Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most commonly diagnosed cancers worldwide, ranking sixth among all
human cancers (1). There are
650,000 new OSCC cases diagnosed and 350,000 deaths due to the
disease reported each year (2).
OSCC has been one of the 10 leading causes of death from cancer in
Taiwan since 1991. According to the Cancer Registry Annual Report
in Taiwan, oral cancer is the sixth leading cause of cancer-related
death in the entire population and fourth in the male population.
The development of oral cancer is highly associated with betal-quid
chewing, cigarette smoking and alcohol consumption in Taiwan
(3). Although technologies for
diagnosis and therapy (surgery, radiation and chemotherapy) have
advanced remarkably, the long-term survival rate of OSCC patients
has not improved significantly for decades (4,5).
The transcription factor NF-κB plays a key role in
several cellular functions, including inflammation, innate and
adaptive immune responses, cell proliferation, survival,
angiogenesis and apoptosis. The constitutive activation of NF-κB is
common in many types of human tumors. Dysregulation of NF-κB
promotes tumor angiogenesis and metastasis as well as resistance to
chemotherapeutic agents and radiation (6). Activation of the NF-κB pathway is
tightly controlled by several feedback mechanisms and is regulated
by ubiquitination (7).
Tumor necrosis factor, α-induced protein 3
(TNFAIP3), which encodes a ubiquitin-modifying enzyme (A20), is one
of the major inhibitors of the NF-κB signaling pathway (8). It is induced by tumor necrosis factor
(TNF) in human endothelial cells (9). A20 has dual functionality; it is not
only able to add ubiquitin moieties to its target protein but can
also cleave K63-linked polyubiquitin chains, preventing the
interaction of receptor interacting serine/threonine protein kinase
1 (RIP1) and NF-κB essential modulator (NEMO) through its
deubiquitination activity mediated by its ovarian tumor (OTU)
domain. A20 also adds K48-linked polyubiquitin chains to RIP1,
targeting it for proteasomal degradation (10).
The TNFAIP3 gene contains eight coding exons
(2–9), along with exon 1 which is non-coding,
and is located on chromosome 6q23. It has been shown to be
inactivated by deletions, point mutations, and/or promoter
methylation in several types of lymphomas, such as B-cell
lymphomas, classical Hodgkin's lymphoma, chronic lymphocytic
leukemia and mucosa-associated lymphoid tissue lymphoma, which
results in loss of the A20 protein (11–14).
All of these lymphomas are characterized by the dysregulation of
the NF-κB signaling pathway. These findings establish
TNFAIP3 as an important tumor-suppressor gene. Human
genome-wide association studies (GWAS) have linked germline single
nucleotide polymorphisms of the TNFAIP3 gene with
susceptibility to human inflammatory and autoimmune pathologies
(15,16).
The high-resolution melting (HRM) analysis is one of
the most effective mutation scanning methodologies. It is a
closed-tube method, such that PCR amplification and subsequent
analysis are performed sequentially in the same tube, which makes
it more convenient than other scanning methodologies. Moreover,
there is no need for processing or separation of PCR products
(17). This study aimed to assess
the utility of HRM analysis using real-time polymerase chain
reaction (PCR) for screening TNFAIP3 mutations.
Materials and methods
Patients and DNA extraction
A total of 81 patients who were recently diagnosed
with OSCC were selected for the present study. Tissue specimens
were stored immediately after resection in liquid nitrogen before
DNA extraction. We included the peripheral blood samples of 50
unaffected individuals from the general population as controls. DNA
was extracted as described previously by Yeh et al (18). The genomic DNA concentration was
assessed using a NanoDrop 1000 spectrophotometer (NanoDrop
Technologies Inc., Wilmington, DE, USA). DNA was stored at −80°C
until use. All tumors were classified according to the TNM
classification system (19). The
present study was approved by the Institutional Review Board of the
China Medical University Hospital (CMUH102-REC1-015).
Design of primers for HRM assay
We used a set of primers for HRM, specific for
TNFAIP3 exons 2–9 that met the requirements of the
LightCycler® 480 System Gene Scanning Assay. The 13
primer pairs for HRM analysis were selected using the Primer3
software (Table I). For exons 2, 7
and 9, more than one pair of primers were used to amplify the exon
in two overlapping segments. All primers synthesized were of
standard molecular biology quality (Protech Technology Enterprise
Co., Ltd., Taiwan).
 | Table IPrimers uses for HRM analysis of
TNFAIP3 gene mutations. |
Table I
Primers uses for HRM analysis of
TNFAIP3 gene mutations.
| Detection for | Sequence | Length of PCR
amplicon
(bp) |
|---|
| Exon 2 | | |
| P2-1 | F:
5′GTCAGGCTAATAGAATGGCTTTTT 3′ | 250 |
| R:
5′ATGATCTCCCGAAACTGAGGAC 3′ | |
| P2-2 | F:
5′TTAAAACCATGCACCGATACACA 3′ | 218 |
| R:
5′CTATCACCCAGGCAAAAGAAACA 3′ | |
| Exon 3 | F:
5′TGGGTCTTACATGCAGATAACTTG 3′ | 293 |
| R:
5′CACCATGGAGCTCTGTTAGTAGAT 3′ | |
| Exon 4 | F:
5′AGGGAGTACAGGATACATTCAAGC 3′ | 245 |
| R:
5′AAGGCTGAAAGCATTTAAGTACAGA 3′ | |
| Exon 5 | F:
5′ATGGAATTTGATGAAAGTCACCTA 3′ | 289 |
| R:
5′AAGGAAAACCCTGATGTTTCAGT 3′ | |
| Exon 6 | F:
5′TGAGATCTACTTACCTATGGCCTTG 3′ | 283 |
| R:
5′GACACAGGAGAGAGCTGAACATAA 3′ | |
| Exon 7 | | |
| P7-1 | F:
5′TGTAAAATCTTGTGTGTGATTTTGTG 3′ | 302 |
| R:
5′CTCTGAGCACTCATGGCATAAAG 3′ | |
| P7-2 | F:
5′CCTTCTTCATGTCTGTGAACACC 3′ | 316 |
| R:
5′CAACGTTCACAAAATCCGTTGT 3′ | |
| P7-3 | F:
5′AGTGAGACCACTGCCATGAAG 3′ | 340 |
| R:
5′TTCCAGCTCTGTGGCAAGAAT 3′ | |
| P7-4 | F:
5′CACCAGCGTTCCAAGTCAGAT 3′ | 301 |
| R:
5′TTCTTAAAGGTCAGGAACAAAACC 3′ | |
| Exon 8 | F:
5′TCTACTGTCAGCATCTCTGTATCG 3′ | 307 |
| R:
5′AGCAAAAAGCATCGAACACAC 3′ | |
| Exon 9 | | |
| P9-1 | F:
5′AGATTTCATTGTGCTCTCCCTAAG 3′ | 215 |
| R:
5′CTGGTTGGGATGCTGACACT 3′ | |
| P9-2 | F:
5′GCCTCCTGCAAGAACATCCT 3′ | 285 |
| R:
5′ATAGCACCATGATGACTGACAGC 3′ | |
HRM techniques
PCR reactions were carried out in a 10-µl
final volume using the LightCycler® 480 High Resolution
Melting Master (Reference 04909631001; Roche Diagnostics) and
contained 1X buffer, Taq polymerase, nucleotides and the
ResoLight dye, and 10 ng of DNA. The primers and MgCl2
were used at 0.25 µM and 2.5 mM, respectively, to detect
TNFAIP3 single-nucleotide polymorphisms.
The PCR program required a SYBR Green I filter (533
nm), and consisted of an initial denaturation activation step at
95°C for 10 min, followed by a 45-cycle program (denaturation at
95°C for 10 sec, annealing at 60°C for 15 sec and elongation at
72°C for 15 sec with reading of the fluorescence; acquisition mode:
single). The melting program included three steps: denaturalization
at 95°C for 1 min, renaturation at 40°C for 1 min and then melting
with a continuous reading from 60°C to 90°C at 25 acquisitions per
°C.
Direct DNA sequencing
After HRM analysis, the samples were purified using
the PCR-M™ clean up system (Viogen, Sunnyvale, CA, USA). The
sequence reaction was performed in a final volume of 10 µl,
including 1 µl of the purified PCR product, 2.5 µM of
one of the PCR primers, 2 µl of the ABI PRISM terminator
cycle sequencing kit v3.1 (Applied Biosystems), and 2 µl of
5X sequencing buffer. The sequencing program followed a 25-cycle
PCR program (denaturation at 96°C for 10 sec, annealing at 50°C for
5 sec, and elongation at 60°C for 4 min). Sequence detection was
performed using an ABI Prism 3130 Genetic Analyzer (Applied
Biosystems) according to standard protocols.
Statistical analysis
Data analysis was performed using the SPSS version
17.0 software (SPSS, Inc., Chicago, IL, USA). The Chi-square test
was used to compare TNFAIP3 genotype distributions between
the case and control groups. Univariate unconditional logistic
regression analyses were used to obtain odds ratios (OR) and
corresponding 95% confidence intervals (CI). A value of P<0.05
was considered to indicate a statistically significant
difference.
Results
Screening of TNFAIP3 mutations in exons
2, 4, 5, 6 and 8
To identify TNFAIP3 gene mutations in exons
2, 4, 5, 6 and 8, we designed primer pairs specific for each exon
region (Table I). HRM analysis of
exons 2, 4, 5, 6 and 8 showed no nucleotide changes in these
regions (data not shown).
Screening of TNFAIP3 mutations in exon
3
We used one primer pair to identify TNFAIP3
mutations in exon 3. The PCR amplification product contained
multiple SNPs. Thus, this PCR primer was not suitable for HRM.
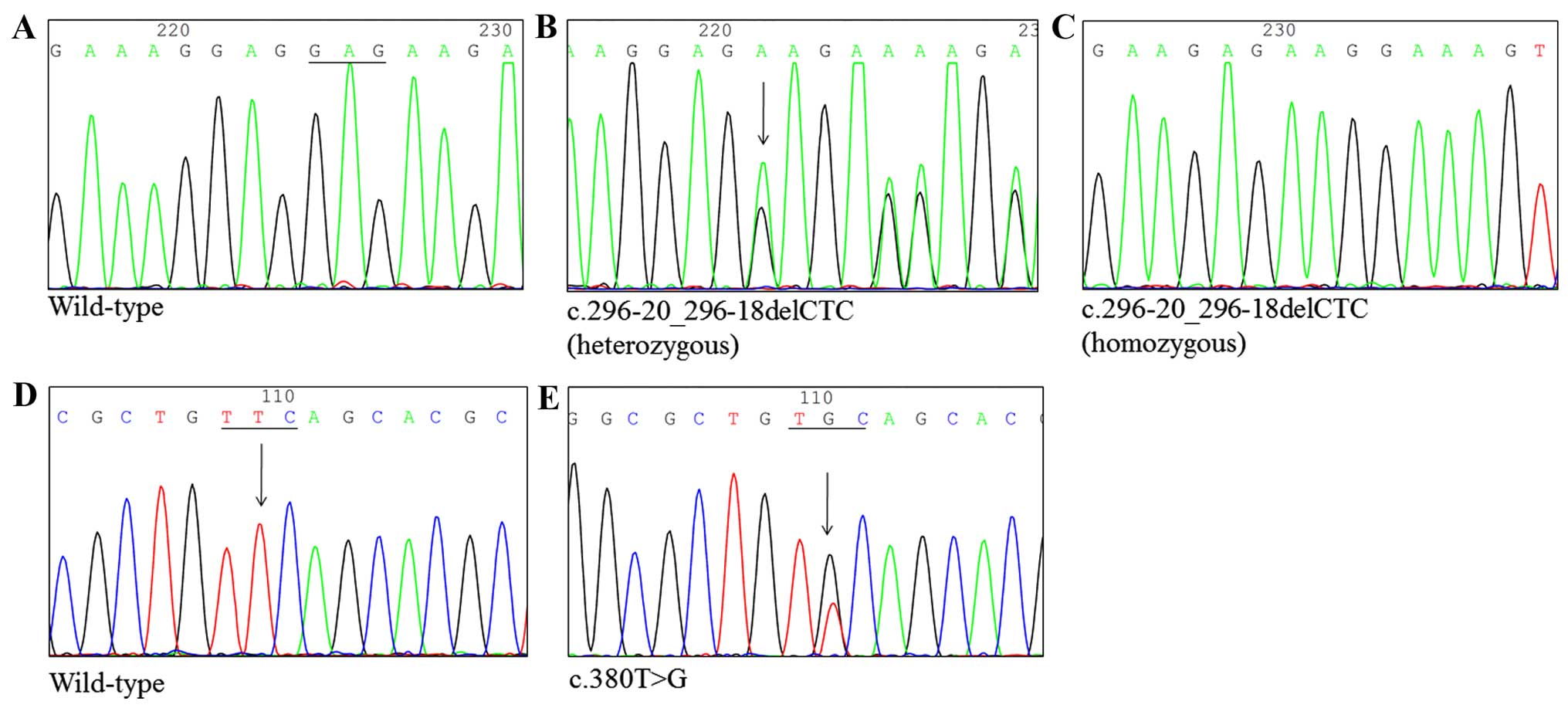
Consequently, this exon was analyzed by direct sequencing. Fig. 1 shows that one SNP resulted in a
change from thymine to guanine at position 380 (c.380T>G), and
another caused a deletion of cytosine, thymine and cytosine (CTC)
at position 296 (c.296-20_296-18delCTC). The c.380T>G change
resulted in a Phe127Cys amino acid substitution. In addition, the
c.380T>G nucleotide change was found in four subjects, for an
incidence of 4.94%, while a three nucleotide deletion
(c.296-20_296-18delCTC) was found in 80 subjects, for the highest
incidence of 98.77%.
Screening of TNFAIP3 mutations in exon
7
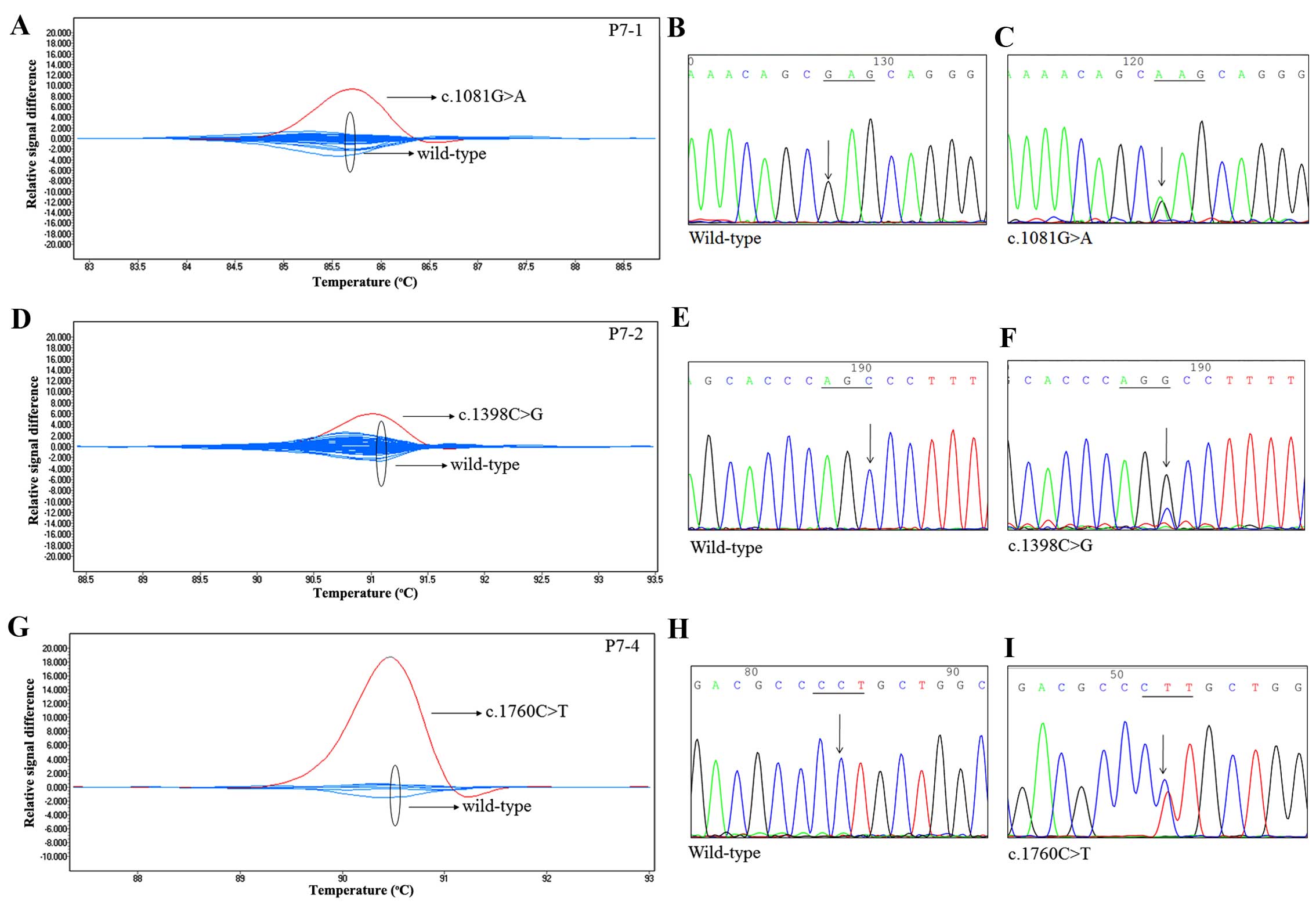
We used four primer pairs (P7-1, P7-2, P7-3 and
P7-4) to identify TNFAIP3 mutations in exon 7. As shown in
Fig. 2, three mutations resulted in
a change from guanine to adenine at position 1081 (c.1081G>A),
from cytosine to guanine at position 1398 (c.1398C>G), and from
cytosine to thymine at position 1760 (c.1760C>T). These were
confirmed by direct sequencing of the codons of interest. The
c.1081G>A, c.1398C>G and c.1760C>T changes resulted in
Glu361Lys, Ser466Arg, and Pro587Leu amino acid substitutions,
respectively. Intriguingly, the mutation p.E361K has not been
reported previously, which the PolyPhen-2 predicted as a benign
mutation. In addition, mutations in exon 7 were found in three
subjects, for an incidence of 3.7%.
Screening of TNFAIP3 mutations in exon
9
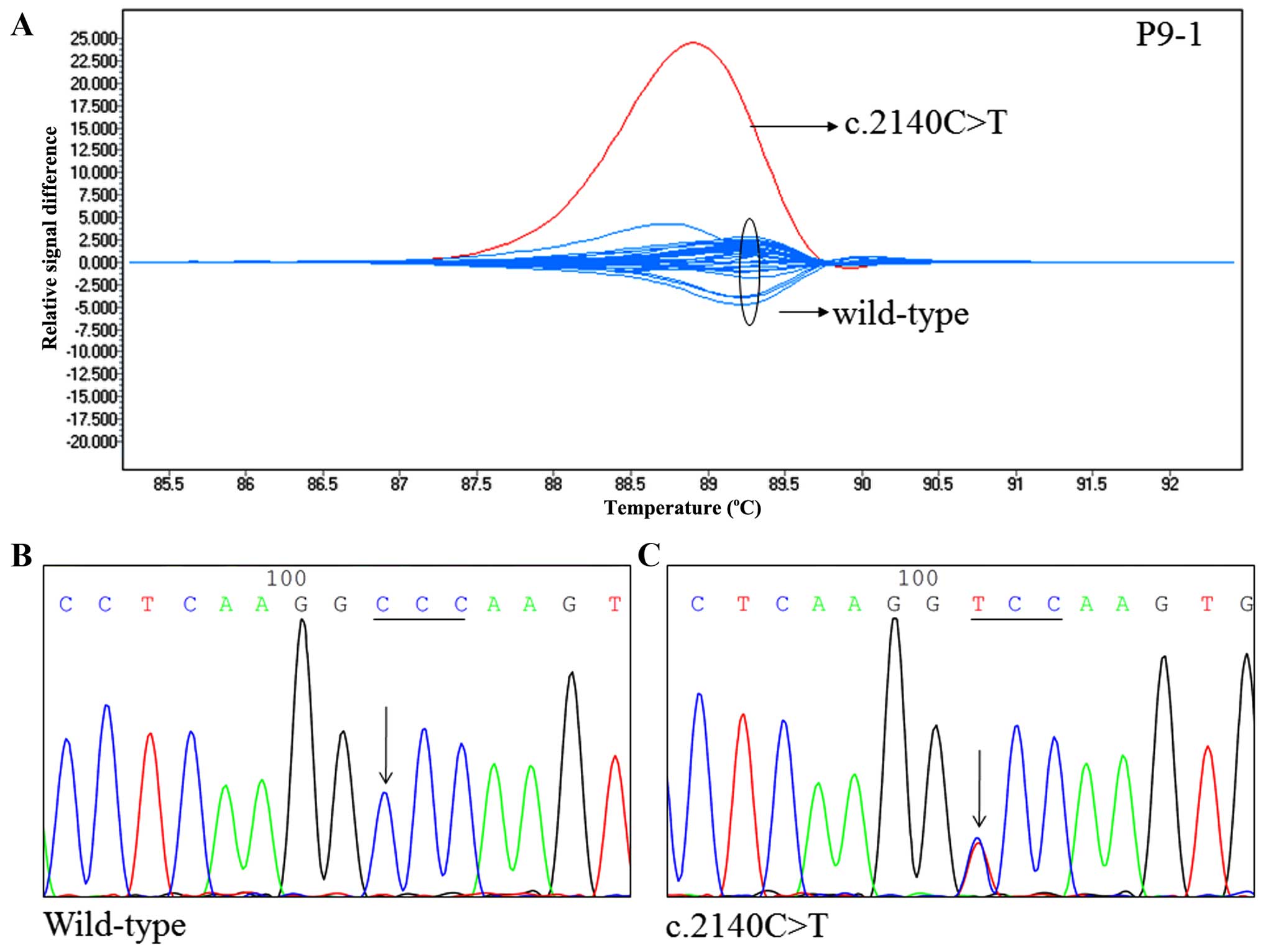
We used two primer pairs (P9-1 and P9-2) to identify
TNFAIP3 mutations in exon 9. One SNP resulted in a change
from cytosine to thymine at position 2140 (c.2140C>T) (Fig. 3). This was confirmed by direct
sequencing. The c.2140C>T change resulted in a Pro714Ser amino
acid substitution. Intriguingly, the SNP p.P714S is similar to the
amino acid change Pro714Ala, which has been reported previously
(rs369155845). In addition, a c.2140C>T nucleotide change was
found in one subject, for an incidence of 1.2%.
TNFAIP3 polymorphisms in normal
controls
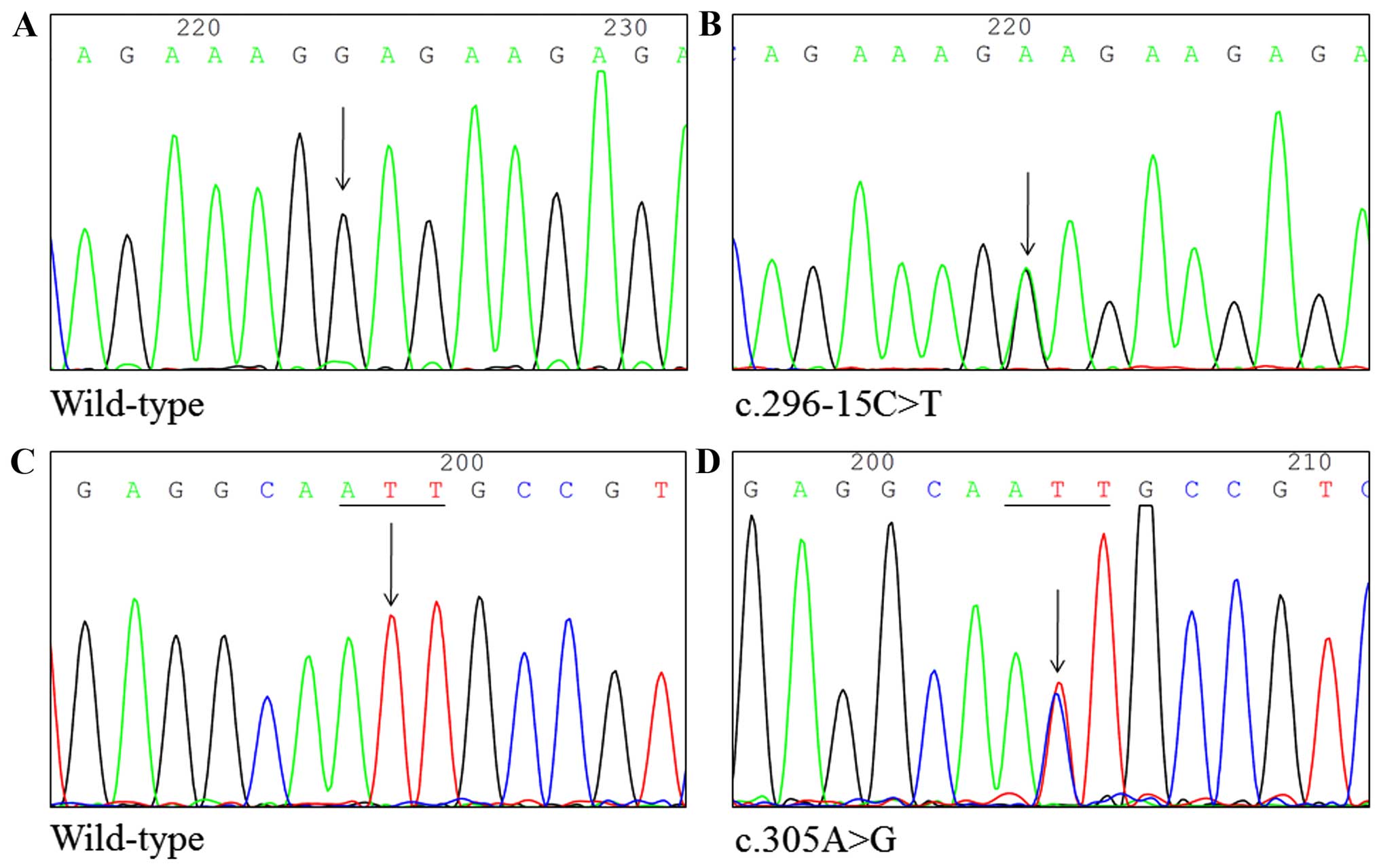
We recruited 50 healthy Taiwanese subjects from the
China Medical University Hospital. DNA from the subjects was
screened for TNFAIP3 mutations. We identified five SNPs by
HRM analysis, which were confirmed by direct DNA sequencing:
c.296-20_296-18delCTC, c.296-15C>T, c.305A>G, c.380T>G and
c.2140C>T (Fig. 4). In addition,
the nucleotide changes with c.296-20_296-18delCTC, c.296-15C>T,
c.305A>G, c.380T>G and c.2140C>T were found in 50, 1, 1,
7, and 1 subject, respectively, for incidences of 100, 2, 2, 14 and
2%, respectively.
Frequencies of the TNFAIP3 haplotype
The rs71670547, rs377482653, rs146534657 and
rs2230926 haplotypes were analyzed to investigate susceptibility to
OSCC. The haplotype CTC-C-A-G was marginally inversely associated
with a high risk of OSCC (OR=0.34; 95% CI=0.10-1.19; P=0.077;
Table II).
 | Table IIHaplotype frequency of
TNFAIP3_rs71670547, rs377482653, rs146534657 and rs2230926
among the OSCC patients. |
Table II
Haplotype frequency of
TNFAIP3_rs71670547, rs377482653, rs146534657 and rs2230926
among the OSCC patients.
| Haplotype | Cases n (%) | Controls n (%) | OR | 95% CI | P-value |
|---|
| del-C-A-T | 149 (92) | 88 (88) | 1 | Ref | |
| CTC-C-A-G | 4 (2) | 7 (7) | 0.34 | 0.10–1.19 | 0.0771 |
| CTC-C-A-T | 9 (6) | 3 (3) | 1.77 | 0.47–6.72 | 0.3946 |
| CTC-C-G-T | 0 (0) | 1 (1) | | | |
| del-T-A-T | 0 (0) | 1 (1) | | | |
Discussion
Several TNFAIP3 mutations involving various
substitutions in multiple tumor types have been reported [Table III, extracted from the Catalogue
of Somatic Mutations in the Cancer (COSMIC) Database]. Many studies
have focused on TNFAIP3 genetic variations in hematopoietic
and lymphoid tissue. According to the COSMIC database, the
TNFAIP3 mutation in OSCC has been reported by only one study
from the USA using whole-exome sequencing (20), and the only mutation (p.Y614C)
identified in that study was not found in the Taiwanese population
of our study. Conversely, we identified three mutations by HRM
analysis (p.E361K, p.S466R and p.P587L) and confirmed them by
direct sequencing. This discrepancy in results may reflect
different lifestyle risk factors. For example, oral cancer patients
from Taiwan are frequently exposed to betel-quid chewing, and the
betel-quid used in Taiwan is different from that used in other
countries.
 | Table IIITNFAIP3 gene mutations in a
variety of tumor types, extracted from the COSMIC database. |
Table III
TNFAIP3 gene mutations in a
variety of tumor types, extracted from the COSMIC database.
| Tumor type | TNFAIP3
Mut | All samples | % | Substitution -
Nonsense | Substitution -
Missense | Substitution -
Synonymous | Insertion -
Frameshift | Deletion -
Inframe | Deletion -
Frameshift | Complex | NonStop
extension | Whole gene
deletion | Unknown |
|---|
| Bone | 1 | 75 | 1.33 | | 1 | | | | | | | | |
| Breast | 8 | 1,390 | 0.58 | 1 | 2 | 5 | | | | | | | |
| Central nervous
system | 1 | 1,212 | 0.08 | | | 1 | | | | | | | |
| Endometrium | 3 | 505 | 0.59 | | 1 | 1 | | | 1 | | | | |
| Haematopoietic and
lymphoid | 200 | 2,720 | 7.35 | 46 | 32 | 4 | 34 | 1 | 62 | 2 | 1 | 25 | 18 |
| Kidney | 1 | 961 | 0.1 | | | 1 | | | | | | | |
| Large
intestine | 64 | 830 | 7.71 | 3 | 35 | 25 | 1 | | 2 | | | | |
| Liver | 6 | 942 | 0.64 | | 5 | | | | | | | | 1 |
| Lung | 21 | 1,505 | 1.4 | | 15 | 6 | | | | | | | |
| NS | 1 | 240 | 0.42 | | 1 | | | | | | | | |
| Oesophagus | 1 | 261 | 0.38 | | 1 | | | | | | | | |
| Ovary | 4 | 823 | 0.49 | | 4 | | | | | | | | |
| Pancreas | 2 | 913 | 0.22 | | 1 | | | | | | | | |
| Prostate | 2 | 503 | 0.4 | | 2 | | | | | | | | |
| Skin | 1 | 334 | 0.3 | | 1 | | | | | | | | |
| Stomach | 2 | 47 | 4.26 | | 2 | | | | | | | | |
| Thyroid | 3 | 32 | 9.38 | | 2 | 1 | | | | | | | |
| Upper aerodigestive
tract | 1 | 244 | 0.41 | | 1 | | | | | | | | |
| Urinary tract | 3 | 366 | 0.82 | | 2 | 1 | | | | | | | |
| Our present
study | 3 | 81 | 3.7 | | 3 | | | | | | | | |
Although the HRM method is a powerful screening
tool, it has some limitations; one of which is that unexpected
polymorphisms present in the mutation of interest may interfere
with genotyping. Therefore, we designed amplicon lengths of 100–300
bp, the suggested ideal size for HRM analysis. Two polymorphisms in
intron 2 (rs71670547 and rs377482653) and two in exon 3
(rs146534657 and rs2230926) caused difficulties in the HRM
analysis; these polymorphisms could not be differentiated using a
melting curve. To avoid this, the designed primers should flank the
exon or intron as closely as possible. When an SNP is close to the
exon or intron boundary, the primer can be placed over the SNP and
a mismatched base with no allelic preference can be introduced at
the SNP position (21). If the
amplicon length is increased, the wild-type and heterozygote curves
become smaller and are more difficult to distinguish. The HRM
method is unable to detect mutations encompassing an entire exon or
deletions of entire genes and exons.
Mice with the A20 deletion in intestinal epithelial
cells (IECs), B cells, myeloid cells, dendritic cells (DCs) and
keratinocytes have been investigated extensively. Specific A20
deletions in B cells exhibit enhanced B-cell proliferation and
survival and autoantibody production (22–24).
Mice with the A20 deletion in all cells of myeloid origin develop
spontaneous polyarthritis with the production of type II collagen
autoantibodies and inflammatory cytokines in serum (25). In addition, mice with a DC-specific
A20 deletion developed either SLE-like symptoms or human
inflammatory bowel disease in independent studies (26,27).
Furthermore, mice with A20-deficient IECs are highly sensitive to
dextran sodium sulfate-induced colitis and TNF due to IEC apoptosis
and loss of barrier integrity (28). Finally, A20 expression was
significantly decreased in human colorectal cancer samples compared
with adjacent non-neoplastic mucosa (29), and mice with A20-deficient
keratinocytes displayed keratinocyte hyperproliferation and
ectodermal organ abnormalities (30). Thus, conditional gene targeting
studies have demonstrated an important role for A20 in controlling
tissue homeostasis.
In conclusion, the HRM DNA screening method provides
a reliable, accurate, and rapid method of identifying
TNFAIP3 mutations for the clinical diagnosis of cancer.
Acknowledgments
This research was supported by grants from the
Taiwan Ministry of Health and Welfare Clinical Trial and Research
Center of Excellence (MOHW105-TDU-B-212-133019 and
MOHW104-TDU-B-212-124-002) and China Medical University Hospital
(DMR-103-121 and DMR-103-116).
References
|
1
|
Molinolo AA, Amornphimoltham P, Squarize
CH, Castilho RM, Patel V and Gutkind JS: Dysregulated molecular
networks in head and neck carcinogenesis. Oral Oncol. 45:324–334.
2009. View Article : Google Scholar :
|
|
2
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar
|
|
3
|
Ko YC, Huang YL, Lee CH, Chen MJ, Lin LM
and Tsai CC: Betel quid chewing, cigarette smoking and alcohol
consumption related to oral cancer in Taiwan. J Oral Pathol Med.
24:450–453. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tanaka T, Tanaka M and Tanaka T: Oral
carcinogenesis and oral cancer chemoprevention: A review. Pathol
Res Int. 2011:4312462011. View Article : Google Scholar
|
|
5
|
Petersen PE: Oral cancer prevention and
control - the approach of the World Health Organization. Oral
Oncol. 45:454–460. 2009. View Article : Google Scholar
|
|
6
|
Karin M and Greten FR: NF-kappaB: Linking
inflammation and immunity to cancer development and progression.
Nat Rev Immunol. 5:749–759. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oeckinghaus A, Hayden MS and Ghosh S:
Crosstalk in NF-κB signaling pathways. Nat Immunol. 12:695–708.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Catrysse L, Vereecke L, Beyaert R and van
Loo G: A20 in inflammation and autoimmunity. Trends Immunol.
35:22–31. 2014. View Article : Google Scholar
|
|
9
|
Dixit VM, Green S, Sarma V, Holzman LB,
Wolf FW, O'Rourke K, Ward PA, Prochownik EV and Marks RM: Tumor
necrosis factor-alpha induction of novel gene products in human
endothelial cells including a macrophage-specific chemotaxin. J
Biol Chem. 265:2973–2978. 1990.PubMed/NCBI
|
|
10
|
Wertz IE, O'Rourke KM, Zhou H, Eby M,
Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al:
De-ubiquitination and ubiquitin ligase domains of A20 downregulate
NF-kappaB signalling. Nature. 430:694–699. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kato M, Sanada M, Kato I, Sato Y, Takita
J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al:
Frequent inactivation of A20 in B-cell lymphomas. Nature.
459:712–716. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nomoto J, Hiramoto N, Kato M, Sanada M,
Maeshima AM, Taniguchi H, Hosoda F, Asakura Y, Munakata W,
Sekiguchi N, et al: Deletion of the TNFAIP3/A20 gene detected by
FICTION analysis in classical Hodgkin lymphoma. BMC Cancer.
12:4572012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Philipp C, Edelmann J, Bühler A, Winkler
D, Stilgenbauer S and Küppers R: Mutation analysis of the TNFAIP3
(A20) tumor suppressor gene in CLL. Int J Cancer. 128:1747–1750.
2011. View Article : Google Scholar
|
|
14
|
Chanudet E, Huang Y, Ichimura K, Dong G,
Hamoudi RA, Radford J, Wotherspoon AC, Isaacson PG, Ferry J and Du
MQ: A20 is targeted by promoter methylation, deletion and
inactivating mutation in MALT lymphoma. Leukemia. 24:483–487. 2010.
View Article : Google Scholar
|
|
15
|
Ma A and Malynn BA: A20: Linking a complex
regulator of ubiquitylation to immunity and human disease. Nat Rev
Immunol. 12:774–785. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Graham RR, Hom G, Ortmann W and Behrens
TW: Review of recent genome-wide association scans in lupus. J
Intern Med. 265:680–688. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Er TK and Chang JG: High-resolution
melting: Applications in genetic disorders. Clin Chim Acta.
414:197–201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yeh KT, Shih MC, Lin TH, Chen JC, Chang
JY, Kao CF, Lin KL and Chang JG: The correlation between CpG
methylation on promoter and protein expression of E-cadherin in
oral squamous cell carcinoma. Anticancer Res. 22:3971–3975.
2002.
|
|
19
|
Sobin LH and Fleming ID: TNM
Classification of Malignant Tumors, 5th edition (1997). Union
Internationale Contre le Cancer and the American Joint Committee on
Cancer. Cancer. 80:1803–1804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stransky N, Egloff AM, Tward AD, Kostic
AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C,
McKenna A, et al: The mutational landscape of head and neck
squamous cell carcinoma. Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shih HC, Er TK, Chang TJ, Chang YS, Liu TC
and Chang JG: Rapid identification of HBB gene mutations by
high-resolution melting analysis. Clin Biochem. 42:1667–1676. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tavares RM, Turer EE, Liu CL, Advincula R,
Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, et al:
The ubiquitin modifying enzyme A20 restricts B cell survival and
prevents autoimmunity. Immunity. 33:181–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hövelmeyer N, Reissig S, Xuan NT,
Adams-Quack P, Lukas D, Nikolaev A, Schlüter D and Waisman A: A20
deficiency in B cells enhances B-cell proliferation and results in
the development of autoantibodies. Eur J Immunol. 41:595–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chu Y, Vahl JC, Kumar D, Heger K, Bertossi
A, Wójtowicz E, Soberon V, Schenten D, Mack B, Reutelshöfer M, et
al: B cells lacking the tumor suppressor TNFAIP3/A20 display
impaired differentiation and hyperactivation and cause inflammation
and autoimmunity in aged mice. Blood. 117:2227–2236. 2011.
View Article : Google Scholar
|
|
25
|
Matmati M, Jacques P, Maelfait J,
Verheugen E, Kool M, Sze M, Geboes L, Louagie E, Mc Guire C,
Vereecke L, et al: A20 (TNFAIP3) deficiency in myeloid cells
triggers erosive polyarthritis resembling rheumatoid arthritis. Nat
Genet. 43:908–912. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kool M, van Loo G, Waelput W, De Prijck S,
Muskens F, Sze M, van Praet J, Branco-Madeira F, Janssens S, Reizis
B, et al: The ubiquitin-editing protein A20 prevents dendritic cell
activation, recognition of apoptotic cells, and systemic
autoimmunity. Immunity. 35:82–96. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hammer GE, Turer EE, Taylor KE, Fang CJ,
Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, et
al: Expression of A20 by dendritic cells preserves immune
homeostasis and prevents colitis and spondyloarthritis. Nat
Immunol. 12:1184–1193. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vereecke L, Sze M, Mc Guire C, Rogiers B,
Chu Y, Schmidt-Supprian M, Pasparakis M, Beyaert R and van Loo G:
Enterocyte-specific A20 deficiency sensitizes to tumor necrosis
factor-induced toxicity and experimental colitis. J Exp Med.
207:1513–1523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ungerbäck J, Belenki D, Jawad ul-Hassan A,
Fredrikson M, Fransén K, Elander N, Verma D and Söderkvist P:
Genetic variation and alterations of genes involved in
NFκB/TNFAIP3- and NLRP3-inflammasome signaling affect
susceptibility and outcome of colorectal cancer. Carcinogenesis.
33:2126–2134. 2012. View Article : Google Scholar
|
|
30
|
Lippens S, Lefebvre S, Gilbert B, Sze M,
Devos M, Verhelst K, Vereecke L, Mc Guire C, Guérin C, Vandenabeele
P, et al: Keratinocyte-specific ablation of the NF-κB regulatory
protein A20 (TNFAIP3) reveals a role in the control of epidermal
homeostasis. Cell Death Differ. 18:1845–1853. 2011. View Article : Google Scholar : PubMed/NCBI
|