm; Corning, New York, NY, USA). RPMI-1640
medium (200 µl) without fetal calf serum containing
5.0×104 cells were placed into the upper chamber, while
700 µl RPMI-1640 with 5.0% fetal calf serum was placed in
the lower chamber. After culturing for 24 h, the cells in the lower
chamber were stained with Giemsa, and the number of visible cells
was counted in six random views at a ×200 magnification.
Luciferase reporter assays
Wild-type (WT) EIF4A1 3′UTR, mutated (MUT) EIF4A1
3′UTR, negative (NC) EIF4A1 3′UTR luciferase reporter vector and
miR-1284 mimic, and NC miRNA plasmid were purchased from GeneChem.
The WT 3′UTR and MUT 3′UTR luciferase reporter vector were
co-transfected with miR-1284 mimic while the NC 3′UTR + miR-1284
mimic was used as the control. The 293T cells were seeded into
24-well plates (2.0×104 cells/well). After 24 h, the
cells were separated into groups and trans-fected with 0.1
µg WT 3′UTR + 0.3 µg miR-1284 mimic, 0.1 µg
MUT 3′UTR + 0.3 µg miR-1284 mimic, and 0.1 µg NC
3′UTR + 0.3 µg miR-1284 mimic. NC miRNA and the same 3′UTR
were co-transfected as a reference in each group. After 48 h,
firefly luciferase luminescence activity was assayed using the Dual
Luciferase Reporter Assay system (Promega, Madison, WI, USA).
Renilla luciferase luminescence was then detected after
adding Stop & Glo® reagent (Promega) to each well.
Luminescence was calculated as follows: Relative luciferase
activity = firefly luciferase luminescence/Renilla
luceriferase luminescence. Each evaluation was processed in
triplicate.
Western blot analysis
Proteins were extracted using cell lysate extraction
(Solarbio, Beijing, China), separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, and then shifted onto
nitrocellulose membranes. The membranes were immersed with the
antibody (1:1,000) overnight at 4°C and then washed with
Tris-buffered saline and Tween-20. The membrane was immersed with a
dilution of infrared-labeled secondary antibody (1:10,000) for 1 h
and Odyssey (LI-COR Biosciences) was used to analyze the optical
density. In addition, the expression of GAPDH (1:1,000) was used as
a reference.
Effect of miR-1284 on GC cells in
vivo
BALBC/c nude mice, aged 5–6 weeks, were provided by
Guangxi Animal Center (Nanning, China), retained in
specific-pathogen free surroundings, and cared for by following the
instructions of the Ethics Committee of Guangxi Medical University.
Tumors were implanted by injecting 4.0×107 SGC-7901/VCR
cells resuspended in 100 µl phosphate-buffered saline (PBS;
Beyotime Institute of Biotechnology, Shanghai, China) into the
armpit region of the mice. After 4 days, the tumors increased to
~5.0 mm in diameter and the animals were separated into the
following three groups (6 mice/group): LV-miR-1284-GFP, LV-GFP, and
control. The tumors were injected with LV-miR-1284-GFP or LV-GFP at
a titer of 5.0×106 TU in 150 µl PBS; a similar
volume of PBS was injected into the control group. VCR (200 ng/kg)
were injected into the peritoneum. Subsequent to the first surgery,
the mice were given the same treatment every 2 days. Tumor size was
calculated every 4 days using a Vernier caliper and the length
diameter (a) and the width diameter (b) were surveyed (tumor volume
= a × b2/2). The relative tumor volume (RTV) was
estimated by RTV = Vt/V0 (V0, the
tumor volume at time of intraperitoneal injection; Vt,
the tumor volume at the next measurement). After 20 days of
feeding, the animals were sacrificed and the tumors were evaluated.
The tumors were immersed in 4% formaldehyde, water was removed
using an ethanol slope, and tumors were inlayed in paraffin. Tumor
segments were dewaxed, rehydrated, and stained with hematoxylin and
eosin. Sections were observed under a microscope on middle power
(×200).
Statistical analyses
SPSS 13.0 (SPSS Inc., Chicago, IL, USA) was used to
analyze the data. Data are shown as the mean ± SE and deemed to
have statistical significance when P<0.05 using Student's
t-test, one-way analysis of variance, or χ2 test.
Results
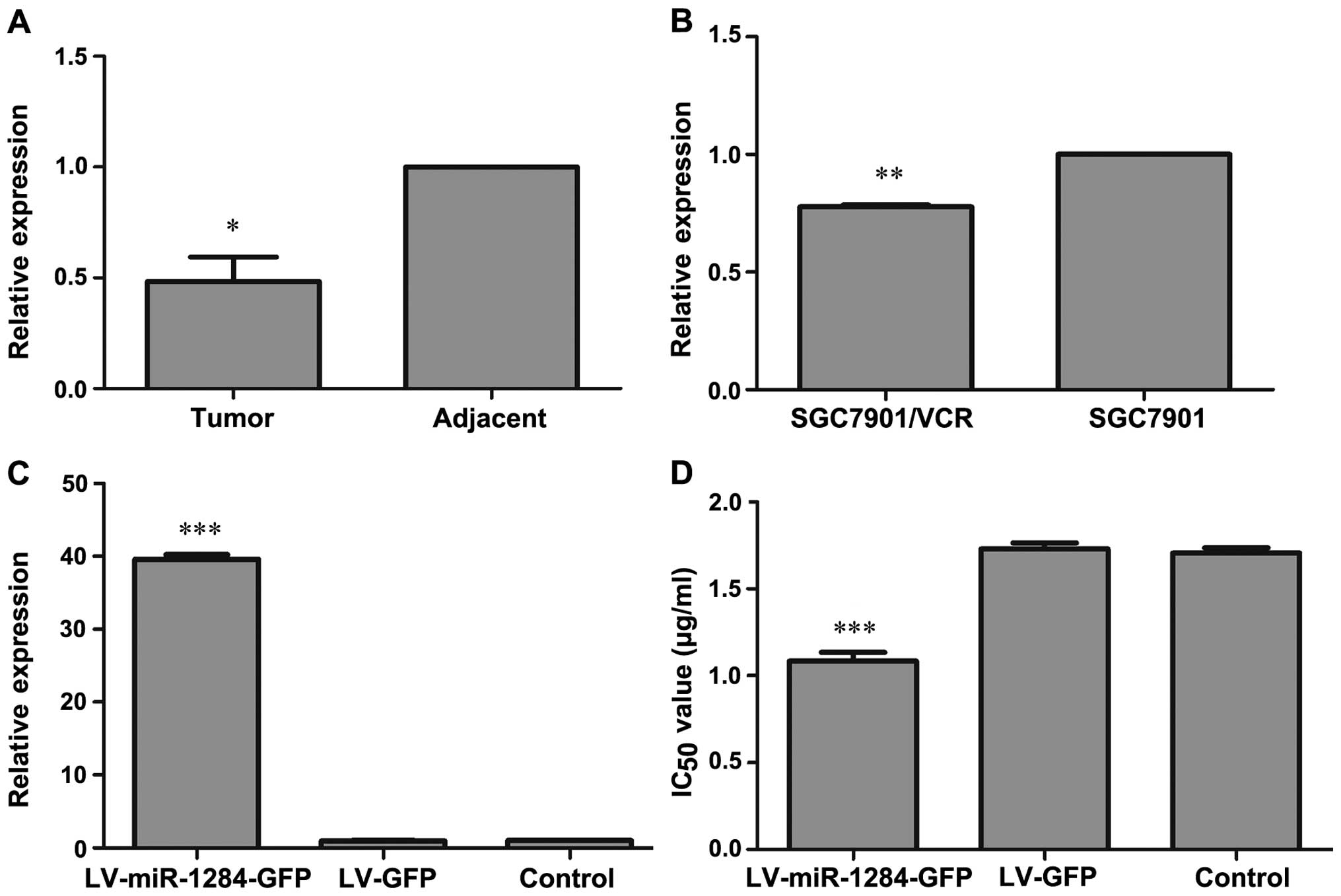
miR-1284 decreases in gastric cancer
tissue specimens and drug resistant GC cells
To determine whether miR-1284 is associated with the
evolution of GC with distant metastasis and MDR in GC cells, we
detected miR-1284 expression in GC tissue specimens from patients
with distant metastasis and compared it to those with adjacent
non-tumor tissues. The data from 16 gastric cancer patients showed
significant downregulation of miR-1284 in GC tissue specimens
(P<0.05) (Fig. 1A). In addition,
the data showed that miR-1284 expression in SGC7901/VCR were
different from that of SGC7901 with significant downregulation of
miR-1284 in SGC7901/VCR (P<0.05) (Fig. 1B). Our data indicated that miR-1284
may be related to GC with distant metastasis and MDR in GC
cells.
miR-1284 recombinant lentiviral vectors
leads to overexpression of miR-1284
To test the hypothesis that miR-1284 may overcome
MDR in GC cells, we first established GC cells that stably
overexpressed miR-1284. LV-miR-1284-GFP and LV-GFP were transfected
into SGC7901/VCR cells, respectively. The expression of miR-1284 in
the LV-miR-1284-GFP group was substantially upregulated (P<0.05)
but there was no difference in miR-1284 levels between the LV-GFP
and control groups (P>0.05) (Fig.
1C). The data demonstrated that LV-miR-1284-GFP could
upregulate miR-1284 expression in SGC7901/VCR.
miR-1284 overcomes MDR in GC cells
We next examined the impact of miR-1284
overexpression on MDR in GC cells. The sensitivity of SGC7901/VCR
cells and IC50 values of each group were measured by
CCK-8 assay. The LV-miR-1284-GFP group demonstrated greatly
enhanced sensitivity to vincristine, as indicated by decreased
IC50 values (P<0.05) (Fig. 1D). These results proved that
miR-1284 overexpression is associated with the GC MDR
phenotype.
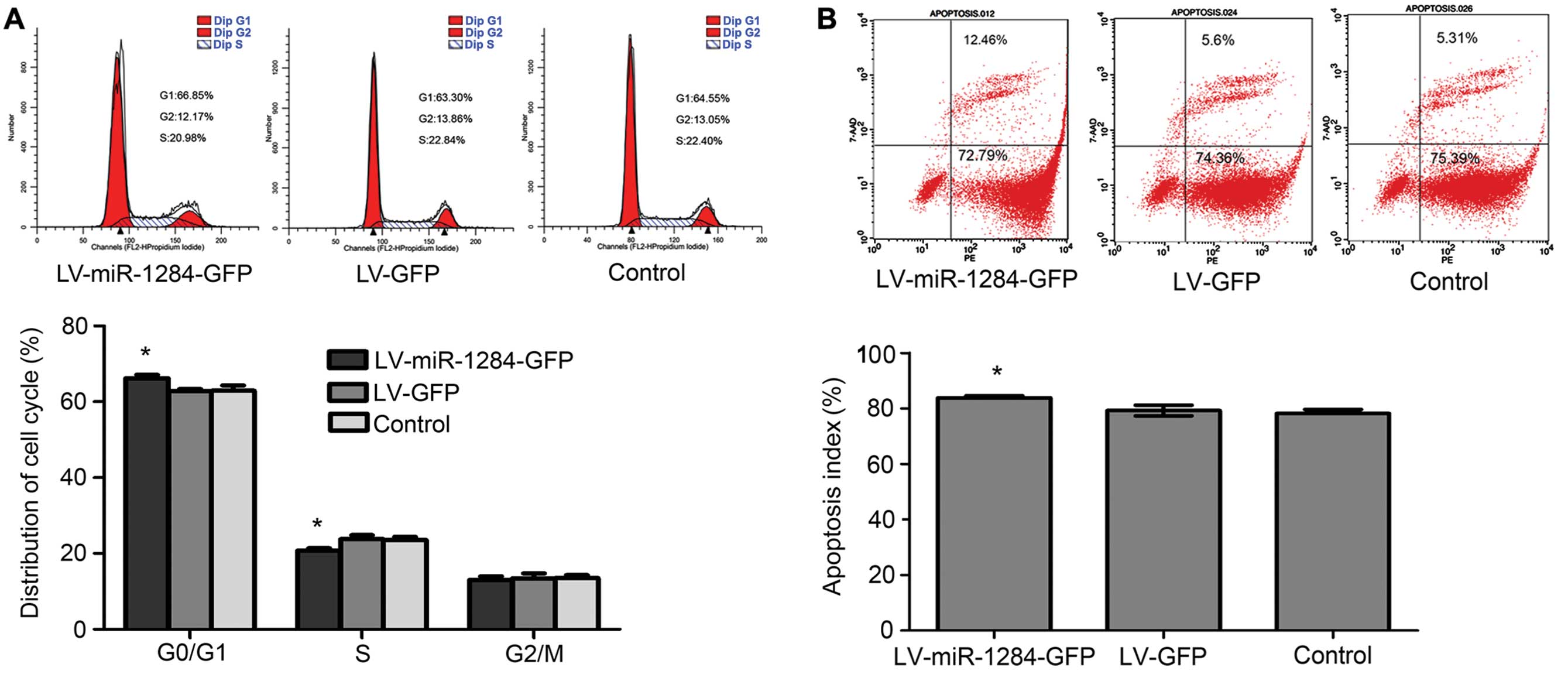
miR-1284 prevents cells from entering the
S phase
To determine whether miR-1284 overexpression can
reverse MDR by effecting the cell cycle, we counted the cells in
specific phases. The data showed that cell counts in the G0/G1
phase observably increased, while those in the S phase decreased in
the LV-miR-1284-GFP group (P<0.05) (Fig. 2A). By miR-1284 preventing GC MDR
cells from entering into the S phase, the GC MDR cells are
reduced.
miR-1284 accelerates drug-induced
apoptosis
Changes in drug-induced apoptosis can influence the
efficacy of chemotherapy drugs; therefore, we looked into the
influence of miR-1284 on apoptosis of GC cells induced by
chemotherapy. After incubation, VCR cells were detected by flow
cytometry. The data show that after miR-1284 overexpression, the
apoptosis rate significantly increased (P<0.05) (Fig. 2B). These results suggested that
miR-1284 may increase the apoptosis rate of SGC7901/VCR cells
induced by chemotherapy drugs.
miR-1284 decreases the migration of GC
cells
To further determine how miR-1284 influences the
migration of SGC7901/VCR cells, we investigated the migratory
ability of the GC cells in each group using the wound-healing
assay. After 96 h, the cells in the LV-miR-1284-GFP group showed
lower migratory ability than those in the LV-GFP and control groups
(relative motility rate of cells in the LV-miR-1284-GFP vs. LV-GFP
and control groups: 51.02±2.18% vs. 82.21±1.25% and 82.55±4.35%,
respectively; P<0.05) (Fig. 2C).
The data demonstrated that miR-1284 decreases the migration of GC
cells.
miR-1284 decreases the invasion of GC
cells
After determining that miR-1284 inhibits the
migratory ability of SGC7901/VCR cells, we evaluated whether it may
also suppress cell invasion. The Transwell assay demonstrated that
fewer cells passed through the membrane of the Matrigel chamber in
the LV-miR-1284-GFP group than through the chamber of the LV-GFP
and control groups. The number of LV-GFP and control group cells
that passed through the membrane was 3.5- and 3.3-fold greater,
respectively, than that of the LV-miR-1284-GFP (P<0.05)
(Fig. 2D). The data demonstrated
that miR-1284 decreases the invasion of GC cells.
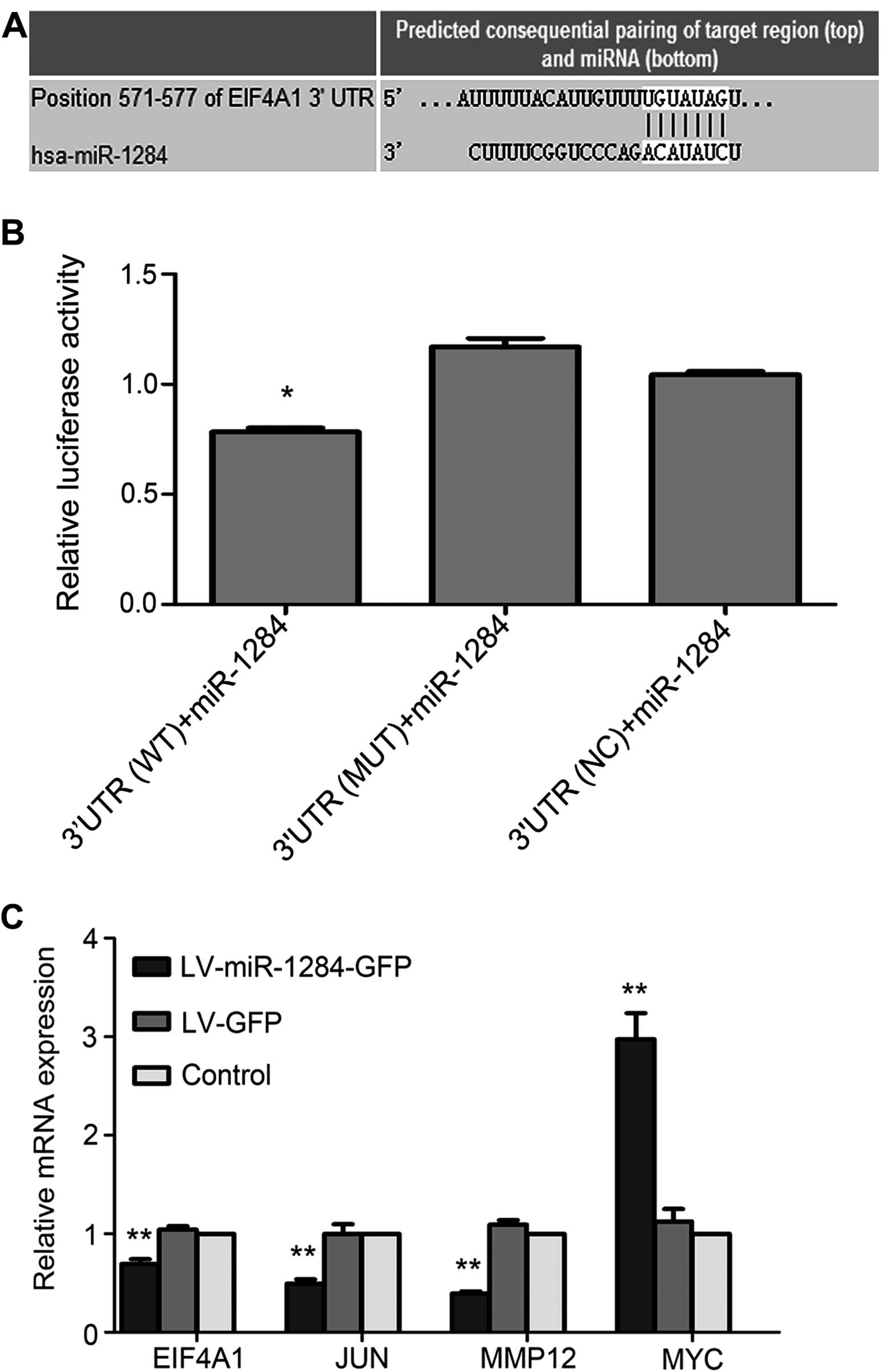
EIF4A1 is the direct target gene of
miR-1284
The results of TargetScan 6.2 indicated that EIF4A1
is one of the underlying targets of miR-1284 (Fig. 3A). To confirm this, the relative
luciferase activity of each group was assayed. The data showed that
the relative luciferase activity in the WT 3′UTR + miR-1284 mimic
group declined in relation to that in the NC 3′UTR + miR-1284 and
MUT 3′UTR + miR-1284 groups (P<0.05) (Fig. 3B). The results suggested that
miR-1284 modulates MDR by targeting EIF4A1.
miR-1284 modulates MDR by reducing the
expression of EIF4A1, JUN, and MMP12 and enhancing the expression
of MYC
To investigate the mechanism by which miR-1284
reverses MDR in SGC7901/VCR, we used qRT-PCR and western blotting
to assess the expression levels of genes that regulate apoptosis,
metastasis, and MDR. In the LV-miR-1284-GFP group, the expressions
of MYC mRNA and protein were higher, and the expression of EIF4A1,
JUN, and MMP12 lower than those in the LV-GFP and control groups
(P<0.05) (Fig. 3C and D).
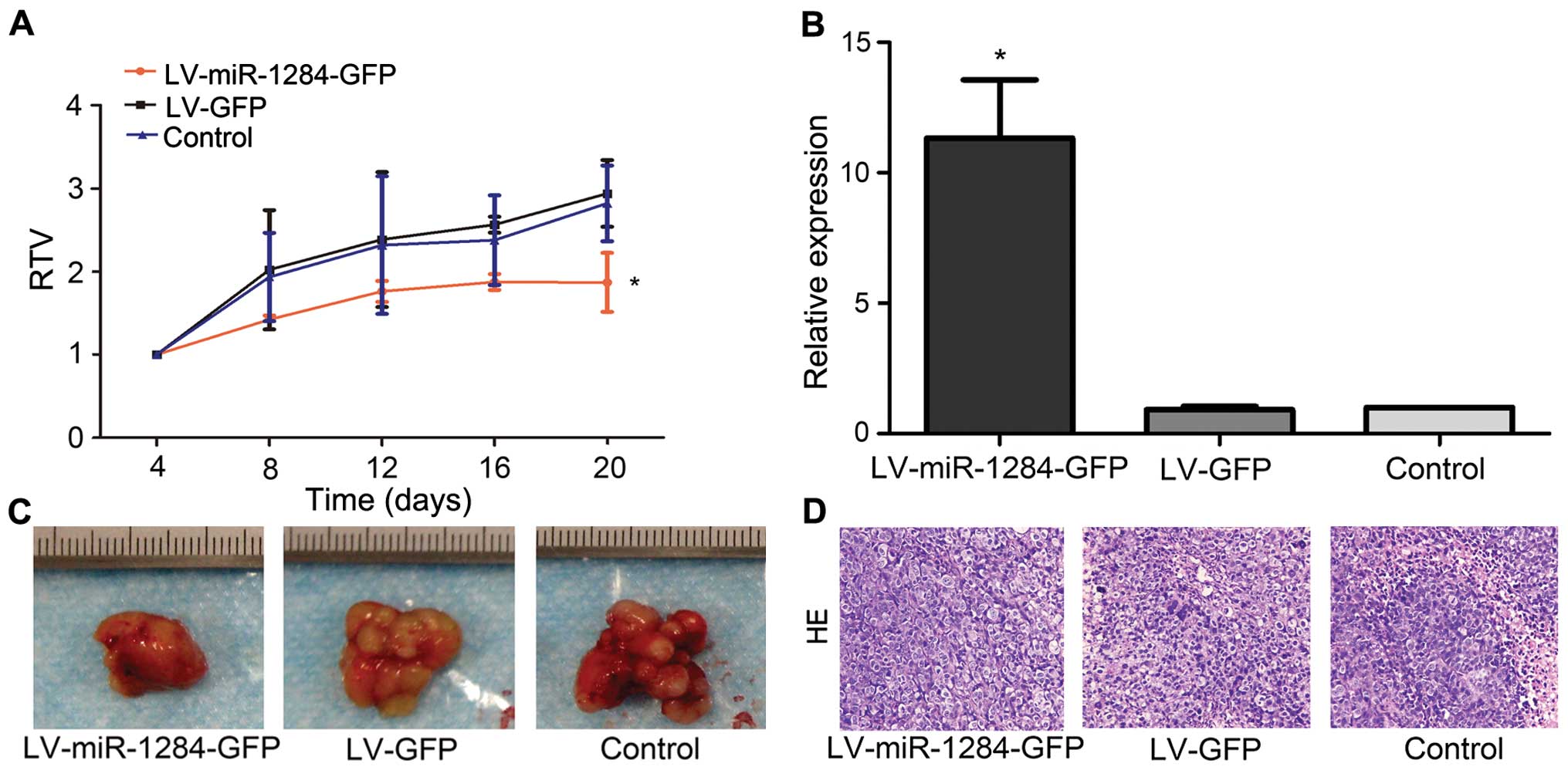
miR-1284 reverses MDR in GC in vivo
We also explored the influence of miR-1284 on
SGC7901/VCR cells in vivo by subcutaneously transplanting
tumors in nude mice. After 4 days, the mice were randomly divided
into the LV-miR-1284-GFP group, LV-GFP group, and control group.
Each group was injected with LV-miR-1284-GFP, LV-GFP, or PBS,
respectively, and given an intratumoral vincris-tine treatment.
After 20 days of treatment, the RTV in the LV-miR-1284-GFP group
was significantly smaller than that in either the LV-GFP or control
group (P<0.05) (Fig. 4A).
qRT-PCR confirmed that the expression of miR-1284 in the
LV-miR-1284-GFP group was greater than that in either the LV-GFP or
control group (P<0.05) (Fig.
4B). The data demonstrated that miR-1284 can reverse MDR in GC
in vivo.
Discussion
GC is the second leading cause of carcinoma death
worldwide (16). Due to lack of
efficacious methods of diagnosing the early stages of the disease
and tumorigenesis, patients who suffer from metastasis lose their
best opportunity for surgery, and chemotherapy becomes the
preferred treatment; however, routine chemotherapy often fails
because of MDR in GC cells (17).
Many mechanisms underlying MDR have been extensively explored,
including ejecting several drugs into the cells, modifying drug
targets, destroying the balance of damaging or repairing DNA, and
deactivating drug-induced apoptosis pathways (18–22),
however, the crucial factors of this phenomenon remain largely
unclear.
Recently, increasing number of studies have revealed
that miRNAs modulate the development of MDR in GC (23–25).
Chen et al (15) reported
that compared with primary GC, miR-1284 is downregulated in GC with
lymph node metastases. Even so, the properties and the potential
mechanisms by which miR-1284 affects MDR have not been
investigated. The current study showed that miR-1284 was reduced in
GC specimens with distant metastasis and in SGC7901/VCR cells
compared with that in the controls, which agree with a previous
study (15); however, there are no
previous studies that compared the connection between miR-1284 and
tumorigenesis in GC. In addition, our data showed that modulation
of miR-1284 overexpression could overcome MDR in SGC7901/VCR,
prevent cells from entering into the S phase, and induce cell
apoptosis. miR-1284 overexpression also decreases the migration and
invasion of SGC7901/VCR and reverses MDR in vivo, as
demonstrated by suspended tumor growth in nude mice. The results
confirm that miR-1284 may function as a new regulator to reverse
MDR in GC cell line SGC7901/VCR in vitro and in
vivo.
Nonetheless, the accurate method by which miR-1284
induces these events is obscure. MDR is reversed by cell death and
is associated with EIF4F, a compound that induces ribosomes to
gather to mRNA templates, which comprise EIF4E, EIF4G, and EIF4A
subunits (26). This study proved
that miR-1284 directly both regulates EIF4A1 and suppresses EIF4A1
expression. EIF4A1 is a member of the translation initiation
composite EIF4A, which controls protein synthesis, and has been
reported to have an indispensable function in tumorigenesis
(27). As a DEAD-box helicase,
EIF4A1 plays an important role in unwinding integrated RNA
components within 5′UTR to gather ribosomes and participate in mRNA
translation (28). Increasing
number of studies have confirmed that EIF4A1 has multiple functions
and is correlated with the genesis and progression of
tumor-promoting proteins. Overexpression of EIF4A1 has been proven
in many cancers, such as liver cancer, melanoma, and melanocytic
nevi (29,30). In addition, recent research has
shown that the presence of EIF4A1 can predict an independently
adverse outcome in breast cancer and that reducing the expression
of EIF4A1 diminishes cell proliferation and prevents cells from
entering the S phase (31).
Bordeleau et al (26)
demonstrated that the absence of EIF4A1 decreases drug resistance
to doxorubicin, which is associated with PI3K/mTOR activation. This
is consistent with our findings that miR-1284 reverses MDR in GC by
targeting EIF4A1. To our knowledge, this study is the first to
demonstrate that EIF4A1 is immediately downregulated by miR-1284 in
GC MDR cells.
By exploring the mechanisms of synergistic
interaction we demonstrated that miR-1284 overexpression can
regulate the response of SGC7901/VCR cells to chemotherapeutic
resistance by targeting EIF4A1, reducing JUN and MMP12, and
increasing MYC. Previous studies have shown that EIF4A1 is
inhibited by PDCD4, while PDCD4 regulates the MYC and JUN pathways
(32,33). This identified an uncommon
integrated network, the miR-1284/EIF4A1/JUN/MYC signaling pathway.
Reducing the permeability glycoprotein (P-gp), a multidrug
resistance protein that is created by MDR1, and downregulating MDR1
play an indispensable role in circumventing MDR in cancer. Studies
have shown that inhibiting JUN significantly reduces the degree of
MDR by decreasing MDR1 and P-gp (34), leading to inefficient transport that
keeps a lower concentration of drugs inside the cancer cells, which
reverses MDR. As previously described, another primary reason for
MDR is to reduce susceptibility to drug-induced apoptosis (21,22).
Fu et al (35) confirmed
that a reduced expression of JUN contributes to an extended G1
phase and induces apoptosis. In addition, activating MYC has been
proved to generate apoptosis by triggering cytochrome c
before activating caspase (36).
JUN, a binding complex that is correlated with the
MMP12 booster, directly activities MMP12 and belongs to the matrix
metalloproteinases (MMPs) that have been known to act in an
indispensable manner in tumor invasiveness and metastasis (37,38).
MMP12 has been reported to be connected to tumor metastasis, such
as lung carcinoma and squamous carcinomas of the head and neck
(39,40). A recent study also showed that
silencing MMP12 suppresses the proliferation and invasion of lung
cancer cells (41). Interestingly,
JUN increases tumor migration and invasion activity (42), and GC cell metastasis is suppressed
by reducing JUN and MMP12. Primary tumors and tumors with
metastasis are often different from MDR cells with metastasis
because of the stronger drug-resistance (43). Inhibiting metastasis may be another
key mechanism by which MDR in tumor cells is circumvented.
In this study, miR-1284 was regulated to reverse MDR
in GC cells by downregulating EIF4A1 in vitro and in
vivo. This study is the first to demonstrate a connection
between miR-1284 and EIF4A1, an important member of the EIF family,
and demonstrates the power of miR-1284 in regulating MDR
development. Moreover, this study draws our attention to EIF4A1 as
an underlying therapeutic target for GC and proposes new concepts
whereby miR-1284 can suppress cancer progression.
Abbreviations:
|
MDR
|
multidrug resistance
|
|
GC
|
gastric cancer
|
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (no. 30860273; no.
81060201); the Key Health Science Foundation of Guangxi (no.
14124004-1-9), the Natural Science Foundation of Guangxi (no.
2015GXNSFDA227001) and Innovation Project of Guangxi Graduate
Education.
References
|
1
|
Baba H, Kuwabara K, Ishiguro T, Kumamoto
K, Kumagai Y, Ishibashi K, Haga N and Ishida H: Prognostic factors
for stage IV gastric cancer. Int Surg. 98:181–187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan D, Zhang X, Chen X, Mou Z, Hu J, Zhou
S, Ding J and Wu K: Bird's-eye view on gastric cancer research of
the past 25 years. J Gastroenterol Hepatol. 20:360–365. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fodale V, Pierobon M, Liotta L and
Petricoin E: Mechanism of cell adaptation: When and how do cancer
cells develop chemore-sistance? Cancer J. 17:89–95. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dai Z, Huang Y and Sadée W: Growth factor
signaling and resistance to cancer chemotherapy. Curr Top Med Chem.
4:1347–1356. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng T, Wang J, Chen X and Liu L: Role of
microRNA in anticancer drug resistance. Int J Cancer. 126:2–10.
2010. View Article : Google Scholar
|
|
9
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar
|
|
10
|
Ma J, Dong C and Ji C: MicroRNA and drug
resistance. Cancer Gene Ther. 17:523–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
An Y, Zhang Z, Shang Y, Jiang X, Dong J,
Yu P, Nie Y and Zhao Q: miR-23b-3p regulates the chemoresistance of
gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis.
6:e17662015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu W, Shan X, Wang T, Shu Y and Liu P:
miR-181b modulates multidrug resistance by targeting BCL2 in human
cancer cell lines. Int J Cancer. 127:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang
B, Shu Y and Liu P: miR-497 modulates multidrug resistance of human
cancer cell lines by targeting BCL2. Med Oncol. 29:384–391. 2012.
View Article : Google Scholar
|
|
14
|
Patnaik SK, Yendamuri S, Kannisto E,
Kucharczuk JC, Singhal S and Vachani A: MicroRNA expression
profiles of whole blood in lung adenocarcinoma. PLoS One.
7:e460452012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen W, Tang Z, Sun Y, Zhang Y, Wang X,
Shen Z, Liu F and Qin X: miRNA expression profile in primary
gastric cancers and paired lymph node metastases indicates that
miR-10a plays a role in metastasis from primary gastric cancer to
lymph nodes. Exp Ther Med. 3:351–356. 2012.PubMed/NCBI
|
|
16
|
Yasui W, Sentani K, Sakamoto N, Anami K,
Naito Y and Oue N: Molecular pathology of gastric cancer: Research
and practice. Pathol Res Pract. 207:608–612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu D, Xiao Z, Wang W, Xu Y, Gao S, Deng L,
He W, Yang Y, Guo X and Wang X: Down regulation of CIAPIN1 reverses
multidrug resistance in human breast cancer cells by inhibiting
MDR1. Molecules. 17:7595–7611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rebucci M and Michiels C: Molecular
aspects of cancer cell resistance to chemotherapy. Biochem
Pharmacol. 85:1219–1226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baguley BC: Multiple drug resistance
mechanisms in cancer. Mol Biotechnol. 46:308–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Butler EB, Zhao Y, Muñoz-Pinedo C, Lu J
and Tan M: Stalling the engine of resistance: Targeting cancer
metabolism to overcome therapeutic resistance. Cancer Res.
73:2709–2717. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: A link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rabik CA and Dolan ME: Molecular
mechanisms of resistance and toxicity associated with platinating
agents. Cancer Treat Rev. 33:9–23. 2007. View Article : Google Scholar :
|
|
23
|
Wu Q, Yang Z, Xia L, Nie Y, Wu K, Shi Y
and Fan D: Methylation of miR-129–5p CpG island modulates
multi-drug resistance in gastric cancer by targeting ABC
transporters. Oncotarget. 5:11552–11563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li
K, Zhou L, Sun Y, Li M, Zhou J, et al: miR-508–5p regulates
multidrug resistance of gastric cancer by targeting ABCB1 and
ZNRD1. Oncogene. 33:3267–3276. 2014. View Article : Google Scholar
|
|
25
|
Wang Y, Gu X, Li Z, Xiang J, Jiang J and
Chen Z: microRNA expression profiling in multidrug resistance of
the 5-FU-induced SGC-7901 human gastric cancer cell line. Mol Med
Rep. 7:1506–1510. 2013.PubMed/NCBI
|
|
26
|
Bordeleau ME, Robert F, Gerard B,
Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco
JA Jr, et al: Therapeutic suppression of translation initiation
modulates chemosensitivity in a mouse lymphoma model. J Clin
Invest. 118:2651–2660. 2008.PubMed/NCBI
|
|
27
|
Hagner PR, Schneider A and Gartenhaus RB:
Targeting the translational machinery as a novel treatment strategy
for hematologic malignancies. Blood. 115:2127–2135. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Svitkin YV, Pause A, Haghighat A, Pyronnet
S, Witherell G, Belsham GJ and Sonenberg N: The requirement for
eukaryotic initiation factor 4A (elF4A) in translation is in direct
proportion to the degree of mRNA 5′ secondary structure. RNA.
7:382–394. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shuda M, Kondoh N, Tanaka K, Ryo A,
Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, et al:
Enhanced expression of translation factor mRNAs in hepatocellular
carcinoma. Anticancer Res. 20:2489–2494. 2000.PubMed/NCBI
|
|
30
|
Eberle J, Krasagakis K and Orfanos CE:
Translation initiation factor eIF-4A1 mRNA is consistently
overexpressed in human melanoma cells in vitro. Int J Cancer.
71:396–401. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Modelska A, Turro E, Russell R, Beaton J,
Sbarrato T, Spriggs K, Miller J, Gräf S, Provenzano E, Blows F, et
al: The malignant phenotype in breast cancer is driven by
eIF4A1-mediated changes in the translational landscape. Cell Death
Dis. 6:e16032015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lankat-Buttgereit B and Göke R: The tumour
suppressor Pdcd4: Recent advances in the elucidation of function
and regulation. Biol Cell. 101:309–317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S,
Long X, Jiang Q, Song Y, Cheng C, et al: Tumor suppressor PDCD4
modulates miR-184-mediated direct suppression of C-MYC and BCL2
blocking cell growth and survival in nasopharyngeal carcinoma. Cell
Death Dis. 4:e8722013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sui H, Zhou S, Wang Y, Liu X, Zhou L, Yin
P, Fan Z and Li Q: COX-2 contributes to P-glycoprotein-mediated
multidrug resistance via phosphorylation of c-Jun at Ser63/73 in
colorectal cancer. Carcinogenesis. 32:667–675. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu Y, Lin Y, Yang Z, Yang G, Li G, Liu Y,
Tan X, Huang Y, Wu X, Wang Y, et al: FBXW7 overexpression
suppresses renal cancer cell proliferation and induces apoptosis.
Med Oncol. 32:2152015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Juin P, Hueber AO, Littlewood T and Evan
G: c-Myc-induced sensitization to apoptosis is mediated through
cytochrome c release. Genes Dev. 13:1367–1381. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu L, Tanimoto A, Murata Y, Sasaguri T,
Fan J, Sasaguri Y and Watanabe T: Matrix metalloproteinase-12 gene
expression in human vascular smooth muscle cells. Genes Cells.
8:225–234. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie S, Issa R, Sukkar MB, Oltmanns U,
Bhavsar PK, Papi A, Caramori G, Adcock I and Chung KF: Induction
and regulation of matrix metalloproteinase-12 in human airway
smooth muscle cells. Respir Res. 6:1482005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hofmann HS, Hansen G, Richter G, Taege C,
Simm A, Silber RE and Burdach S: Matrix metalloproteinase-12
expression correlates with local recurrence and metastatic disease
in non-small cell lung cancer patients. Clin Cancer Res.
11:1086–1092. 2005.PubMed/NCBI
|
|
40
|
Kim JM, Kim HJ, Koo BS, Rha KS and Yoon
YH: Expression of matrix metalloproteinase-12 is correlated with
extracapsular spread of tumor from nodes with metastasis in head
and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol.
270:1137–1142. 2013. View Article : Google Scholar
|
|
41
|
Lv FZ, Wang JL, Wu Y, Chen HF and Shen XY:
Knockdown of MMP12 inhibits the growth and invasion of lung
adeno-carcinoma cells. Int J Immunopathol Pharmacol. 28:77–84.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Toft DJ, Rosenberg SB, Bergers G, Volpert
O and Linzer DI: Reactivation of proliferin gene expression is
associated with increased angiogenesis in a cell culture model of
fibrosarcoma tumor progression. Proc Natl Acad Sci USA.
98:13055–13059. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
O'Shaughnessy J: Extending survival with
chemotherapy in metastatic breast cancer. Oncologist. 10(Suppl 3):
20–29. 2005. View Article : Google Scholar : PubMed/NCBI
|