Introduction
Nasopharyngeal carcinoma (NPC) is a non-lymphomatous
squamous cell carcinoma arising from epithelial cells located in
the nasopharynx (1). NPC is a rare
disease in most parts of the world and the incidence is 0.2–0.5
cases/100,000 people in the USA, but is high in Southern China and
Southeast Asia (2–5). NPC responds well to radiotherapy and
adjuvant chemotherapy; however, the 5-year overall survival rate is
still 70% (6–8). Therefore, it is important to elucidate
the pathogenesis of NPC for discovering new therapeutic
approaches.
MicroRNAs (miRNAs) are a class of short
single-stranded non-coding RNAs, 22–25 nucleotides, repressing
protein translation through directly binding to the 3′-untranslated
region (3′-UTR) of target mRNAs, leading to mRNA degradation or
translational suppression. miRNAs participate in the regulation of
a variety of basic biological processes such as development,
cellular differentiation, proliferation, apoptosis, invasion and
metabolism (9–11). A large number of studies have
demonstrated that the gain or loss of function of miRNAs
contributes to cancer development through silencing of tumor
suppressor genes or upregulation of oncogenes, including in NPC,
such as miR-21, miR-506, miR-223 and miR-142-3p (12–15).
miR-491-5p is deregulated and acts as a tumor suppressor in many
tumors, including cervical and breast cancer, oral squamous cell
and ovarian carcinoma (16–19). However, the function of miR-491-5p
in NPC pathogenesis, as well as the molecular mechanisms by which
miR-491-5p exerts its function and modulates the malignant
phenotypes of NPC cells, is not fully understood.
Notch genes (Notch 1–4) encode single-pass,
heterodimeric transmembrane receptors that serve as receptors for
the Jagged and δ-like ligands (20,21).
The Notch pathway has pivotal roles in cellular processes such as
proliferation, invasion, differentiation and development (22,23).
Notably, Notch signaling plays key roles in the progression of
cancer, and some Notch inhibitors have shown therapeutic efficacy
in preclinical studies (22,24).
In the present study, we aimed to explore the
potential regulatory mechanisms of miR-491-5p in NPC. We showed for
the first time that miR-491-5p is downregulated in NPC, and its
overexpression could suppress proliferation and invasion of NPC
cells through suppression of Notch3 expression.
Materials and methods
Cell culture and clinical sample
preparation
Four human NPC cell lines (C666-1, CNE1, CNE2 and
SUNE-1) were cultured in RPMI-1640 medium (Invitrogen, Carlsbad,
CA, USA) supplemented with 10% fetal bovine serum (FBS). The normal
nasopharyngeal epithelia cell line NP69 was cultured in
keratinocyte/serum-free medium (Invitrogen) supplemented with
bovine pituitary extract (BD Biosciences, San Jose, CA, USA)
(25). HEK293T cells were cultured
in Dulbecco's modified Eagle's medium (DMEM) at 37°C in a
humidified atmosphere with 5% CO2. Twenty NPC tissue
samples and 10 normal nasopharyngeal epithelial specimens were
frozen in liquid nitrogen and stored at −80°C until further use.
Informed consent was obtained from all patients and the present
study was approved by the Ethics Committee of the Jinshan Hospital
Affiliated to Fudan University.
RNA preparation and quantitative reverse
transcriptase polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from cell lines and tissues
using the TRIzol (Invitrogen). RNA was reverse transcribed to cDNA
using PrimeScript First Strand cDNA synthesis kit, qRT-PCR was
performed by SYBR Premix Ex Taq (both from Takara, Dalian, China).
β-actin or U6 was used for normalization. The relative expression
levels of the gene of interest were calculated using the
2−ΔΔCt method. All reactions were carried out in
triplicate and all experiments were performed on three independent
occasions.
Oligonucleotides and plasmid
transfection
miR-491-5p mimic and negative control were
synthesized by RiboBio (Guangzhou, China). The Notch3-3′-UTR
wild-type (WT) sequence was amplified and cloned into psiCHECK-2
luciferase reporter vector (Promega, Madison, WI, USA). Mutant
3/-UTR of Notch3 containing two mutated sequences within the
miR-491-5p target sites was generated. For Notch3 overexpression,
the full-length open reading frame of Notch3 without 3′-UTR was
subcloned into pcDNA3.1 vector (Invitrogen). Cells were transfected
with miR-491-5p mimic or plasmid using Lipofectamine 2000 reagent
(Invitrogen).
Western blot analysis
Cells were lysed in RIPA buffer and the protein
concentrations were evaluated using BCA protein assay kit (both
from Beyotime, Nantong, China). Total protein was prepared and
separated by 10% SDS-PAGE, then transferred onto polyvinylidene
difluoride (PVDF) membranes and blocked with 5% non-fat milk. The
target proteins were detected according to the standard methods
with the following primary antibodies: anti-Notch3 antibody and
goat anti-peroxidase-conjugated secondary antibody (both from Santa
Cruz Biotechnology, Santa Cruz, CA, USA). An anti-β-actin antibody
(Sigma, St. Louis, MO, USA) was used as the loading control.
Lentivirus production and
transduction
miR-491-5p precursor sequences were amplified and
cloned into the lentiviral vector pCDH-CMV-MCS-EF1-copGFP (System
Biosciences, Mountain View, CA, USA). A lentiviral vector that
expressed GFP alone was used as a control. Virus particles were
harvested 48 h after pCDH-CMV-miR-491-5p transfection with the
packaging plasmids psPAX2 and pMD into 293T cells using
Lipofectamine 2000 reagent. Target cells were infected with
recombinant lentivirus-transducing units plus 8 mg/ml Polybrene
(Sigma).
Cell proliferation assay
Cell viability was detected using the
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) assay. Cells were seeded in 96-well plates at
2×103 cells/well and incubated for 24, 48, 72 and 96 h.
Next, 5 µl MTT solution was added to each well and then
terminated by 200 µl dimethyl sulphoxide (DMSO). The
viability of cells was detected and measured at 490 nm using a
microplate reader (Bio-Rad, Richmond, CA, USA). All experiments
were repeated three times.
Cell migration and invasion assays
We used a Transwell chamber assay (24-well plates,
8-mm pore size; Corning, Corning, NY, USA) coated without or with
Matrigel (BD Biosciences), respectively, on the upper surface of
the membrane to determine the effect of miR-491-5p on migration and
invasion in vitro. Cells (2.5×104) were placed in
the top chambers in triplicate. For the invasion assay, the upper
chamber was coated with Matrigel. Cells on the upper surface of the
membrane were removed after 24 h incubation, and cells on the
undersurface were fixed, stained with 0.1% crystal violet and
counted under a light microscope. The Transwell migration assay was
performed in the same way as the invasion assay, but without the
Matrigel coating.
Cell cycle analysis
Cells at 80–90% confluence were transfected with
miR-491-5p or negative control. After 48 h, cells were collected by
trypsinization, washed twice in phosphate-buffered saline (PBS) and
fixed in 70% ethanol. These cells were incubated with DNA-binding
dye propidium iodide and RNase for 30 min at 37°C. Cells were
analyzed on a flow cytometer (FACSCalibur; BD Biosciences).
Luciferase reporter assay
For dual luciferase assays, cells were transiently
transfected with appropriate reporter plasmid and miRNA using
Lipofectamine 2000. After 48 h, cells were collected and luciferase
activity was measured using a Dual Luciferase Reporter Assay kit
(Promega).
In vivo tumor growth model
For tumor growth assay, 106 SUNE-1 cells
stably overexpressing miR-491-5p or negative control were suspended
in 200 µl PBS, and then subcutaneously injected into nude
mice. Four weeks later, the mice were sacrificed, tumors were
dissected and weighed, and tumor volume was calculated every three
days. The volumes were calculated as follows: volume = (D ×
d2)/2, where D was the longest diameter and d the
shortest diameter. All experiments involving animals were
undertaken in accordance with the Animal Experimentation Ethics
Committee Guide for the Care and Use of Laboratory Animals, with
the approval of the Institutional Animal Care and Use Committee of
Fudan University.
Statistical analysis
All statistical analyses were performed using SPSS
version 16.0 software (SPSS, Inc., Chicago, IL, USA) and are
expressed as mean ± SD. Differences between groups were determined
by Student's t-test or by two-way analysis of variance. P<0.05
was considered significant.
Results
miR-491-5p is downregulated in NPC cell
lines and tissues
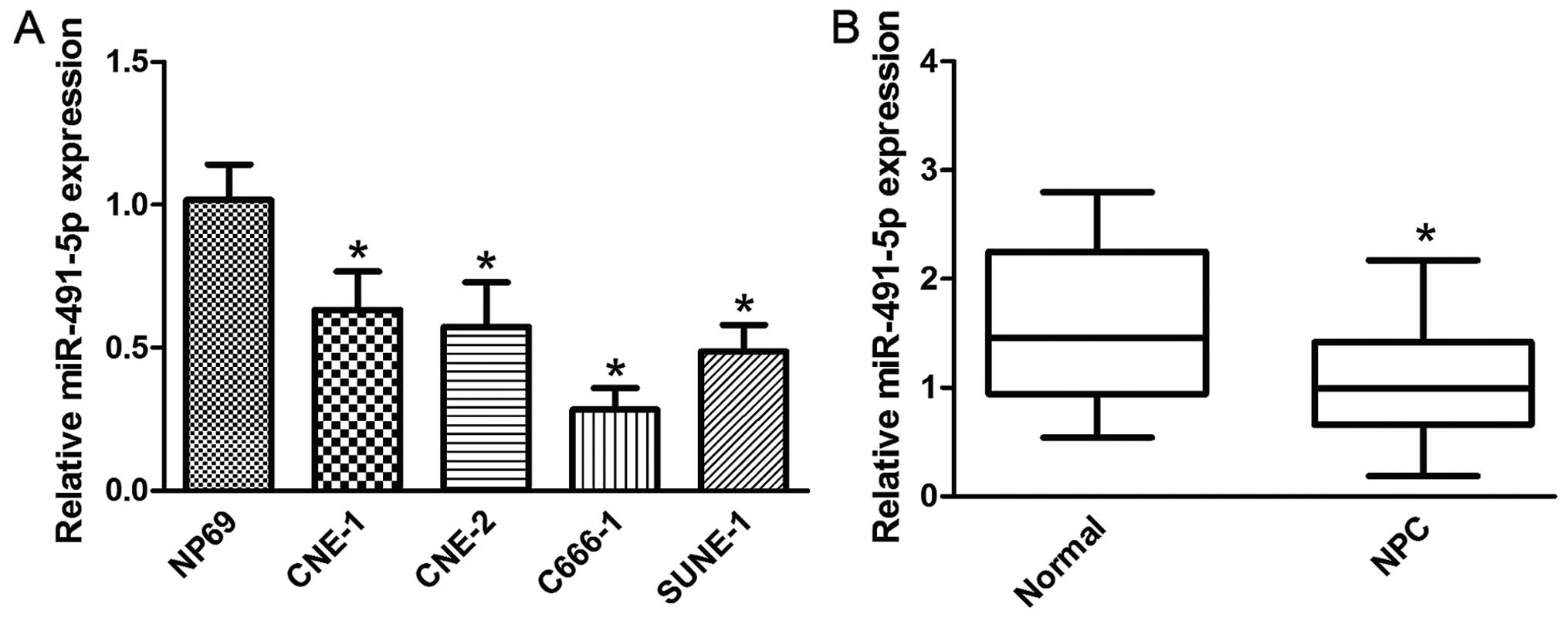
To examine the role of miR-491-5p in NPC, the
expression level of miR-491-5p was detected by qRT-PCR in NPC cell
lines and tissues. As shown in Fig.
1A, miR-491-5p was expressed at markedly lower levels in all
NPC cell lines tested, compared to the normal nasopharyngeal
epithelial cell line NP69. miR-491-5p was also downregulated in NPC
tissues compared with normal nasopharyngeal epithelial tissues
(Fig. 1B).
Effects of miR-491-5p overexpression on
NPC cell proliferation, migration and invasion
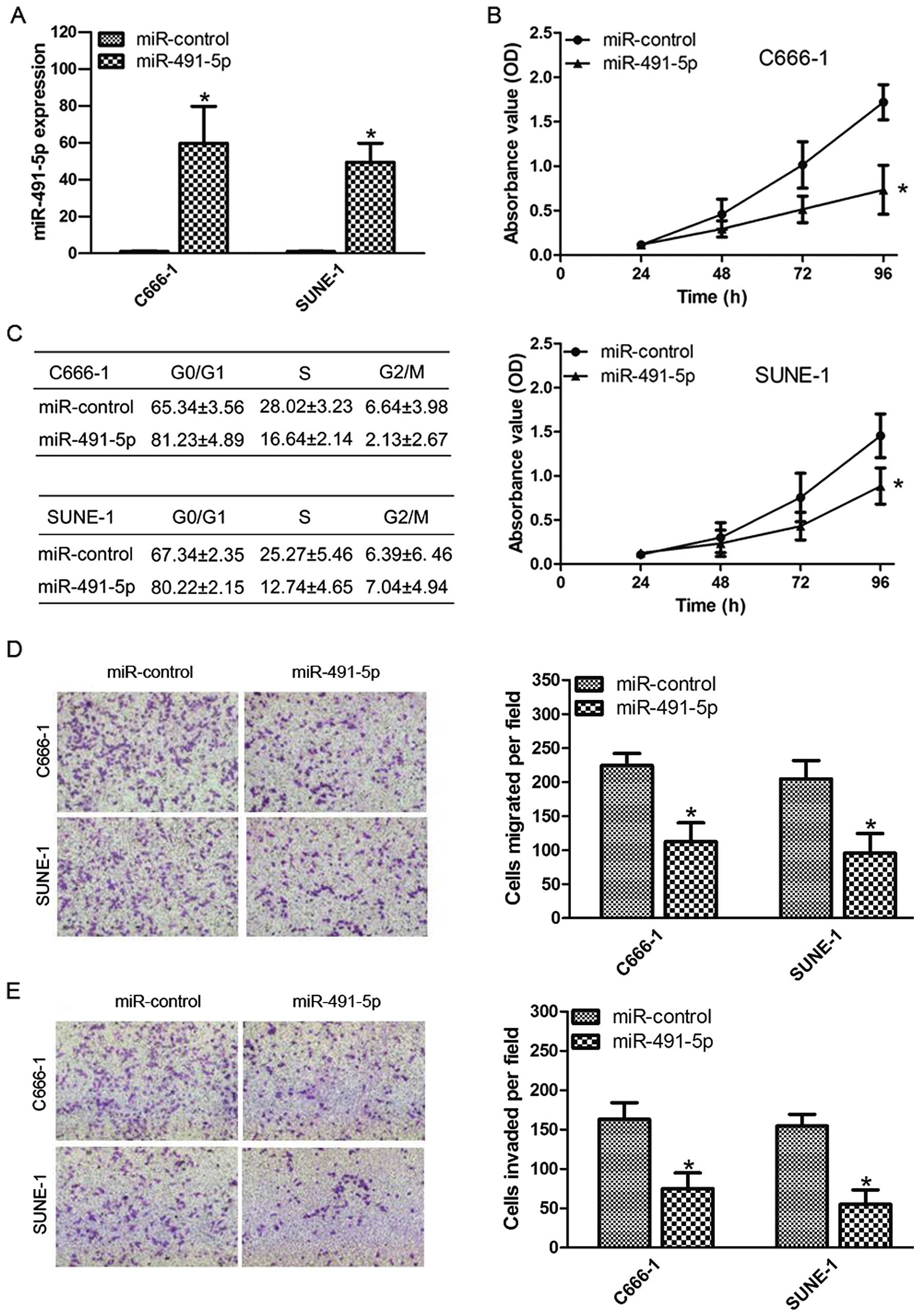
To investigate the effects of miR-491-5p on NPC cell
growth, C666-1 and SUNE-1 cells were transfected with miR-491-5p
mimic or negative control (Fig.
2A). Using the MTT assay, we found that overexpression of
miR-491-5p significantly inhibited cell proliferation compared with
the negative control (Fig. 2B). We
also tested the effects of miR-491-5p on the cell cycle progression
by flow cytometric analysis. Compared with the miR-control group,
miR-491-5p overexpression increased the percentage of cells in
G1/G0 phases and decreased the percentage of cells in S phase
(Fig. 2C). Transwell migration and
invasion assays showed that miR-491-5p overexpression inhibited
cell migration and invasion compared to the negative control
(Fig. 2D and E). These results
demonstrate that miR-491-5p suppresses several malignancy
parameters in human NPC cells.
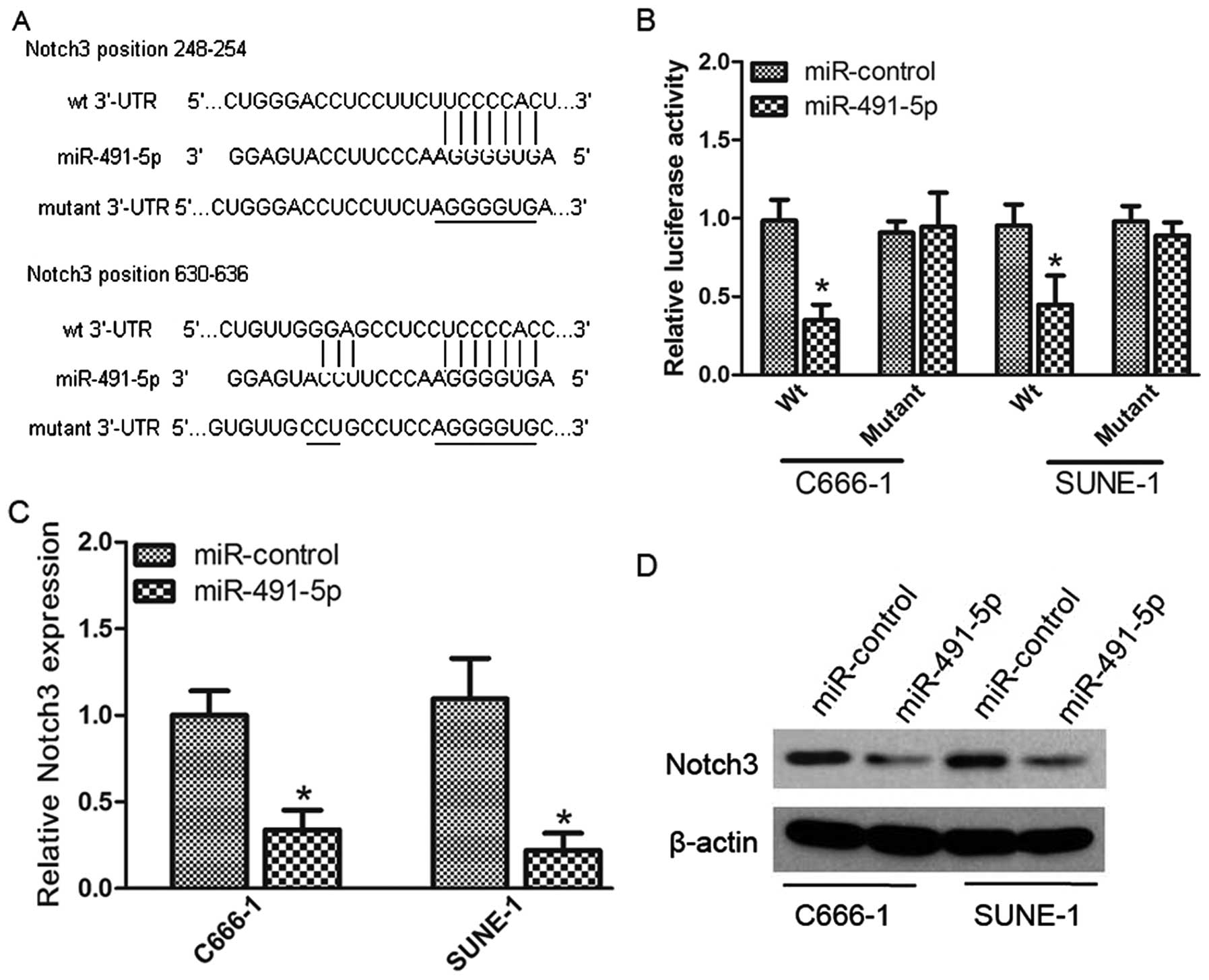
Notch3 is a target of miR-491-5p
The miRNA target analysis tools miRanda and
TargetScan were used to explore potential targets of miR-491-5p and
Notch3 was selected due to its positive roles in cancer cell
proliferation and invasion. Luciferase reporter assay was performed
to determine whether Notch3 was a direct target of miR-491-5p. We
first cloned the WT or mutant miR-491-5p target sequences of the
Notch3 3′-UTR into luciferase reporter vectors (Fig. 3A). After co-transfection with
miR-491-5p mimic, the luciferase activity of the WT 3′-UTR reporter
gene reduced significantly, whereas the activity of the mutant
reporter gene was not affected (Fig.
3B), confirming that miR-491-5p can bind to the Notch3 3′-UTR.
In addition, we analyzed the level of Notch3 expression in C666-1
and SUNE-1 cells transfected with miR-491-5p mimic or miR-control
by qRT-PCR and western blotting. miR-491-5p overexpression
significantly reduced the Notch3 expression at both the mRNA and
protein levels (Fig. 3C and D).
These data indicate that Notch3 was directly and negatively
regulated by miR-491-5p.
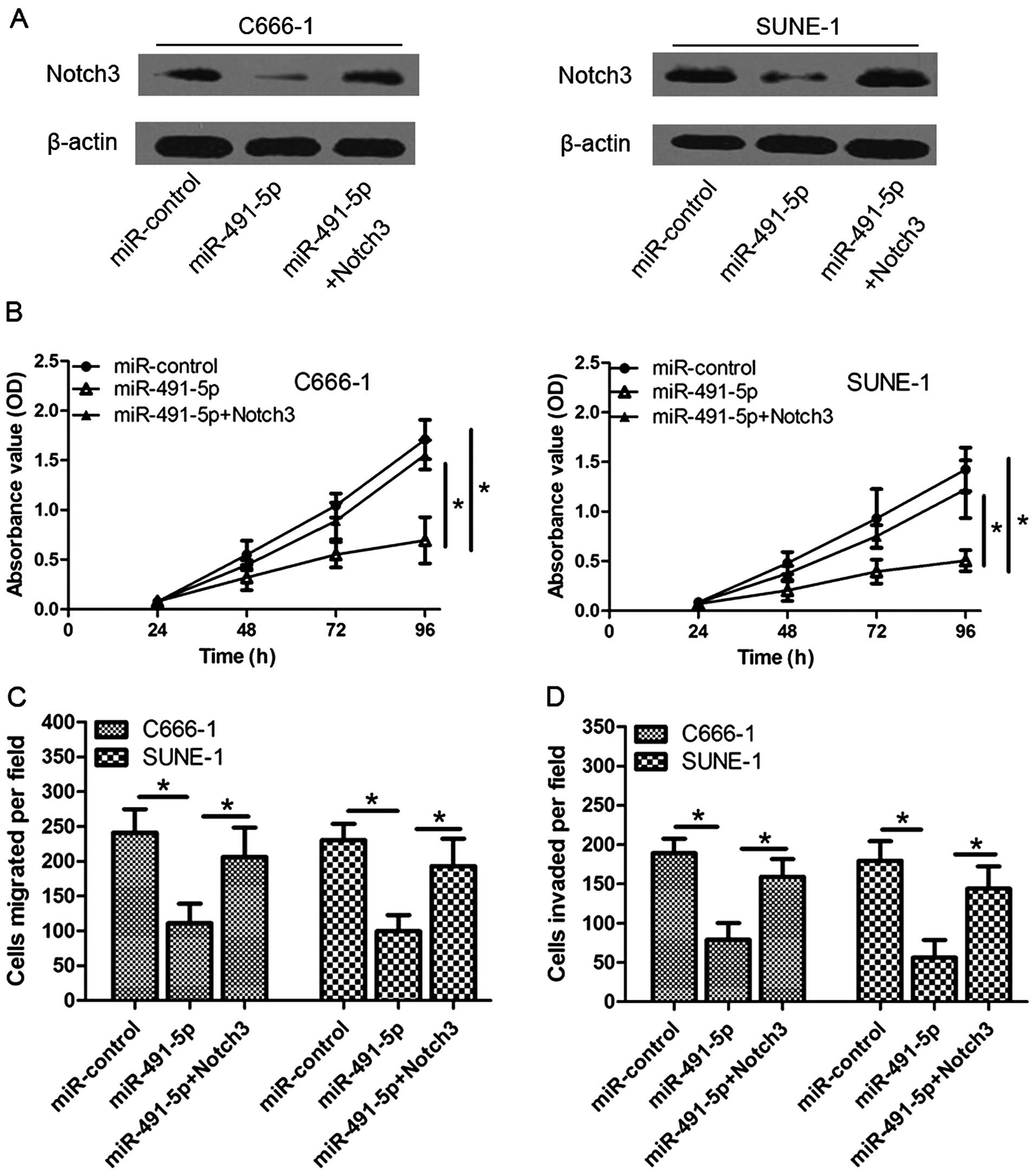
Notch3 attenuates the suppressive effect
of miR-491-5p
Given that miR-491-5p could suppress cell
proliferation, migration and invasion as well as inhibit the
expression of Notch3 through regulating Notch3 3′-UTR, we predicted
that ectopic expression of Notch3 may reverse the suppressive
effects of miR-491-5p. Notch3 ORF lacking 3′-UTR was co-transfected
with miR-491-5p in C666-1 and SUNE-1 cells, and western blotting
validated expression of Notch3 (Fig.
4A). As shown in Fig. 4B–D,
restoration of Notch3 partially rescued proliferative and invasive
ability impaired by miR-491-5p. These data indicate that Notch3 is
critically involved in mediating the tumor-inhibitory function of
miR-491-5p.
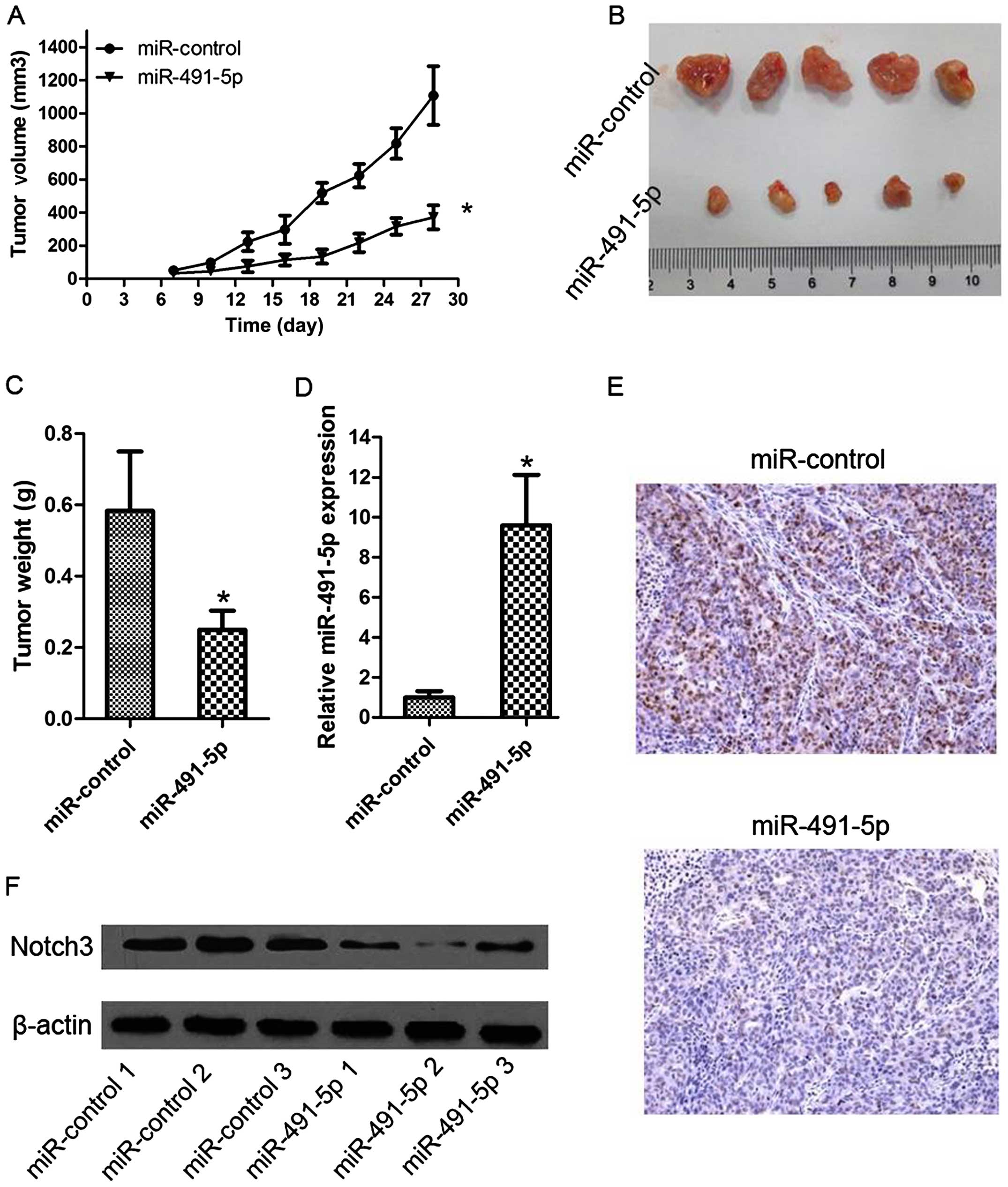
Antitumor effects of miR-491-5p on NPC
xenografts
To explore the function of miR-491-5p on tumor
growth, we constructed a SUNE-1 cell line stably overexpressing
miR-491-5p by lentivirus-mediated transduction. These cells were
injected subcutaneously into the flanks of nude mice, and tumor
growth was measured until sacrifice of the mice after 28 days. The
tumors of the LV-miR-491-5p group had smaller volumes and sizes
than the control tumors (Fig. 5A and
B). The average tumor weight was also significantly lower in
the LV-miR-491-5p group (Fig. 5C).
Furthermore, we examined cell proliferation of NPC and the
expression of miR-491-5p and Notch3 in the subcutaneous tissues.
The results revealed that LV-miR-491-5p increased tumor miR-491-5p
levels, reduced the expression of Notch3 protein, and suppressed
expression of cell proliferation marker Ki67 compared with negative
control (Fig. 5D–F). These data
indicate that miR-491-5p can significantly inhibit NPC cell growth,
and it acts as a negative regulator in NPC.
Discussion
It is now widely accepted that miRNAs contribute to
cancer development by acting as oncogenes or tumor suppressor
genes. Previous studies have shown that miR-491-5p is dysregulated
in various cancers and its potential function has also been partly
explored in several studies (16–18).
For example, Zhao et al reported that miR-491-5p suppressed
cervical cancer cell proliferation, migration and invasion, induced
cell apoptosis and suppressed tumor growth in a mouse model
(16). Zeng et al found that
miR-491-5p functions as a tumor suppressor by targeting JMJD2B in
estrogen-receptor-α-positive breast cancer (17). Huang et al showed that
miR-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis (18). These consistent results suggest that
miR-491-5p functions as a tumor suppressor and affects cell
biological behavior in cancer. However, to date, no functional
evidence of miR-491-5p in NPC has been documented. Therefore,
further extensive investigations are required to identify
miR-419-5p that is involved in the cell growth of NPC.
In the present study, we found that miR-491-5p
expression was markedly downregulated in human NPC cells and
tissues. Increased expression of miR-491-5p inhibited cell
proliferation, migration and invasion and induced cell cycle
arrest. We also explored the role of miR-491-5p in vivo
using a xenograft mouse model and found that tumor growth was
suppressed when miR-491-5p was overexpressed. These data suggest a
tumor-inhibitory role of miR-491-5p in NPC cells both in
vitro and in vivo.
The mechanism of the effect of miR-491-5p on the
development of NPC was also investigated in the present study.
Notch3 was predicted to be a theoretical target gene of miR-491-5p.
We found that miR-491-5p bound to the 3′-UTR of Notch3 mRNA and
repressed its expression. Further studies indicated that
overexpression of Notch3 reversed the tumor suppressive effects of
miR-491-5p on NPC cells. All these data suggest that Notch3 is a
direct and functional target of miR-491-5p in NPC cells.
Notch3 is one of the four different Notch proteins
(Notch1-4), which have emerged as important players in cancer
therapy, and which have been found to affect diverse processes,
such as proliferation, invasion and migration. For instance, Notch3
was upregulated in ovarian cancer and its inhibitor reduced
viability, migration and angiogenesis, and increased apoptosis
(26). Jaskula-Sztul et al
indicated that Notch3 suppressed medullary thyroid cancer cell
proliferation and induced apoptosis (27). Alqudah et al showed that
Notch3 plays a major role in glioma cell proliferation, migration,
invasion and apoptosis (28).
Notch3 is regulated by miRNAs in many cancers. Wang et al
found that miR-206 inhibited tumor proliferation and migration
involving the downregulation of Notch3 in colorectal cancer
(29). Furukawa et al
suggested that the miR-1/Notch3 pathway is important for colorectal
tumor cell migration and may be a promising molecular target for
the treatment of colorectal tumors (30). Previous studies reported that Notch3
is upregulated in NPC cell lines, and targeting Notch3 signaling
may serve as a potential therapeutic approach in patients with
Epstein-Barr-virus-associated NPC (31). Thus, it is particularly important
for the exploration of the regulatory mechanism of Notch3 in NPC
development, and the present study showed for the first time that
Notch3 was a direct downstream target gene of miR-491-5p, affecting
NPC development.
In conclusion, we demonstrated that miR-491-5p is
downregulated in NPC and may function as a tumor suppressor by
directly targeting Notch3. To the best of our knowledge, this is
the first study to demonstrate that the miR-491-5p/Notch3 axis
regulates the proliferation, migration and invasion of NPC cells.
These findings provide a better understanding of the pathogenesis
and development of NPC and may have important implications for
future therapy of NPC.
Acknowledgments
The present study was supported by the Key Specialty
Construction Project of Medical Health System in The Fourth Cycle,
Jinshan, Shanghai (2012-26).
References
|
1
|
Zhang SX, Qiu QH, Chen WB, Liang CH and
Huang B: Celecoxib enhances radiosensitivity via induction of
G2-M phase arrest and apoptosis in nasopharyngeal
carcinoma. Cell Physiol Biochem. 33:1484–1497. 2014. View Article : Google Scholar
|
|
2
|
Kamran SC, Riaz N and Lee N:
Nasopharyngeal carcinoma. Surg Oncol Clin N Am. 24:547–561. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu J, Zhao FP, Peng Z, Zhang MW, Lin SX,
Liang BJ, Zhang B, Liu X, Wang L, Li G, et al: EZH2 promotes
angiogenesis through inhibition of miR-1/Endothelin-1 axis in
nasopharyngeal carcinoma. Oncotarget. 5:11319–11332. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen LX, Zhu LY, Jacob TJ and Wang LW:
Roles of volume-activated Cl− currents and regulatory
volume decrease in the cell cycle and proliferation in
nasopharyngeal carcinoma cells. Cell Prolif. 40:253–267. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo Z, Dai Y, Zhang L, Jiang C, Li Z, Yang
J, McCarthy JB, She X, Zhang W, Ma J, et al: miR-18a promotes
malignant progression by impairing microRNA biogenesis in
nasopharyngeal carcinoma. Carcinogenesis. 34:415–425. 2013.
View Article : Google Scholar
|
|
6
|
Zhang T, Sun Q, Liu T, Chen J, Du S, Ren
C, Liao G and Yuan Y: MiR-451 increases radiosensitivity of
nasopharyngeal carcinoma cells by targeting ras-related protein 14
(RAB14). Tumour Biol. 35:12593–12599. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang GL, Chen ML, Li YZ, Lu Y, Pu XX, He
YX, Tang SY, Che H, Zou Y, Ding C, et al: Association of miR-146a
gene polymorphism with risk of nasopharyngeal carcinoma in the
central-southern Chinese population. J Hum Genet. 59:141–144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang GL, Lu Y, Pu XX, He YX, Chen ML, Li
YZ, Tang SY, Che H and He Z: Association study between miR-149 gene
polymorphism and nasopharyngeal carcinoma. Biomed Rep. 1:599–603.
2013.
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Y, Yan L, Zhang W, Wang H, Chen W, Hu N
and Ou H: miR-21 inhibitor suppresses proliferation and migration
of nasopharyngeal carcinoma cells through down-regulation of BCL2
expression. Int J Clin Exp Pathol. 7:3478–3487. 2014.PubMed/NCBI
|
|
13
|
Zhang Z, Ma J, Luan G, Kang L, Su Y, He Y
and Luan F: MiR-506 suppresses tumor proliferation and invasion by
targeting FOXQ1 in nasopharyngeal carcinoma. PLoS One.
10:e01228512015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang W, Lan X, Li D, Li T and Lu S:
MiR-223 targeting MAFB suppresses proliferation and migration of
nasopharyngeal carcinoma cells. BMC Cancer. 15:4612015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MiR-142-3p suppresses SOCS6 expression and promotes cell
proliferation in nasopharyngeal carcinoma. Cell Physiol Biochem.
36:1743–1752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Q, Zhai YX, Liu HQ, Shi YA and Li XB:
MicroRNA-491-5p suppresses cervical cancer cell growth by targeting
hTERT. Oncol Rep. 34:979–986. 2015.PubMed/NCBI
|
|
17
|
Zeng H, Yiling C, Wenting Y, XuQun H,
ChuanYi Z and Hui L: miR-491-5p functions as a tumor suppressor by
targeting JMJD2B in ERα-positive breast cancer. FEBS Lett.
589:812–821. 2015. View Article : Google Scholar
|
|
18
|
Huang WC, Chan SH, Jang TH, Chang JW, Ko
YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al:
miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis. Cancer Res.
74:751–764. 2014. View Article : Google Scholar
|
|
19
|
Denoyelle C, Lambert B, Meryet-Figuière M,
Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH,
Gauduchon P, et al: miR-491-5p-induced apoptosis in ovarian
carcinoma depends on the direct inhibition of both
BCL-XL and EGFR leading to BIM activation. Cell Death
Dis. 5:e14452014. View Article : Google Scholar
|
|
20
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Baron M: An overview of the Notch
signalling pathway. Semin Cell Dev Biol. 14:113–119. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fre S, Huyghe M, Mourikis P, Robine S,
Louvard D and Artavanis-Tsakonas S: Notch signals control the fate
of immature progenitor cells in the intestine. Nature. 435:964–968.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: A little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan X, Wu H, Xu H, Xiong H, Chu Q, Yu S,
Wu GS and Wu K: Notch signaling: An emerging therapeutic target for
cancer treatment. Cancer Lett. 369:20–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang
L, Xu X, Peng X, Li G, Tian W, et al: miR-9 targets CXCR4 and
functions as a potential tumor suppressor in nasopharyngeal
carcinoma. Carcinogenesis. 35:554–563. 2014. View Article : Google Scholar
|
|
26
|
Kang H, Jeong JY, Song JY, Kim TH, Kim G,
Huh JH, Kwon AY, Jung SG and An HJ: Notch3-specific inhibition
using siRNA knockdown or GSI sensitizes paclitaxel-resistant
ovarian cancer cells. Mol Carcinog. Jul 24–2015.Epub ahead of
print. View
Article : Google Scholar
|
|
27
|
Jaskula-Sztul R, Eide J, Tesfazghi S,
Dammalapati A, Harrison AD, Yu XM, Scheinebeck C, Winston-McPherson
G, Kupcho KR, Robers MB, et al: Tumor-suppressor role of Notch3 in
medullary thyroid carcinoma revealed by genetic and pharmacological
induction. Mol Cancer Ther. 14:499–512. 2015. View Article : Google Scholar :
|
|
28
|
Alqudah MA, Agarwal S, Al-Keilani MS,
Sibenaller ZA, Ryken TC and Assem M: NOTCH3 is a prognostic factor
that promotes glioma cell proliferation, migration and invasion via
activation of CCND1 and EGFR. PLoS One. 8:e772992013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang XW, Xi XQ, Wu J, Wan YY, Hui HX and
Cao XF: MicroRNA-206 attenuates tumor proliferation and migration
involving the downregulation of NOTCH3 in colorectal cancer. Oncol
Rep. 33:1402–1410. 2015.PubMed/NCBI
|
|
30
|
Furukawa S, Kawasaki Y, Miyamoto M,
Hiyoshi M, Kitayama J and Akiyama T: The miR-1-NOTCH3-Asef pathway
is important for colorectal tumor cell migration. PLoS One.
8:e806092013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Man CH, Wei-Man Lun S, Wai-Ying Hui J, To
KF, Choy KW, Wing-Hung Chan A, Chow C, Tin-Yun Chung G, Tsao SW,
Tak-Chun Yip T, et al: Inhibition of NOTCH3 signalling
significantly enhances sensitivity to cisplatin in EBV-associated
nasopharyngeal carcinoma. J Pathol. 226:471–481. 2012. View Article : Google Scholar
|