Introduction
Prostate cancer is among the most common neoplasias
in men and a leading cause of death (1). Although the majority of prostate tumor
patients are diagnosed with localized disease that can be
efficiently treated with surgery and radiation therapy, men
presenting with metastasized prostate tumors have a much bleaker
survival chance. Thus, there is a critical need to develop new
avenues of therapy, which may arise from a better understanding of
the molecular changes in prostate cancer cells (2).
In approximately half of all human prostate tumors,
the gene encoding the ETS transcription factor, ERG, becomes
translocated, resulting in the overexpression of the ERG protein
(3,4). Mimicking this overexpression in the
prostates of mice led to the development of the precursor of
prostate carcinomas, prostatic intraepithelial neoplasia (5,6). One
research group even reported that very high prostate-specific
expression of ERG induced carcinoma formation in approximately half
of the respective transgenic mice at old age (7). Furthermore, when combined with
knockout of the tumor-suppressor phosphatase and tensin homolog
(PTEN), ERG overexpression accelerated prostate cancer development
(8–11). These data implicate a causal role
for ERG in the development of prostate tumors. However, the
molecular details of how ERG overexpression contributes to the
neoplastic transformation of prostate cells are far from
resolved.
In the present study, we identified lysine
demethylase 4A (KDM4A), also known as Jumonji C domain-containing
protein 2A (JMJD2A), as a novel interaction partner of ERG.
Furthermore, we found that ERG and KDM4A cooperated in regulating
the transcription of the Yes-associated protein 1 (YAP1) gene,
which is a downstream effector in the Hippo signaling pathway that
plays important roles in development, homeostasis and cancer
(12,13).
Materials and methods
Luciferase assays
Human VCaP prostate cancer cells were grown in
6-wells at 37°C in a humidified atmosphere containing 5%
CO2 and were transfected utilizing 8 µg
polyethylenimine. In general, 1,500 ng pBluescript KS+
as a carrier and 500 ng of indicated luciferase reporter gene
constructs, which were based on the pGL2-Basic plasmid (Promega)
and contained human YAP1 promoter fragments (amplified out of LNCaP
prostate cancer cells) cloned between the SmaI and
HindIII sites, were used for transfection. In addition,
indicated amounts of ERG expression plasmid, empty vector pEV3S and
Flag-tagged KDM4A (or its H188A mutant) expression vector were
co-transfected. Cells were lysed 36 h after transfection and
luciferase activity was measured as previously reported (14). In the case of human BPH-1 normal
prostate cells, they were grown in 12-wells and transfected with
500 ng pBluescript KS+ and 500 ng YAP1 (−390/+22)
luciferase reporter construct utilizing 2 µg
polyethylenimine. Similarly, human LAP-C4 prostate cancer cells
were grown in 12-wells and transfected with 750 ng pBluescript
KS+ and 250 ng YAP1 (−390/+22) luciferase reporter
plasmid also using 2 µg polyethylenimine, whereas 200 ng
luciferase reporter construct, 800 ng pBluescript KS+, 1
ng ERG expression plasmid or pEV3S, and 2.5 µg
polyethylenimine were employed in the case of human embryonic
kidney 293T cells.
Preparation of protein extracts
Human 293T cells were seeded onto
poly-L-lysine-coated 6-cm dishes (15) and transiently transfected by the
calcium phosphate coprecipitation method (16) with 4.5 µg pBluescript
KS+ and either 4.5 µg empty vector pEV3S or
ERG-Myc-Flag expression plasmid. Thirty-six hours after
transfection, cells were washed once with phosphate-buffered saline
and cells were detached by a 5-min incubation in 40 mM HEPES (pH
7.4), 150 mM NaCl, 10 mM EDTA, after which cells were sprayed off
by pipetting. Then, cells were collected by centrifugation and
resuspended in 150 µl of 10 mM Tris, 30 mM
Na4P2O7 (pH 7.1), 175 mM NaCl, 50
mM NaF, 2 mM dithiothreitol, 1% Triton X-100, 1 mM
phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 2
µg/ml aprotinin, 1 µg/ml pepstatin, lysed for 30 min
on ice and debris was removed by centrifugation (17). Extracts were frozen in liquid
nitrogen and then stored at −80°C before use in DNA-binding assays.
The presence of ERG in these extracts was assessed by western
blotting (18) utilizing rabbit
monoclonal ERG antibody (EPR3864; ab92513; Abcam), while total
actin was detected with a rabbit polyclonal antibody (A2066;
Sigma).
Electrophoretic mobility shift assay
Wild-type or mutated E74 oligonucleotides, which
were previously described (19) or
the below listed pairs of DNA oligonucleotides were hybridized to
obtain double-stranded oligonucleotides. Then, they were
radioactively labeled with 32P-dATP by filling in
5′-overhanging ends with Klenow DNA polymerase (20). Binding of 0.1 µl protein
extract to ~0.25 ng 32P-labeled oligonucleotides
occurred in 10 µl of 20 mM HEPES (pH 7.4), 25 mM NaCl, 0.5
mM EDTA, 0.1 µg/µl bovine serum albumin, 0.05
µg/µl poly(dI-dC)•poly(dI-dC), 2 mM dithiothreitol,
0.01% Tween-20 and 12% glycerol (21). As indicated, 0.05 µl of
anti-Myc (9E10 mouse monoclonal antibody; M4439; Sigma) or anti-HA
(12CA5 mouse monoclonal antibody; ab16918; Abcam) antibody was
added. For competition experiments, 0.05 µl unlabeled
oligonucleotide (12.5 ng) was additionally added. After a 30-min
incubation on ice, the binding reactions were electrophoresed at
4°C on 4% native acrylamide gels as previously described (19). After drying, the gels were exposed
to film at −80°C (20). The
sequence of the oligonucleotide pairs used was as follows: '1/2',
5′-AGCGGAGCGGAAGAACTTCCTGCAGCCA-3′ and 5′-CTTGGCTGCAGGAAGTTCTTC
CGCTCCGCT-3′; 'm1/2', 5′-AGCGGAGCGGTAGAACTTC CTGCAGCCA-3′ and
5′-CTTGGCTGCAGGAAGTTCTACC GCTCCGCT-3′; '1/m2',
5′-AGCGGAGCGGAAGAACTACCTGCAGCCA-3′ and
5′-CTTGGCTGCAGGTAGTTCTTCCGCTCCGCT-3′; '3',
5′-GTTCGGACCCGGATTGGACCC-3′ and 5′-GATGGGTCCAATCCGGGTCCGA-3′; '4',
5′-AGTGTGCAGGAATGTAGCA-3′ and 5′-AGTTGCTACATTCCTGCAC-3′; '5',
5′-CTTGCAGCGAAAAGTTTCCCT GCGCTG-3′ and 5′-CAGCGCAGGGAAACTTTTCGCT
GCA-3′; '6/7', 5′-GCGCAGAGGAAGGAAGAGCCGAG-3′ and
5′-CTCTCGGCTCTTCCTTCCTCTGCGC-3′; 'm6/7',
5′-GCGCAGACGAAGGAAGAGCCGAG-3′ and 5′-CTCTCGGCTCTTCCTTCGTCTGCGC-3′;
'6/m7', 5′-GCGCAGAGGAAGGACGAGCCGAG-3′ and
5′-CTCTCGGCTCGTCCTTCCTCTGCGC-3′; '8', 5′-GCCGCCAGGGAAAAGAA-3′ and
5′-CTTTCTTTTCCCTGGCGGC-3′.
Protein binding assays
ERG was fused C-terminally of GST (glutathione
S transferase) and was expressed in Escherichia coli
(22). The resulting GST-ERG fusion
protein was purified by employing glutathione agarose beads, and
then was dialyzed against 20 mM HEPES (pH 7.4), 50 mM NaCl, 10%
glycerol, 0.2 mM phenylmethylsulfonyl fluoride and 1 mM
dithiothreitol (23). To produce
human KDM4A protein, its cDNA was cloned into a derivative of
pFastBac™ 1 (Invitrogen, Carlsbad, CA, USA), which added a combined
Flag/6His-tag onto the KDM4A N-terminus. The Bac-to-Bac system
(Invitrogen) was used to generate KDM4A recombinant baculovirus
according to the recommendations of the manufacturers, and Sf9
insect cells were infected with this virus and subsequently grown
at 27°C in a spinner culture for 4 days. The His-tagged KDM4A
protein was then affinity-purified with the help of
Ni2+-nitrilotriacetic acid agarose (Qiagen) and dialyzed
as previously described (24).
Then, binding reactions were set up in 600 µl of 25 mM HEPES
(pH 7.4), 25 mM NaCl, 0.01% Tween-20, 1 mM dithiothreitol, 0.2 mM
phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 2
µg/ml aprotinin, 1 µg/ml pepstatin by first binding
GST or GST-ERG to ~20 µl of glutathione agarose beads
followed by challenge with purified Flag/6His-KDM4A. After three
washes in binding buffer, bound proteins were boiled off with
Laemmli sample buffer, subjected to SDS polyacrylamide gel
electrophoresis and then revealed by either Coomassie staining or
anti-Flag (M2 mouse monoclonal antibody; F3165; Sigma) western
blotting (25).
Coimmunoprecipitation
Human embryonic kidney 293T cells, which were grown
in 6-cm dishes, were transfected using the calcium phosphate
coprecipitation method (26) with
expression plasmids encoding Myc-tagged ERG and HA-tagged KDM4A.
Thirty-six hours after transfection, cells were lysed and
immunoprecipitations with anti-Myc mouse monoclonal antibodies
(9E10; M4439; Sigma) were performed as previously described
(27). Thereafter, the
immunoprecipitates were subjected to SDS polyacrylamide gel
electrophoresis. Then, western blotting was performed using anti-HA
mouse monoclonal antibodies (12CA5; ab16918; Abcam) for detection
of coprecipitated KDM4A (28).
Chromatin immunoprecipitation assay
Human 293T cells were grown in 10-cm dishes and
transiently transfected with 3 µg YAP1 (−496/+22) luciferase
reporter gene, 25 µg pBluescript KS+ and 0, 10 or
50 ng ERG expression plasmid using the calcium phosphate
coprecipitation method (29). Two
days after transfection, the cells were treated with 1%
formaldehyde for 12 min at room temperature (30). Lysis of cells, sonication of
resultant extracts and chromatin immunoprecipitations were then
performed as previously described (31). The following rabbit polyclonal
antibodies were employed: H3K4me3 (2.4 µg;
ab8580; Abcam), H3K9me3 (3 µg; 07-442),
H3K27me3 (4 µg; 07-449) (both from Upstate) and
H3K36me3 (2 µg; ab9050; Abcam).
Immunoprecipitated DNA fragments were amplified by PCR using the
GoTaq DNA polymerase kit (M3008; Promega, Fitchburg, WI, USA)
according to the manufacturer's recommendation and with the
following temperature program (32): 2 min at 98°C; 8 cycles at 98°C for
25 sec, 65°C (−1°C/cycle) for 25 sec, 72°C for 25 sec; 25 cycles
(or 20 cycles for input DNA) at 98°C for 25 sec, 57°C for 25 sec,
72°C for 25 sec (+1 sec/cycle); 72°C for 4 min as a final
additional extension step. Primers used were: YAP1-2561-f
(5′-GGCGAACTGGAAGCGCCTTTCC-3′) and YAP1-2989-r
(5′-GAGACAGAAACTCGCCTCAAACGC-3′), yielding a 429-bp PCR product.
Please note that these two primers can potentially amplify both the
endogenous YAP1 promoter as well as the YAP1 promoter fragment in
the YAP1 (−496/+22) luciferase reporter; however the utilized PCR
cycle number was too low to detect any endogenous YAP1 promoter
signals. In case of input DNA, the alternative primers: pGL2, sense
(5′-CACTGCATTCTAGTTGTGGTTTGTCC-3′) and YAP1-2845-r
(5′-CGCTGCAAGTTGCTACATTCCTGC-3′) were utilized that yielded a
419-bp PCR product. All PCR products were separated on 1.5% agarose
gels and visualized with ethidium bromide staining (33).
Knockdown experiments
Oligonucleotides encoding shRNAs were inserted into
the pSIREN-RetroQ (Clontech, Mountain View, CA, USA) retroviral
vector and targeted the following human sequences: ERG #1
(5′-GCAGCTACATGGAGGAG AA-3′), ERG #3 (5′-GGGAAGGAACTGTGCAAGA-3′),
KDM4A #3 (5′-GTTGAGGATGGTCTTACCT-3′), KDM4A #4
(5′-CACAGTTATTGACCATACT-3′), YAP1 #2 (5′-GCTTATAAGGCATGAGACA-3′)
and YAP1 #3 (5′-AGT AATAGTTGGTTGTGAA-3′). Retrovirus was generated
as previously described (34) and
used to thrice infect VCaP cells followed by selection with 1
µg/ml puromycin (35). Cell
growth was then measured with the PrestoBlue cell viability kit
(Invitrogen) according to the recommendations of the manufacturer.
For this, cells were seeded into 96-well plates, grown for 1–5
days, treated with PrestoBlue reagent for 1 h, excited with 530 nm
light and fluorescence was measured at 590 nm.
Statistical analysis
Averages with standard deviations of at least three
experiments were calculated, and statistical significance was
assessed by performing an unpaired, two-tailed t-test. P≤0.05 was
considered to indicate statistical significance.
Results
Activation of the human YAP1 promoter by
ERG
Previously, it was shown that the ETS transcription
factor GABP can bind to and stimulate the promoter of the mouse
YAP1 gene, whereas ERG seemingly was incapable of doing so
(36). In contrast, a recent report
indicated that ERG promotes transcription of the human YAP1 gene
(7). To clarify this discrepancy,
we employed a luciferase reporter gene controlled by the human YAP1
promoter and transfected it into three different human cell lines:
VCaP, a prostate cancer cell line characterized by the TMPRSS2-ERG
translocation (3), another prostate
cancer cell line (LAP-C4) that is devoid of such a translocation
(37), and benign BPH-1 prostate
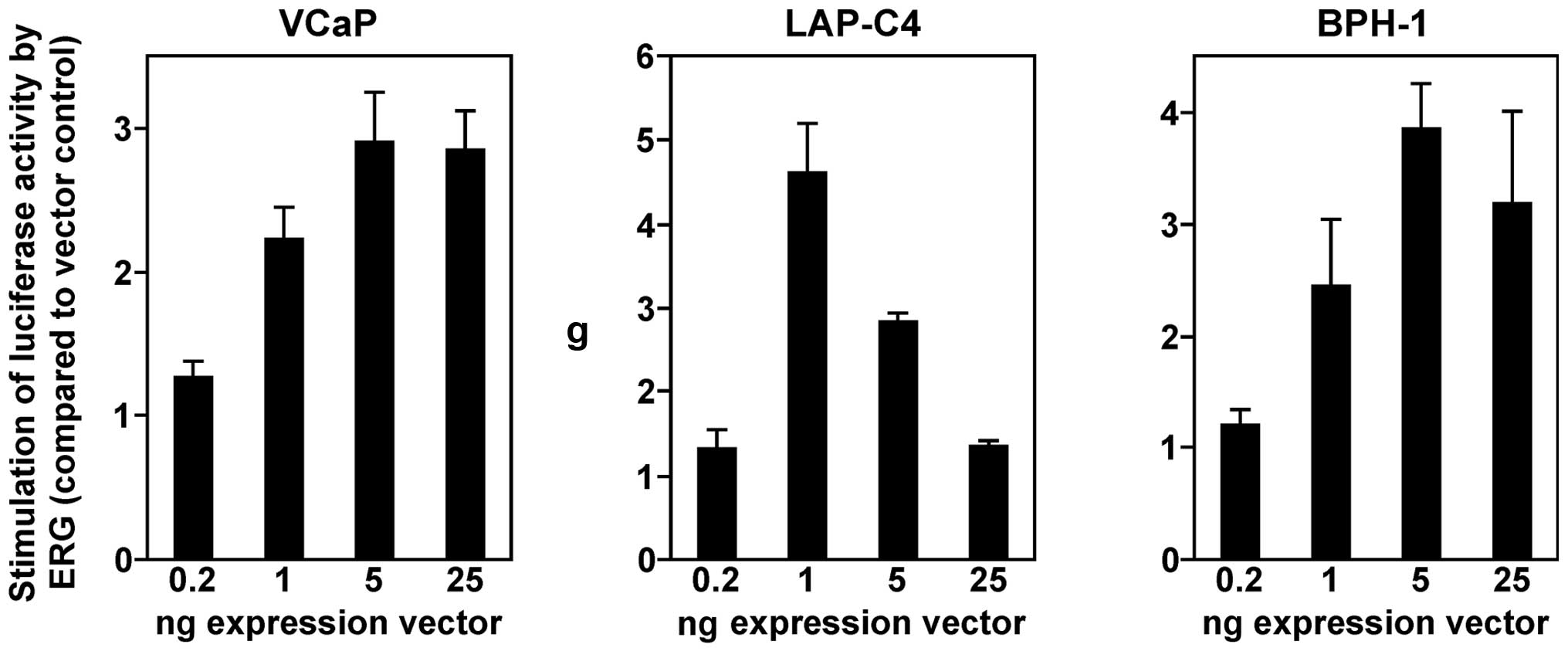
cells. In all three cell lines, we observed a dose-dependent
activation of the YAP1 gene promoter by ERG (Fig. 1). In VCaP and BPH-1 cells, 5 ng ERG
expression vector was sufficient to elicit a maximal response, and
25 ng expression vector led to a slight reduction of promoter
activity. In contrast, LAP-C4 cells displayed the highest YAP1
promoter activity at 1 ng ERG expression vector, and larger amounts
led progressively to greatly reduced luciferase activities. Such a
behavior is likely due to squelching, the titration of limiting
cofactors by an abundance of ERG. Regardless, our data showed that
ERG is capable of stimulating the human YAP1 gene promoter in
various prostate cell lines.
Direct binding of ERG to ETS sites within
the human YAP1 promoter
Although ERG was found to interact with the human
YAP1 promoter in chromatin immunoprecipitation assays (7), it has remained unresolved whether this
is due to direct DNA-binding of ERG. Analysis of the human YAP1
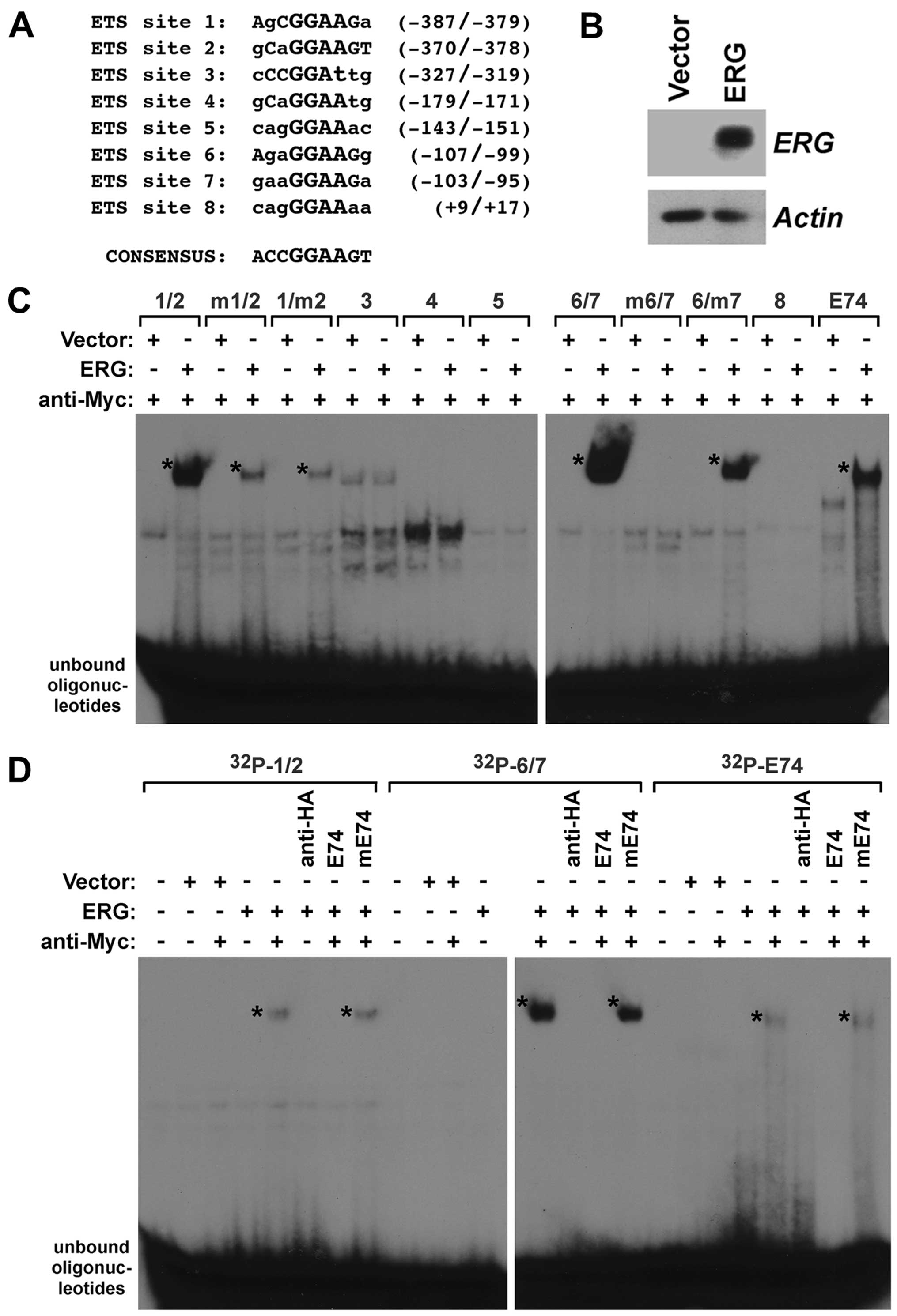
promoter revealed the presence of eight potential ETS binding sites
(Fig. 2A) that are characterized by
a 5′-GGAA/T-3′ core sequence (38)
and may be bound by ERG. Thus, we expressed Myc-tagged ERG in human
293T cells that have no detectable endogenous ERG (Fig. 2B), and prepared cell extracts to
probe for a potential binding of ERG to the ETS sites in the YAP1
gene promoter. To this end, we generated 32P-labeled
oligonucleotides encompassing these ETS sites and incubated them
with control lysate or lysate from ERG-transfected 293T cells. In
preliminary experiments (data not shown, but see also Fig. 2D), we observed that ERG alone did
not bind to any of the ETS sites within the YAP1 promoter. However,
it is known that ERG DNA-binding is auto-inhibited and this
inhibition may be relieved by interaction with other proteins
(39). Since our ERG expression
construct contained a C-terminal Myc-tag, we employed anti-Myc
antibodies to emulate such a protein-protein interaction and
indeed, this resulted into noticeable DNA-binding (Fig. 2C); please note that we cannot
exclude other explanations why the anti-Myc antibodies promoted
DNA-binding of ERG, such as the disruption of binding of an ERG
inhibitor that is present in lysates from 293T cells. In
particular, we observed binding to oligonucleotides encompassing
the juxtaposed ETS sites 1 and 2 as well as ETS sites 6 and 7.
Binding to ETS sites 6 and 7 appeared to be stronger than to ETS
sites 1 and 2 (Fig. 2C; and more
visible in the shorter exposures of autoradiograms shown in
Fig. 2D). No binding to ETS sites
3, 4, 5 and 8 was detected (Fig.
2C), consistent with those four ETS sites being very divergent
to the ERG consensus site of 5′-ACCGGAAGT-3′ (40). In addition, as a positive control,
we observed DNA-binding to the E74 site, a paradigmatic ETS binding
site that was shown to interact with various ETS proteins (19,41,42).
To determine whether ERG binds to both ETS sites 1
and 2, we mutated each one individually in the '1/2'
oligonucleotide. Mutation of either ETS site 1 or 2 resulted in
similarly reduced ERG binding (Fig.
2C), indicating that ERG can interact with ETS sites 1 and 2
with comparable affinity. Likewise, we observed that mutation of
either ETS site 6 or 7 reduced ERG binding to the
32P-labeled '6/7' oligonucleotide (Fig. 2C). However, whereas mutation of ETS
site 7 somewhat reduced DNA-binding, mutation of ETS site 6
completely abolished DNA-binding, suggesting that ERG binding to
ETS site 7 is dependent on the integrity of ETS site 6. Lastly, we
assessed the specificity of the observed DNA-binding. To this end,
we made use of the unlabeled E74 oligonucleotide. An excess of this
oligonucleotide suppressed binding to the 32P-labeled
'1/2' and '6/7' oligonucleotides (Fig.
2D). In contrast, a mutated E74 oligonucleotide that no longer
binds to ETS proteins was unable to compete for binding. In
conclusion, our data show that ERG can directly bind to several ETS
sites within the human YAP1 gene promoter.
Importance of ETS sites 6 and 7
Next, we started to evaluate which of the ERG
binding sites in the YAP1 promoter are crucial for its activity.
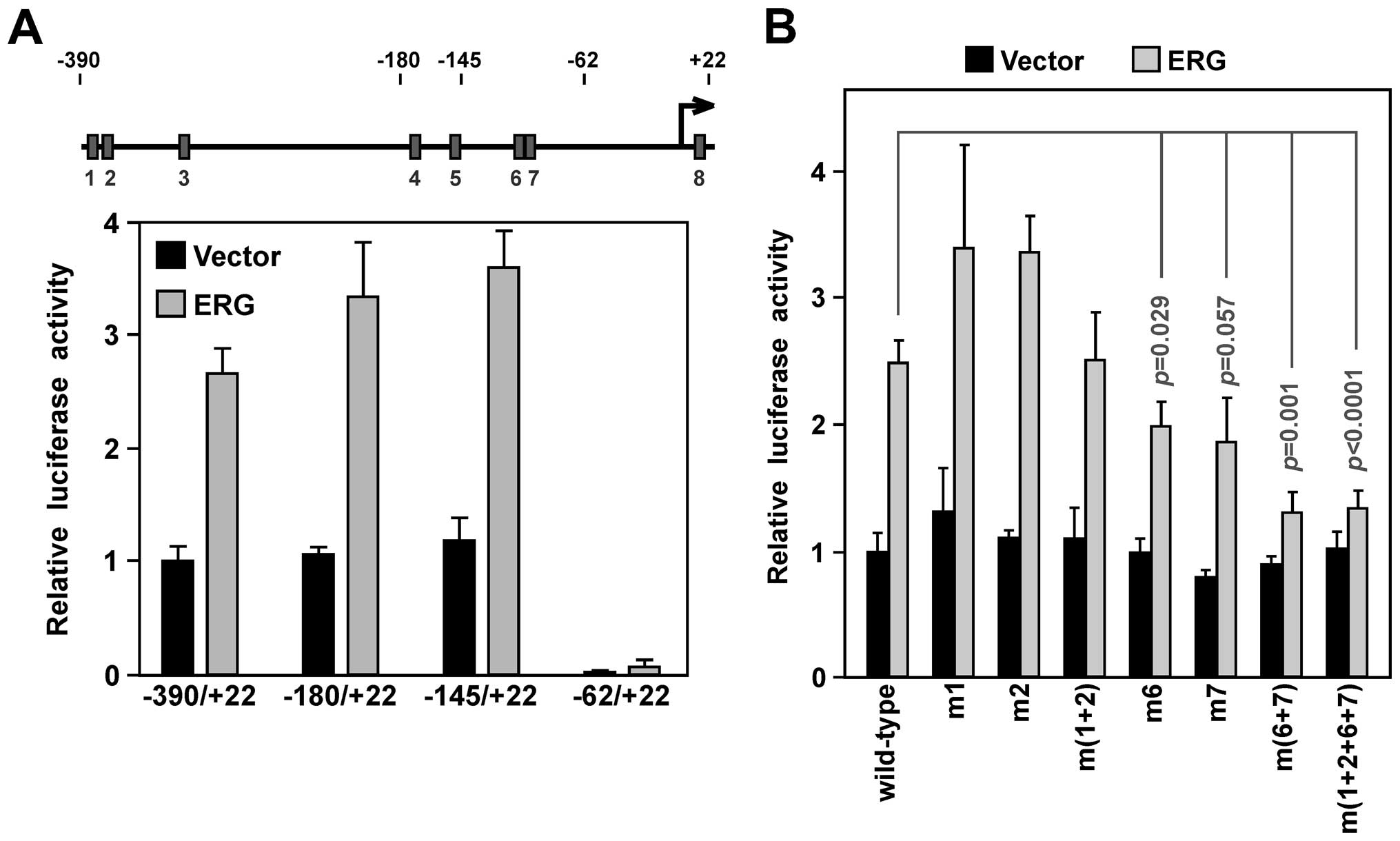
First, we employed promoter truncations. The −180/+22 truncation,
in which ETS sites 1–3 become deleted, and the −145/+22 truncation,
in which additionally ETS sites 4 and 5 become removed, were at
least as active as the longest YAP1 promoter (−390/+22) fragment in
the absence or presence of ectopic ERG in the VCaP prostate cancer
cells (Fig. 3A), suggesting that
ETS sites 1–5 are not important for ERG-dependent YAP1 promoter
upregulation. When ETS sites 1–7 were removed in the −62/+22
promoter construct, promoter activity was vastly reduced,
suggesting that ETS sites 6 and 7 are crucial for YAP1 promoter
activity. This would be consistent with the fact that ETS sites 6
and 7 were most avidly bound by ERG as shown above.
To corroborate this, we mutated ETS sites in the
−390/+22 YAP1 promoter construct. Neither mutation of ETS site 1 or
2 resulted in decreased promoter activity, and even joint mutation
of these two ETS sites had no significant effect (Fig. 3B). In contrast, mutation of ETS site
6 or 7 reduced YAP1 promoter activity. Joint mutation of ETS site 6
and 7 resulted in even more reduction of transcription: ERG was
only able to increase luciferase activity by ~1/3, compared to
2.5-fold for the wild-type promoter. Lastly, joint mutation of ETS
sites 1, 2, 6 and 7 was no different from mutation of ETS sites 6
and 7 (Fig. 3B). We conclude that
ETS sites 6 and 7 are crucial for ERG-dependent stimulation of YAP1
promoter transcription.
Cooperation between ERG and KDM4A
Since our laboratory is interested in the
(de)methylation of histone H3 on lysine residues, we analyzed how
ERG would affect trimethylation on four prominent H3 lysine
residues. For this, we utilized human embryonic kidney 293T cells,
since: i) they can be efficiently transfected in contrast to VCaP
cells; and ii) they also display activation of the YAP1 promoter by
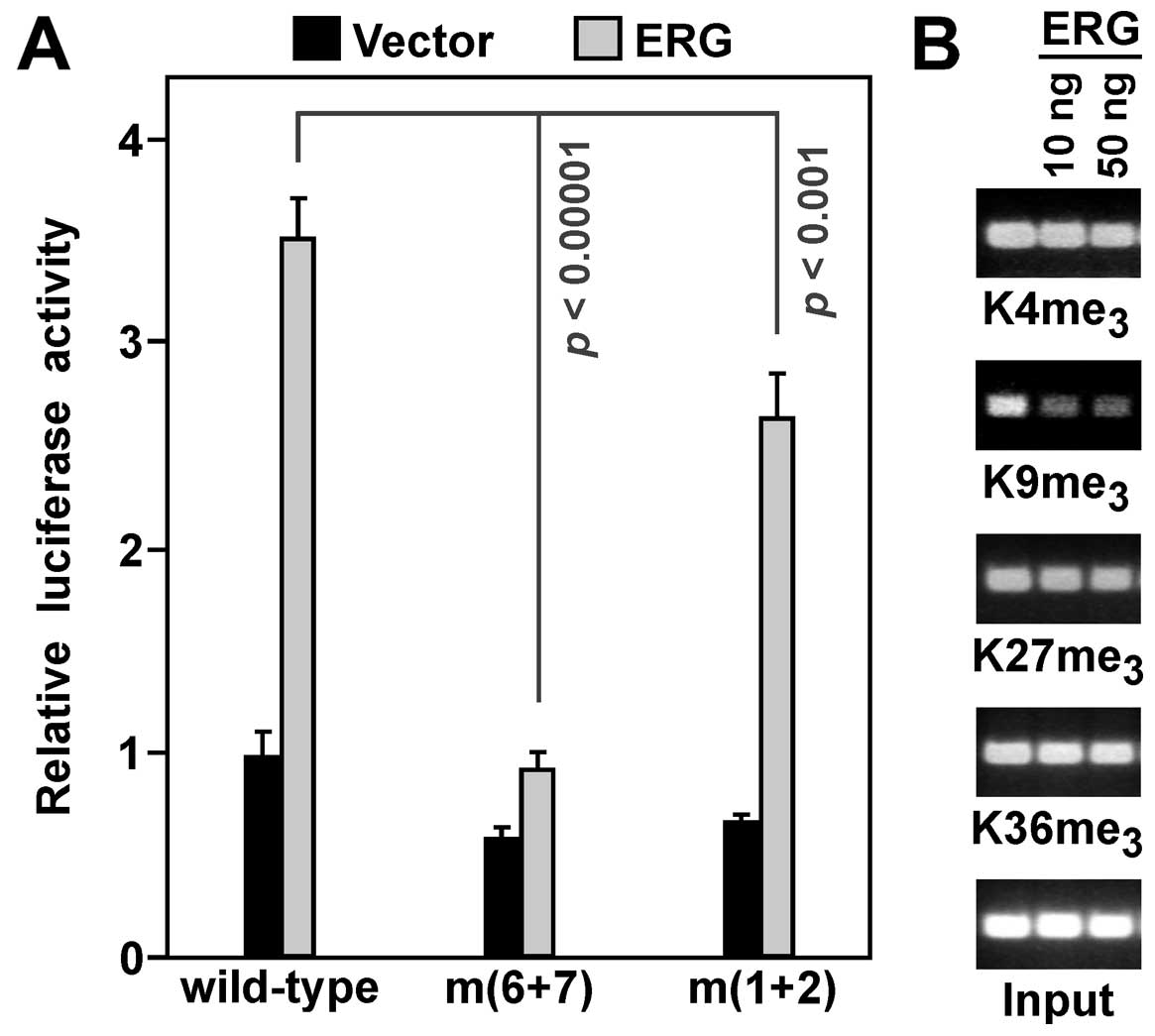
ERG (Fig. 4A). Moreover, we
observed that ETS sites 6 and 7 were crucial for ERG-dependent
transcription in 293T cells, while mutation of ETS sites 1 and 2
had, in contrast to VCaP cells (compare to Fig. 3B), also a small effect (Fig. 4A). This suggests that in some cell
lines, ETS sites 1 and 2 may contribute to ERG-dependent YAP1
upregulation. Utilizing chromatin immunoprecipitation assays, we
observed that expression of ERG led to no significant changes of
trimethylation on histone H3 lysines K4, K27 and K36, but
K9me3 levels were reduced on the YAP1 luciferase
reporter (Fig. 4B).
H3K9me3 is normally a marker for transcriptional
repression (43), thus, it would be
consistent that its removal contributes to ERG-mediated
transcriptional activation.
This result led us to speculate that ERG may recruit
a histone demethylase targeting trimethylated H3K9. Only one
subclass of histone demethylases, the KDM4 proteins, is known for
demethylating H3K9me3 (44). Hence, we focused on the protagonist
of this family, KDM4A (45), and
tested whether it would interact with ERG. To this end, we
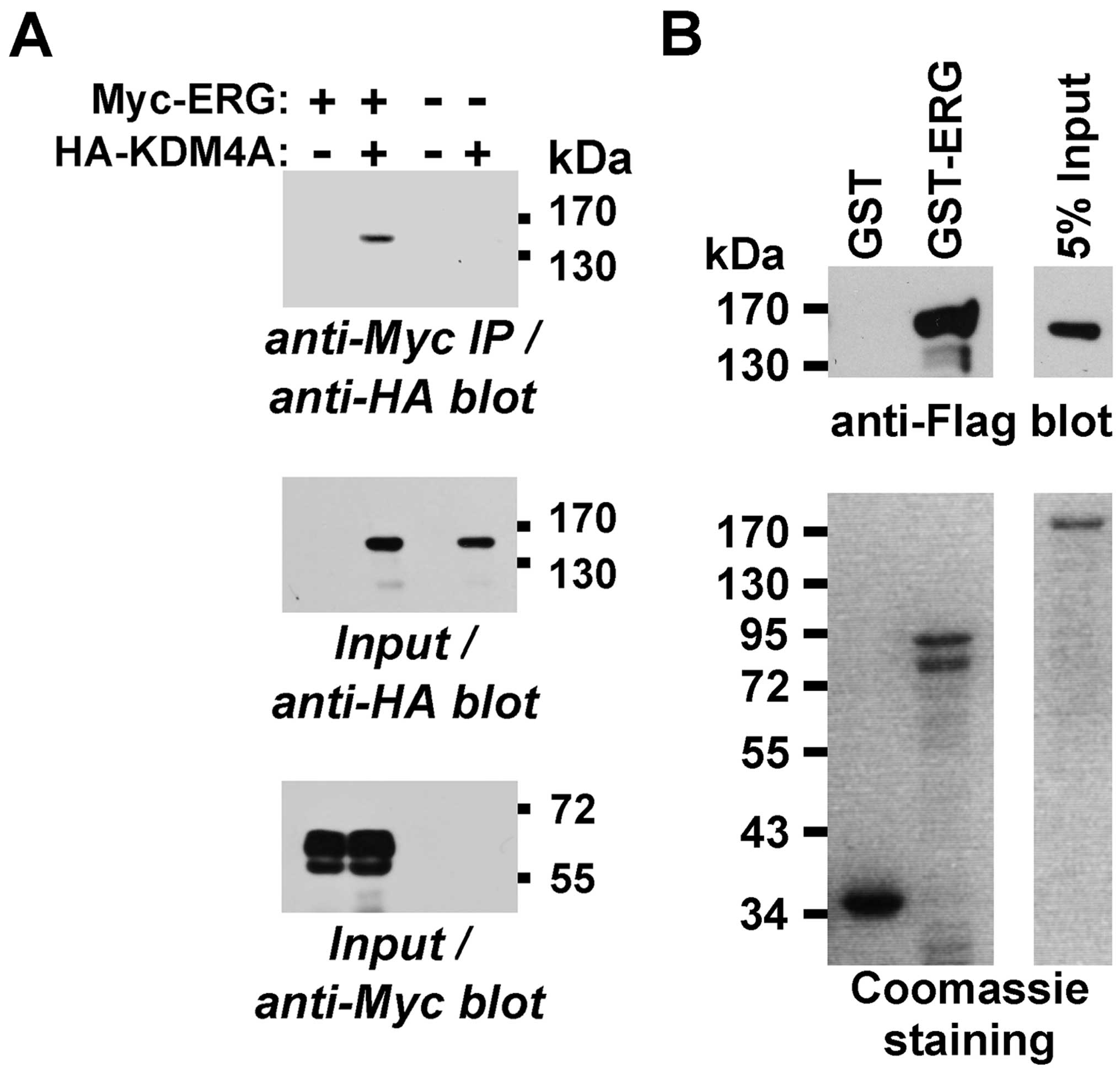
coexpressed Myc-tagged ERG and HA-tagged KDM4A in 293T cells and
observed that KDM4A coprecipitated with ERG (Fig. 5A). Moreover, we produced a GST-ERG
fusion protein in bacteria and challenged it with KDM4A purified
from baculovirus. Whereas GST-ERG bound KDM4A, GST did not
(Fig. 5B), indicating that ERG and
KDM4A can directly bind to each other.
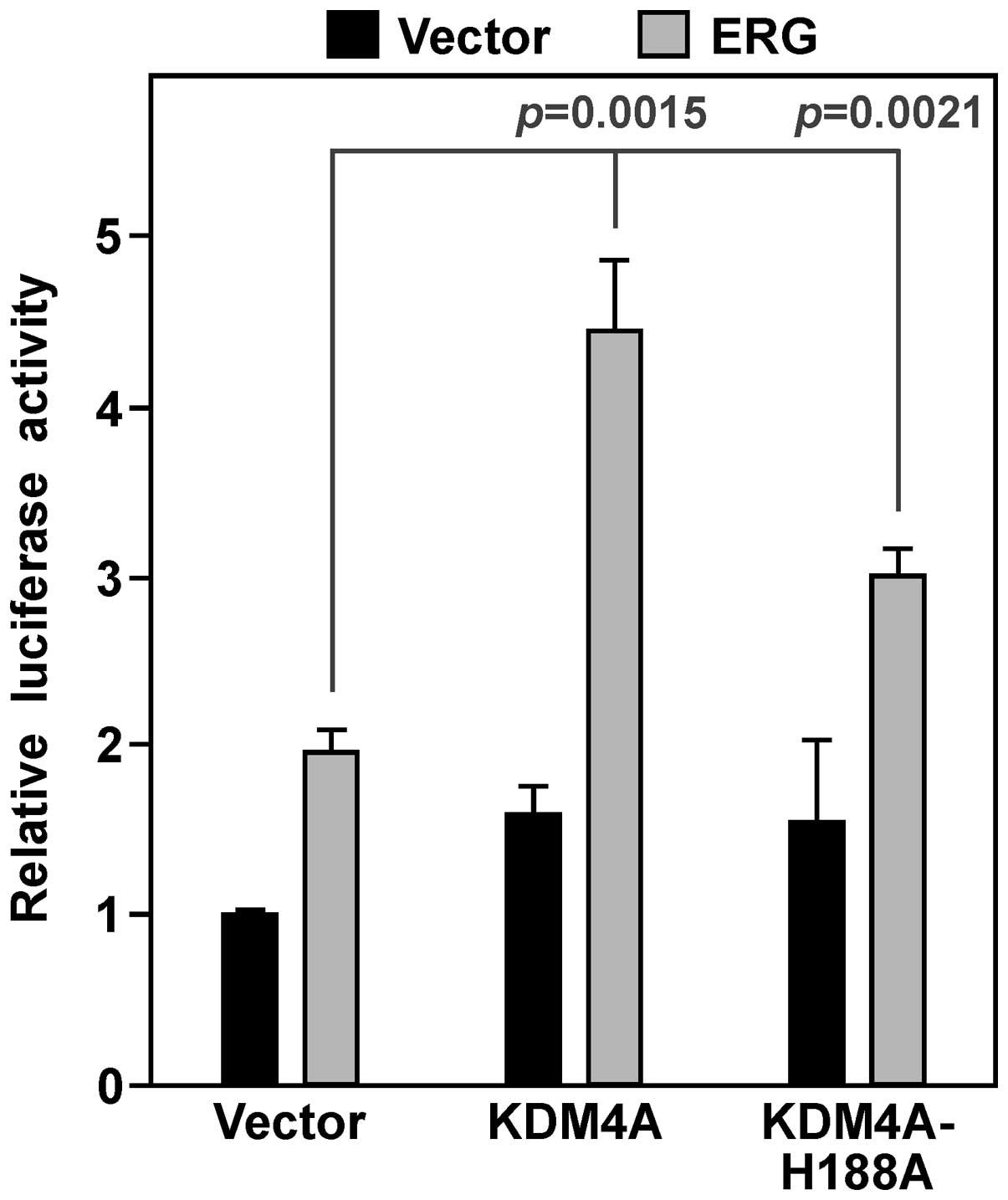
We then determined whether KDM4A would cooperate
with ERG in activating YAP1 gene transcription. On its own, KDM4A
had a modest impact on YAP1 luciferase activity in VCaP cells, but
combined with ERG it caused a synergistic activation of the YAP1
promoter (Fig. 6). We also employed
a catalytically inactive KDM4A protein, the H188A point mutant
(46,47). This mutant was less active compared
to wild-type KDM4A, yet still significantly raised ERG-mediated
YAP1 luciferase activity (Fig. 6).
These data suggest that KDM4A is a coactivator that stimulates ERG
in a manner dependent and independent of its catalytic
activity.
Relationship between ERG/KDM4A and YAP1
in VCaP cells
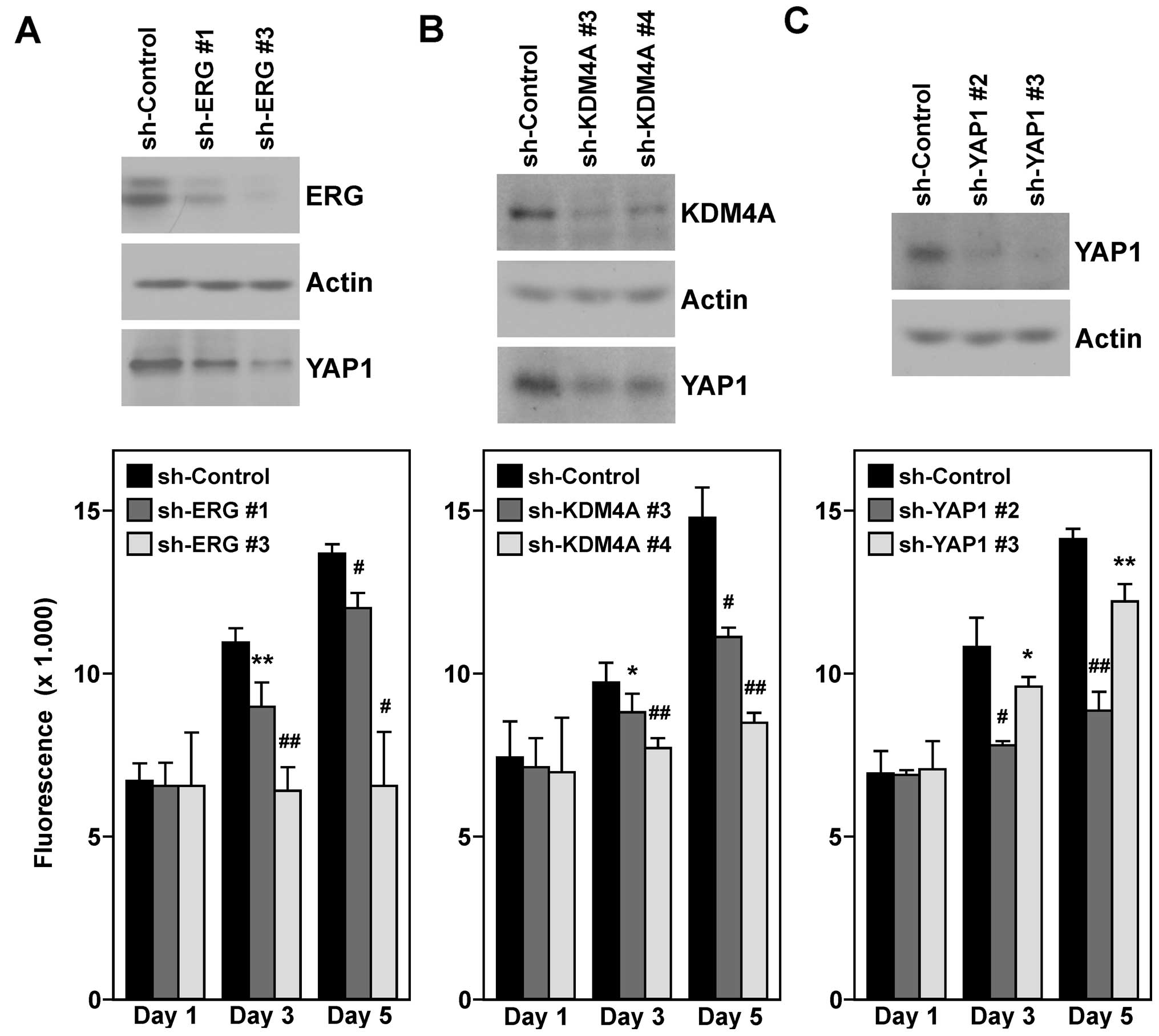
Next, we wished to confirm that YAP1 is a target
gene of ERG and KDM4A in VCaP prostate cancer cells. To this end,
we downregulated either ERG or KDM4A with two different shRNAs and
observed that YAP1 protein levels were reduced (Fig. 7A and B, top panels). This suggested
that both ERG and KDM4A are required for maximal YAP1 gene
transcription in VCaP cells.
Furthermore, we assayed VCaP cell growth upon ERG
and KDM4A downregulation. As previously reported (48,49),
ERG knockdown led to a robust decrease in VCaP cell growth
(Fig. 7A, bottom panel). Notably,
the same was observed upon KDM4A knockdown (Fig. 7B, bottom panel), highlighting a role
of KDM4A in cell proliferation. We then reasoned that whether YAP1
is a seminal downstream target of both ERG and KDM4A, its
downregulation should phenocopy the observed reduction in cell
growth upon ERG/KDM4A knockdown. In addition, indeed, we observed
that YAP1 was required for maximal VCaP cell proliferation
(Fig. 7C).
Discussion
In the present study, we uncovered a new mechanism
by which ERG may exert its oncogenic function. This mechanism
involves a physical interaction of ERG with the histone demethylase
KDM4A that could lead to pleiotropic changes in the transcriptome,
including an upregulation of YAP1 gene transcription. Since ERG
overexpression is found in approximately half of all prostate tumor
patients (4), our findings
particularly pertain to prostatic malignancies.
YAP1 is a transcriptional cofactor that can be
recruited to chromatin by several DNA-binding proteins. Frequently,
YAP1 expression is enhanced in various human tumors and may
correlate with poor prognosis, and its oncogenic potential was
confirmed both in vitro as well as in transgenic mouse
models (12,13). However, recent studies suggest that
YAP1 may also exert growth suppressive actions in the colon and
hematological cancers (50,51), suggesting that YAP1
context-dependently acts as an oncogene or tumor suppressor.
However, the fact that YAP1 is overexpressed in human prostate
tumors (52) indicates that it
functions as an oncogene in this organ, which is consistent with
prostate-specific overexpression of YAP1 leading to the development
of prostatic neoplasias in mice (7). All this stresses that YAP1 may serve
as a target for therapy particularly in ERG-overexpressing prostate
tumors. Notably, small molecules as well as a peptide that suppress
YAP1 function have been identified (53,54),
which could be harnessed for future avenues of therapeutic
interference. A caveat is that our report does not establish
whether YAP1 is the only crucial downstream effector of ERG. Given
that ERG downregulation seems to be more detrimental to VCaP cell
proliferation than YAP1 downregulation (see Fig. 7), it is likely that YAP1
upregulation is not the sole reason why ERG overexpression induces
prostate tumors. Yet, even partially blunting ERG's oncogenic
potential through YAP1 inhibition would still have therapeutic
value.
KDM4A is the protagonist of the KDM4 family of
histone demethylases that are encoded by six different genes in the
human genome (45,55). It is particularly competent in
demethylating trimethylated lysine 9 on histone H3 and lysine 26 on
histone H1.4 that are regarded as repressive chromatin marks
(46,47,56,57).
Accordingly, KDM4A may function as a transcriptional coactivator at
least in part by removing these repressive marks. However, we
observed that catalytically inactive KDM4A was still capable,
albeit at a much reduced rate compared to wild-type KDM4A, to
cooperate with ERG in stimulating the YAP1 promoter. This suggests
that KDM4A coactivates ERG both in a manner dependent on and
independent of its catalytic activity. Likewise, Drosophila
KDM4A has been shown to often affect gene transcription independent
of its catalytic activity (58) and
also mammalian KDM4A can impact DNA repair without involving its
catalytic activity (59),
corroborating that KDM4A may act both as an enzyme and in
non-enzymatic ways.
However, in case of stimulating ERG, our data
suggest that KDM4A is mostly acting through its enzymatic activity.
If so, inhibition of its catalytic center may prove beneficial in
the treatment of prostate cancer patients that are afflicted by an
ERG chromosomal translocation. Several small molecules have been
uncovered that can inhibit KDM4A enzymatic activity (60–66).
However, the specificity of these inhibitors, their selectivity for
suppressing tumor vs. normal cells, their toxicity,
pharmacokinetics and pharmacodynamics need to be further explored
before any of these inhibitors can enter clinical trials.
Similar to ERG, KDM4A seems to be overexpressed in
prostate tumors (67), which would
be alike to breast and lung tumors that display overexpression of
KDM4A (68–71). This may suggest that KDM4A is
oncogenic in its own right in the prostate, breast or lung.
Furthermore, it is unlikely that KDM4A exclusively promotes
prostate tumorigenesis as a coactivator of ERG. For instance, KDM4A
can also stimulate the androgen receptor or repress the p53 tumor
suppressor thereby leading to abnormal cell growth (72,73).
Moreover, KDM4A is capable of inducing copy number gains in cells,
which may represent another mechanism by which it contributes to
the development of cancer (74).
In conclusion, the present study has provided more
mechanistic insight into how ERG overexpression due to chromosomal
translocations can induce prostate cancer formation. Despite its
obvious validity as a drug target in prostate cancer, no effective
ERG inhibitors have surfaced in the clinic, which may be due to the
difficulty of targeting a DNA-binding transcription factor. The
present study suggests two alternative targets to blunt the ERG
oncogenic activity, KDM4A and YAP1, both of which can in principal
be inhibited by small molecules and may therefore merit more
research.
Acknowledgments
The present study was in part funded by a grant from
the National Institutes of Health/National Cancer Institute (R01
CA154745) to R.J. The content is solely the responsibility of the
authors and does not necessarily represent the official views of
the granting agencies.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ryan CJ and Tindall DJ: Androgen receptor
rediscovered: The new biology and targeting the androgen receptor
therapeutically. J Clin Oncol. 29:3651–3658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clark JP and Cooper CS: ETS gene fusions
in prostate cancer. Nat Rev Urol. 6:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Klezovitch O, Risk M, Coleman I, Lucas JM,
Null M, True LD, Nelson PS and Vasioukhin V: A causal role for ERG
in neoplastic transformation of prostate epithelium. Proc Natl Acad
Sci USA. 105:2105–2110. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomlins SA, Laxman B, Varambally S, Cao X,
Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al:
Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia.
10:177–188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nguyen LT, Tretiakova MS, Silvis MR, Lucas
J, Klezovitch O, Coleman I, Bolouri H, Kutyavin VI, Morrissey C,
True LD, et al: ERG activates the YAP1 transcriptional program and
induces the development of age-related prostate tumors. Cancer
Cell. 27:797–808. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carver BS, Tran J, Gopalan A, Chen Z,
Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino
PT, et al: Aberrant ERG expression cooperates with loss of PTEN to
promote cancer progression in the prostate. Nat Genet. 41:619–624.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
King JC, Xu J, Wongvipat J, Hieronymus H,
Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et
al: Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation
in prostate oncogenesis. Nat Genet. 41:524–526. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baena E, Shao Z, Linn DE, Glass K, Hamblen
MJ, Fujiwara Y, Kim J, Nguyen M, Zhang X, Godinho FJ, et al: ETV1
directs androgen metabolism and confers aggressive prostate cancer
in targeted mice and patients. Genes Dev. 27:683–698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Chi P, Rockowitz S, Iaquinta PJ,
Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, et al:
ETS factors reprogram the androgen receptor cistrome and prime
prostate tumorigenesis in response to PTEN loss. Nat Med.
19:1023–1029. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harvey KF, Zhang X and Thomas DM: The
Hippo pathway and human cancer. Nat Rev Cancer. 13:246–257. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Varelas X: The Hippo pathway effectors TAZ
and YAP in development, homeostasis and disease. Development.
141:1614–1626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dowdy SC, Mariani A and Janknecht R:
HER2/Neu- and TAK1-mediated up-regulation of the transforming
growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol
Chem. 278:44377–44384. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mooney SM, Grande JP, Salisbury JL and
Janknecht R: Sumoylation of p68 and p72 RNA helicases affects
protein stability and transactivation potential. Biochemistry.
49:1–10. 2010. View Article : Google Scholar
|
|
16
|
Oh S, Shin S, Lightfoot SA and Janknecht
R: 14-3-3 proteins modulate the ETS transcription factor ETV1 in
prostate cancer. Cancer Res. 73:5110–5119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu J and Janknecht R: Regulation of the
ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1
and protein kinase A. J Biol Chem. 277:42669–42679. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Papoutsopoulou S and Janknecht R:
Phosphorylation of ETS transcription factor ER81 in a complex with
its coactivators CREB-binding protein and p300. Mol Cell Biol.
20:7300–7310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Haro L and Janknecht R: Cloning of the
murine ER71 gene (Etsrp71) and initial characterization of its
promoter. Genomics. 85:493–502. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
DiTacchio L, Bowles J, Shin S, Lim DS,
Koopman P and Janknecht R: Transcription factors ER71/ETV2 and SOX9
participate in a positive feedback loop in fetal and adult mouse
testis. J Biol Chem. 287:23657–23666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Haro L and Janknecht R: Functional
analysis of the transcription factor ER71 and its activation of the
matrix metal-loproteinase-1 promoter. Nucleic Acids Res.
30:2972–2979. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mooney SM, Goel A, D'Assoro AB, Salisbury
JL and Janknecht R: Pleiotropic effects of p300-mediated
acetylation on p68 and p72 RNA helicase. J Biol Chem.
285:30443–30452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Knebel J, De Haro L and Janknecht R:
Repression of transcription by TSGA/Jmjd1a, a novel interaction
partner of the ETS protein ER71. J Cell Biochem. 99:319–329. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shin S and Janknecht R: Concerted
activation of the Mdm2 promoter by p72 RNA helicase and the
coactivators p300 and P/CAF. J Cell Biochem. 101:1252–1265. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goel A and Janknecht R:
Acetylation-mediated transcriptional activation of the ETS protein
ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 23:6243–6254.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goel A and Janknecht R: Concerted
activation of ETS protein ER81 by p160 coactivators, the
acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J
Biol Chem. 279:14909–14916. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berry WL, Kim TD and Janknecht R:
Stimulation of β-catenin and colon cancer cell growth by the KDM4B
histone demethylase. Int J Oncol. 44:1341–1348. 2014.PubMed/NCBI
|
|
28
|
Janknecht R: Regulation of the ER81
transcription factor and its coactivators by mitogen- and
stress-activated protein kinase 1 (MSK1). Oncogene. 22:746–755.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shin S, Bosc DG, Ingle JN, Spelsberg TC
and Janknecht R: Rcl is a novel ETV1/ER81 target gene upregulated
in breast tumors. J Cell Biochem. 105:866–874. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Goueli BS and Janknecht R: Regulation of
telomerase reverse transcriptase gene activity by upstream
stimulatory factor. Oncogene. 22:8042–8047. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shin S, Rossow KL, Grande JP and Janknecht
R: Involvement of RNA helicases p68 and p72 in colon cancer. Cancer
Res. 67:7572–7578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shin S, Oh S, An S and Janknecht R: ETS
variant 1 regulates matrix metalloproteinase-7 transcription in
LNCaP prostate cancer cells. Oncol Rep. 29:306–314. 2013.
|
|
33
|
Goueli BS and Janknecht R: Upregulation of
the catalytic telomerase subunit by the transcription factor ER81
and oncogenic HER2/Neu, Ras, or Raf. Mol Cell Biol. 24:25–35. 2004.
View Article : Google Scholar :
|
|
34
|
Shin S, Kim TD, Jin F, van Deursen JM,
Dehm SM, Tindall DJ, Grande JP, Munz JM, Vasmatzis G and Janknecht
R: Induction of prostatic intraepithelial neoplasia and modulation
of androgen receptor by ETS variant 1/ETS-related protein 81.
Cancer Res. 69:8102–8110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim TD, Oh S, Shin S and Janknecht R:
Regulation of tumor suppressor p53 and HCT116 cell physiology by
histone demethylase JMJD2D/KDM4D. PLoS One. 7:e346182012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu H, Xiao Y, Zhang S, Ji S, Wei L, Fan F,
Geng J, Tian J, Sun X, Qin F, et al: The Ets transcription factor
GABP is a component of the hippo pathway essential for growth and
antioxidant defense. Cell Reports. 3:1663–1677. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Haffner MC, Aryee MJ, Toubaji A, Esopi DM,
Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al:
Androgen-induced TOP2B-mediated double-strand breaks and prostate
cancer gene rearrangements. Nat Genet. 42:668–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hollenhorst PC, McIntosh LP and Graves BJ:
Genomic and biochemical insights into the specificity of ETS
transcription factors. Annu Rev Biochem. 80:437–471. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Regan MC, Horanyi PS, Pryor EE Jr, Sarver
JL, Cafiso DS and Bushweller JH: Structural and dynamic studies of
the transcription factor ERG reveal DNA binding is allosterically
autoinhibited. Proc Natl Acad Sci USA. 110:13374–13379. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei GH, Badis G, Berger MF, Kivioja T,
Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al:
Genome-wide analysis of ETS-family DNA-binding in vitro and in
vivo. EMBO J. 29:2147–2160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bosc DG, Goueli BS and Janknecht R:
HER2/Neu-mediated activation of the ETS transcription factor ER81
and its target gene MMP-1. Oncogene. 20:6215–6224. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Janknecht R, Monté D, Baert JL and de
Launoit Y: The ETS-related transcription factor ERM is a nuclear
target of signaling cascades involving MAPK and PKA. Oncogene.
13:1745–1754. 1996.PubMed/NCBI
|
|
43
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kooistra SM and Helin K: Molecular
mechanisms and potential functions of histone demethylases. Nat Rev
Mol Cell Biol. 13:297–311. 2012.PubMed/NCBI
|
|
45
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demeth-ylases: Epigenetic regulators in cancer cells.
Cancer Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Whetstine JR, Nottke A, Lan F, Huarte M,
Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al:
Reversal of histone lysine trimethylation by the JMJD2 family of
histone demethylases. Cell. 125:467–481. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shin S and Janknecht R: Diversity within
the JMJD2 histone demethylase family. Biochem Biophys Res Commun.
353:973–977. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun C, Dobi A, Mohamed A, Li H,
Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan
G, Whitman E, et al: TMPRSS2-ERG fusion, a common genomic
alteration in prostate cancer activates C-MYC and abrogates
prostate epithelial differentiation. Oncogene. 27:5348–5353. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang J, Cai Y, Yu W, Ren C, Spencer DM and
Ittmann M: Pleiotropic biological activities of alternatively
spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res.
68:8516–8524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Barry ER, Morikawa T, Butler BL, Shrestha
K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, et
al: Restriction of intestinal stem cell expansion and the
regenerative response by YAP. Nature. 493:106–110. 2013. View Article : Google Scholar :
|
|
51
|
Cottini F, Hideshima T, Xu C, Sattler M,
Dori M, Agnelli L, ten Hacken E, Bertilaccio MT, Antonini E, Neri
A, et al: Rescue of Hippo coactivator YAP1 triggers DNA
damage-induced apoptosis in hematological cancers. Nat Med.
20:599–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim
J, Xie J, Ikenoue T, Yu J, Li L, et al: Inactivation of YAP
oncoprotein by the Hippo pathway is involved in cell contact
inhibition and tissue growth control. Genes Dev. 21:2747–2761.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu-Chittenden Y, Huang B, Shim JS, Chen
Q, Lee SJ, Anders RA, Liu JO and Pan D: Genetic and pharmacological
disruption of the TEAD-YAP complex suppresses the oncogenic
activity of YAP. Genes Dev. 26:1300–1305. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiao S, Wang H, Shi Z, Dong A, Zhang W,
Song X, He F, Wang Y, Zhang Z, Wang W, et al: A peptide mimicking
VGLL4 function acts as a YAP antagonist therapy against gastric
cancer. Cancer Cell. 25:166–180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Labbé RM, Holowatyj A and Yang ZQ: Histone
lysine demeth-ylase (KDM) subfamily 4: Structures, functions and
therapeutic potential. Am J Transl Res. 6:1–15. 2013.
|
|
56
|
Klose RJ, Yamane K, Bae Y, Zhang D,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: The
transcriptional repressor JHDM3A demethylates trimethyl histone H3
lysine 9 and lysine 36. Nature. 442:312–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Trojer P, Zhang J, Yonezawa M, Schmidt A,
Zheng H, Jenuwein T and Reinberg D: Dynamic histone H1 isotype 4
methylation and demethylation by histone lysine methyltransferase
G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. J
Biol Chem. 284:8395–8405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Crona F, Dahlberg O, Lundberg LE, Larsson
J and Mannervik M: Gene regulation by the lysine demethylase KDM4A
in Drosophila. Dev Biol. 373:453–463. 2013. View Article : Google Scholar
|
|
59
|
Mallette FA, Mattiroli F, Cui G, Young LC,
Hendzel MJ, Mer G, Sixma TK and Richard S: RNF8- and
RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1
recruitment to DNA damage sites. EMBO J. 31:1865–1878. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hamada S, Kim TD, Suzuki T, Itoh Y,
Tsumoto H, Nakagawa H, Janknecht R and Miyata N: Synthesis and
activity of N-oxalylglycine and its derivatives as Jumonji
C-domain-containing histone lysine demethylase inhibitors. Bioorg
Med Chem Lett. 19:2852–2855. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hamada S, Suzuki T, Mino K, Koseki K,
Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, et
al: Design, synthesis, enzyme-inhibitory activity, and effect on
human cancer cells of a novel series of jumonji domain-containing
protein 2 histone demethylase inhibitors. J Med Chem. 53:5629–5638.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Rose NR, Woon EC, Kingham GL, King ON,
Mecinović J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U,
McDonough MA, et al: Selective inhibitors of the JMJD2 histone
demethylases: Combined nondenaturing mass spectrometric screening
and crystallographic approaches. J Med Chem. 53:1810–1818. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
King ON, Li XS, Sakurai M, Kawamura A,
Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Beswick P, et al:
Quantitative high-throughput screening identifies
8-hydroxyquinolines as cell-active histone demethylase inhibitors.
PLoS One. 5:e155352010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Luo X, Liu Y, Kubicek S, Myllyharju J,
Tumber A, Ng S, Che KH, Podoll J, Heightman TD, Oppermann U, et al:
A selective inhibitor and probe of the cellular functions of
Jumonji C domain-containing histone demethylases. J Am Chem Soc.
133:9451–9456. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang L, Chang J, Varghese D, Dellinger M,
Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, et al: A
small molecule modulates Jumonji histone demethylase activity and
selectively inhibits cancer growth. Nat Commun.
4:20352013.PubMed/NCBI
|
|
66
|
Kim TD, Fuchs JR, Schwartz E, Abdelhamid
D, Etter J, Berry WL, Li C, Ihnat MA, Li PK and Janknecht R:
Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon
cancer cells and identification of curcuminoids as JMJD2
inhibitors. Am J Transl Res. 6:236–247. 2014.PubMed/NCBI
|
|
67
|
Cloos PA, Christensen J, Agger K, Maiolica
A, Rappsilber J, Antal T, Hansen KH and Helin K: The putative
oncogene GASC1 demethylates tri- and dimethylated lysine 9 on
histone H3. Nature. 442:307–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Patani N, Jiang WG, Newbold RF and Mokbel
K: Histone-modifier gene expression profiles are associated with
pathological and clinical outcomes in human breast cancer.
Anticancer Res. 31:4115–4125. 2011.PubMed/NCBI
|
|
69
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012.PubMed/NCBI
|
|
70
|
Slee RB, Steiner CM, Herbert BS, Vance GH,
Hickey RJ, Schwarz T, Christan S, Radovich M, Schneider BP,
Schindelhauer D, et al: Cancer-associated alteration of
pericentromeric heterochromatin may contribute to chromosome
instability. Oncogene. 31:3244–3253. 2012. View Article : Google Scholar
|
|
71
|
Mallette FA and Richard S: JMJD2A promotes
cellular transformation by blocking cellular senescence through
transcriptional repression of the tumor suppressor CHD5. Cell
Reports. 2:1233–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shin S and Janknecht R: Activation of
androgen receptor by histone demethylases JMJD2A and JMJD2D.
Biochem Biophys Res Commun. 359:742–746. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kim TD, Shin S, Berry WL, Oh S and
Janknecht R: The JMJD2A demethylase regulates apoptosis and
proliferation in colon cancer cells. J Cell Biochem. 113:1368–1376.
2012. View Article : Google Scholar
|
|
74
|
Black JC, Manning AL, Van Rechem C, Kim J,
Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al:
KDM4A lysine demethylase induces site-specific copy gain and
rereplication of regions amplified in tumors. Cell. 154:541–555.
2013. View Article : Google Scholar : PubMed/NCBI
|