Introduction
Breast cancer is the most frequently diagnosed
female cancer worldwide and remains the top cause of cancer death
in females, although the diagnostic techniques and treatment
modalities advanced greatly (1,2).
Triple-negative breast cancer (TNBC) is a heterogeneous group of
breast cancers, characterized by the loss of estrogen receptor
(ER), progesterone receptor (PR) and human epidermal growth factor
receptor 2 (HER2) gene expression (3). As the representative of the most
aggressive subtype, it accounts for 10–20% of all invasive breast
cancers (4). Since there is no
targeted therapy available at present, one-third of patients with
this disease develop recurrence within 3 years, even when receiving
adjuvant therapy (5,6). Hence, understanding the mechanisms
underlying distant metastasis and early relapse in TNBC will be key
to discover new therapeutic targets and improve clinical treatment
of these patients.
The Ras pathway is one of the most commonly
deregulated pathways in human cancer (7), as one-third of human cancers have been
observed to harbor high frequency of mutational activation of Ras
proteins (8). Ras proteins,
together with guanine nucleotide exchange factors (GEFs) and
GTPase-activating proteins (GAPs), constitute cellular binary
switches cycling between 'on' and 'off' conformations determined by
the loading of GTP or GDP, respectively. GEFs stimulate GDP for GTP
exchange, and therefore activates the Ras pathway, whereas GAPs
terminates the activation status by promoting GTP to GDP hydrolysis
(9). RASAL2, function as a GAP, and
has been shown to be implicated in the establishment and metastasis
of several types of tumors, such as lung, ovarian, thyroid and
breast cancer (10–13). A recent report showed that RASAL2
was oncogenic in TNBC and drives mesenchymal invasion and
metastasis. Moreover, RASAL2 expression was tightly associated with
the poor prognosis of patients suffering of TNBC (14).
MicroRNAs (miRNA) are a class of small, endogenous,
non-coding RNAs that regulate gene expression
post-transcriptionally (15).
Accumulating studies have demonstrated that miRNA-regulated
transcriptional dynamics was a critical step in tumor initiation,
promotion and progression (16,17).
In human cancers, many miRNAs were identified to function as
potential tumor suppressors and their downregulation leads to
overexpression of oncogenic genes (18–20).
miR-136 was recently identified to be tightly associated with
tumorigenesis and metastasis. It was first characterized to be
upregulated in human and murine lung cancers by miRNA microarray
expression profiling (21). miR-136
was also reported to be predominately overexpressed in the Jurkat
cell line (22) and was found to
target tumor suppressor PTEN in breast cancer cells (23), indicating a possible significance in
cancer development and progression. Further studies showed that it
may act as a cancer promoting gene in human non-small cell lung
cancer (NSCLC) (24) and it was
found to be downregulated in human glioma and promotes apoptosis of
glioma cells by targeting AEG-1 and Bcl-2 (25), indicating it may also act as an
tumor suppressor. The precise role of miR-136 in breast cancer,
especially in TNBC, remains largely unknown. In the present study
we report that miR-136 may act as an tumor suppressor in TNBC.
Decreased expression of miR-136 was observed in TNBC correlating
with the pathological grades, and overexpressed miR-136 restrained
the migration and invasion of MDA-MB-231 cells (a highly invasive
TNBC cell line). We further demonstrated that the anti-invasive
effect of miR-136 was mediated through targeting RASAL2, a
cancer-promoting gene in TNBC. Our results demonstrated that
miRNA-136 was a key anti-invasive miRNA and further confirmed the
oncogenic role of RASAL2 in TNBC.
Materials and methods
Cell culture, plasmids and
transfection
MCF10A, MCF7, ZR751 and MDA-MB-231 cells were
cultivated in Leibovitz's L-15 Medium containing 10% fetal bovine
serum plus 2 mM L-glutamine. The cells were split before confluence
and incubated at 37°C in a humidified incubator with 5%
CO2. miR-136 mimics and antisense oligonucleotides (ASO)
were purchased from Shanghai GenePharma Co., Ltd. (Shanghai,
China). RASAL2 full length CDS were cloned to pCMV2-myc.
Transfection of miRNA mimics and ASO was carried out using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions.
Human tissue samples
Forty TNBC samples or adjacent normal mucosa tissues
were obtained from patients with TNBC. Detailed pathological and
clinical data were collected for all samples including WHO tumor
grade, invasion and metastasis. The diagnoses of these samples were
verified by pathologists. The collection of human tissue samples
was approved and supervised by the Ethics Committee of Harbin
Medical University.
Mouse xenograft model
Female immune-deficient nude mice (strain BALB/c
nu/nu; 4–5 weeks old) used in the present study were bred at the
Department of Pathology, Harbin Medical University. For orthotopic
injection, 1×107 MDA-MB-231 cells were injected into the
mammary gland fat pad of each CB-17 SCID mouse in a volume of 0.1
ml. miR-136 mimics and its negative control miRNA were injected
into the tumor every 2 days from day 7 of the graft. Twenty-one
days later, the tumors were collected and sectioned for
immunohistochemical analysis.
Immunohistochemistry analysis
Tumor tissues of xenograft mice, human colon cancer
samples or adjacent normal mucosa tissues were fixed in 10% neutral
buffered formalin for 24 h and then embedded in paraffin. Sections
(4 µm) were cut and stained for histological examination.
E-cadherin (BD Biosciences; cat. BD 610182), RASAL2 (Santa Cruz
Biotechnology; cat. sc-67935) antibody was diluted in PBS with 1%
(wt/vol) BSA. Images were obtained with a Nikon DP70 camera mounted
on a Nikon Bx60 microscope with Cell-F imaging software (Soft
Imaging System).
RNA preparation and quantitative PCR
RNA was extracted from cells or tissue samples using
the mirVana miRNA isolation kit (Ambion, Foster City, CA, USA)
according to the manufacturer's instructions. Small RNA fraction
(<200 nt) was separated and purified according to the procedure.
cDNA was obtained using M-MLV (Promega, Madison, WI, USA) and 1
µg RNA. The relative level of miR-136 was detected by
stem-loop RT-PCR with the following conditions: denaturing the DNA
at 94°C for 4 min, followed by 40 cycles of amplification: 94°C for
60 sec, 58°C for 60 sec, 72°C for 60 sec for data collection. U6
snRNA was used as an endogenous control. Quantitative PCR was
performed on an ABI 7500 thermocycler (Applied Biosystems) using
SYBR® Premix Ex Taq™ (perfect real-time) kits (Takara
Bio, Inc., Shiga, Japan) according to the manufacturer's
instructions.
Wound-healing assay
Equal number of MDA-MB-231 cells transfected with
miR-136 mimics and control miRNA were seeded on BioCoat™ collagen I
coated 6-well tissue culture dishes (BD Biosciences; cat. 354400)
respectively, and allowed to grow to confluent for 48 h. Then
scratches were made using p200 pipette tips, however, floating
cells were carefully washed away with fresh growth medium. The
wound-healing of the scratch regions were monitored and imaged at
designated time-points.
Cell migration and invasion assays
A total of 5×104 MDA-MB-231 cells (in 0.2
ml RPMI-1640 with 5% FBS) were seeded into the upper part of a
Transwell chamber (Corning Life Sciences, Corning, NY, USA). For
migration invasion assay, the chamber was pre-coated with 1 mg/ml
Matrigel (Growth Factor Reduced BD Matrigel™ Matrix) for 2 h. In
the lower part of the chamber, 0.6 ml RPMI-1640 with 20% FBS was
added. After incubating for 30 h, chambers were disassembled and
the membranes were stained with 2% crystal violet for 10 min and
placed on a glass slide. Then, cells on the bottom of the membranes
were counted in 5 random visual fields under a light microscope.
All assays were performed in triplicate and independently performed
three times.
Fluorescent reporter assays
The human RASAL2 3′UTR harboring three miR-136
potential target-binding sequences was synthesized by Shanghai
GenePharma. Luciferase constructs were made by ligating the
synthesized 3′UTR as well as the seed-sequence mutated version
after the lucORF in the pMIR-Report luciferase vector (Ambion). For
the fluorescent reporter assay, cells were seeded in a 48-well
plate the day before transfection. The cells were co-transfected
with miRNA mimics or ASO as well as the controls and RASAL2-3UTR or
RASAL2-3UTRmut. The cells were lyzed 48 h later and the intensity
of luciferase was detected.
Western blotting
Western blotting was performed to determine protein
expression and the GAPDH was used as the internal control. Total
protein from cells were lysed by RIPA buffer and then measured by
Micro BCA protein assay kit (Pierce Biotechnology). Protein (50
µg/lane) was resolved on sodium dodecyl
sulfate-polyacrylamide gel followed by transferred to a
nitrocellulose membrane (Life Technologies, Carlsbad, CA, USA). The
nitrocellulose membrane was incubated with polyclonal rabbit
anti-human RASAL2 (1:3,000) and alkaline phosphatase-conjugated
goat anti-rabbit antibody, respectively. After incubation the
nitrocellulose membrane was evaluated by ECL.
Immunofluorescent cell staining
assay
MDA-MB-231 cells were seeded at 4×105
cells/well in 6-well culture plates. Twenty-four hours later, the
attached cells were transfected with 30 µM microRNA mimics
and allowed to grow further for 72 h. Then the post-treatment cells
were trypsinized and re-seeded at a density of 1.5×105
cells/well on 8-mm coverslips in 12-well plates. After additional
48 h, coverslips with cells were fixed in methanol, and probed with
primary E-cadherin (BD Biosciences; cat. BD 610182), or vimentin
(Santa Cruz Biotechnology; cat. SC-6260) antibodies in 1:100 to
1:1,000, dilution then subsequently with florescent labeled
secondary antibodies. Cell nuclei were stained with DAPI. After
cell staining the coverslips were mounted with FluorSave reagent
(Calbiochem, Darmstadt, Germany). Cells were imaged using Zeiss
Meta upright microscope under 63X oil objective.
Statistical analysis
Student's t-test was performed to analyze the
significance of differences between the sample means obtained from
three independent experiments. One-way ANOVA was used for multiple
group comparisons. Differences were considered statistically
significant at P<0.05.
Results
miR-136 expression is downregulated in
TNBC and negatively associated with the WHO grades
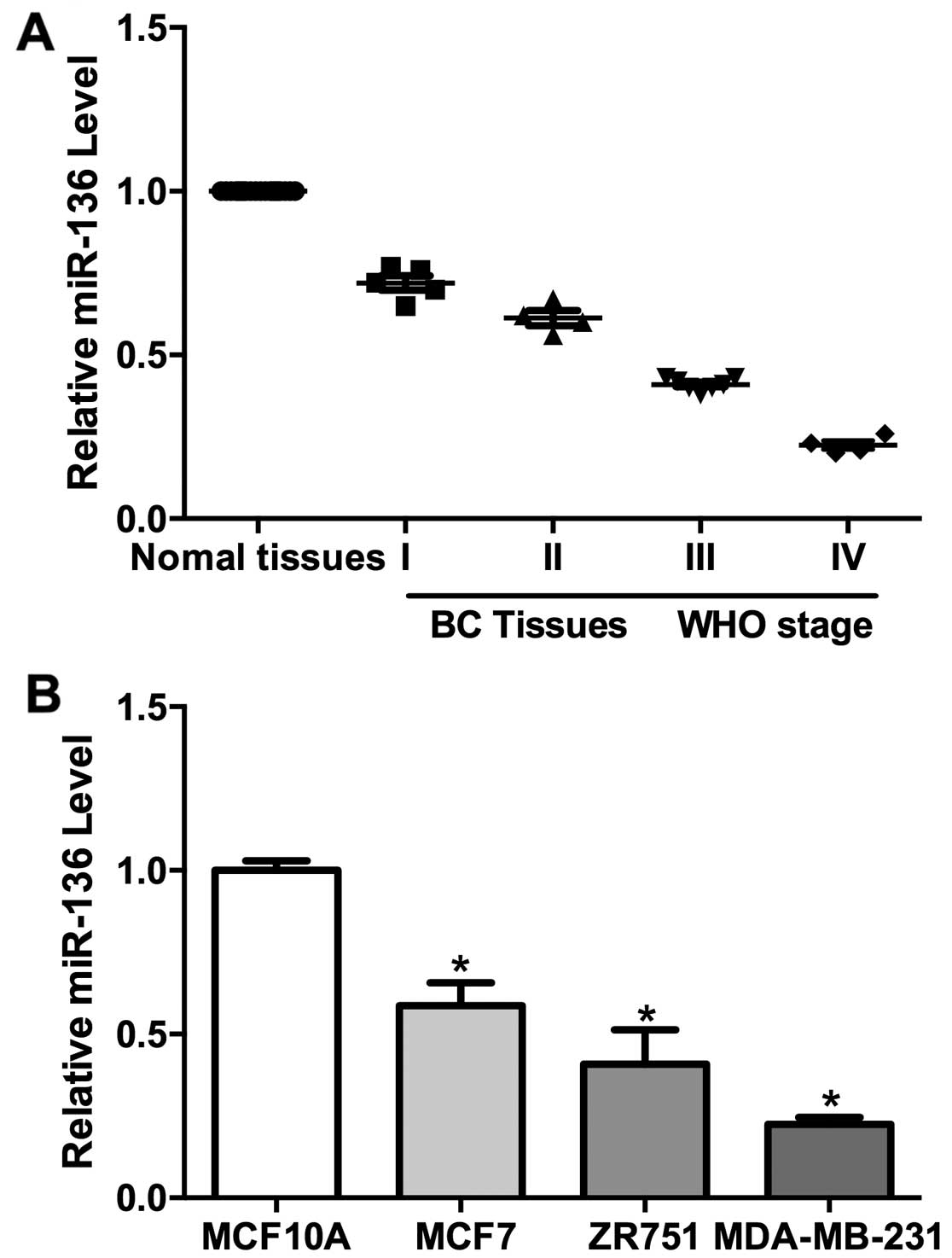
qPCR analysis revealed that miR-136 transcripts were
reduced in tumor tissues compared to normal tissues (Fig. 1A). Further analysis showed that the
miR-136 expression were negatively correlated with the WHO grades
(Fig. 1A). We also determined
miR-136 levels in the immortalized mammary epithelial cells
(MCF10A) and three breast cancer cell lines (MCF7, ZR751 and
MDA-MB-231). The level of miR-136 is much lower in the TNBC cell
lines (MDA-MB-231, ZR751 and MCF7) than control cells (Fig. 1B). However, the miR-136 expression
is negatively correlated with the invasive ability of these breast
cancer cell lines (Fig. 1B). These
results indicated a role for miR-136 in the development and/or
metastasis of TNBC.
miR-136 suppresses cell migration and
invasion
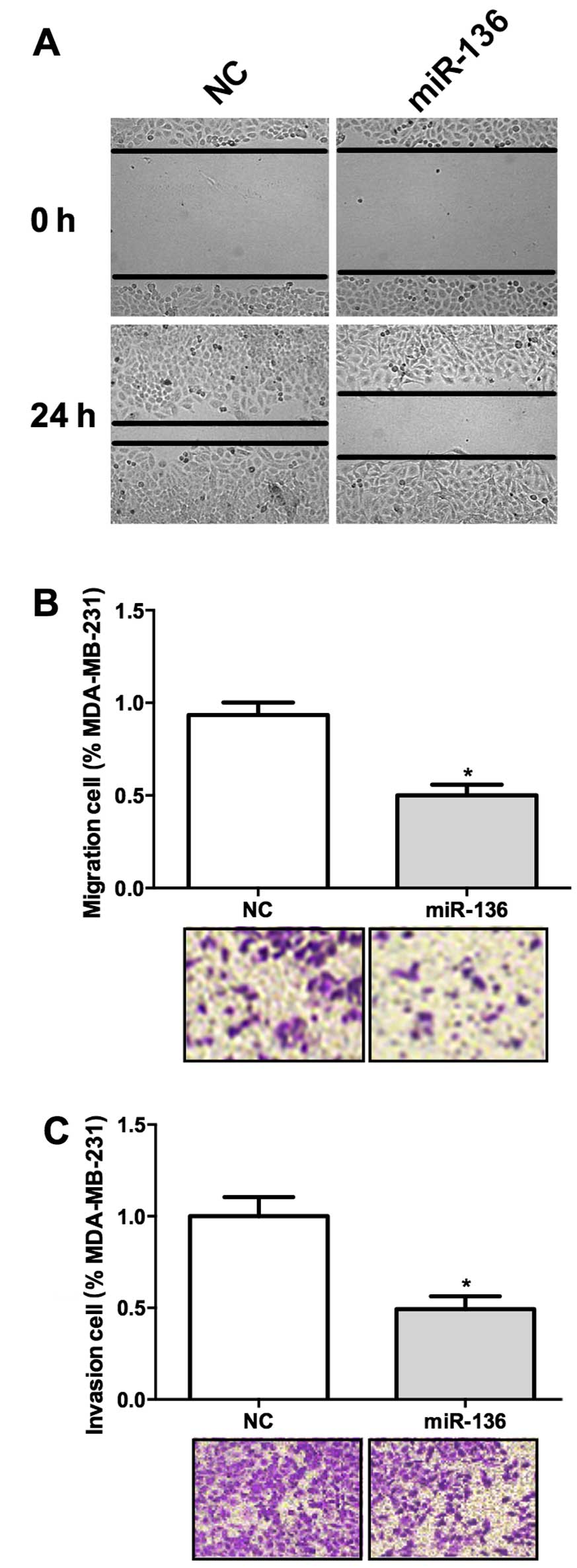
We have shown that miR-136 expression was negatively
correlated with WHO grade in TNBC, we wonder whether it was
involved in the invasion of this cancer. Wound healing assay result
suggested that overexpression of miR-136 significantly hampered the
migration of these cells (Fig. 2A).
Transwell assay also revealed that miR-136 overexpression decreased
the migration ability of MDA-MB-231 cells (Fig. 2B). We used a Matrigel coated
Transwell assay system in vivo to further test the function
of miR-136 in cancer invasion. MDA-MB-231 cells transfected with
miR-136 showed a decreased ability of invasion through the Matrigel
(Fig. 2C). These results suggested
that miR-136 may act as a suppressor of tumor invasion.
miR-136 suppresses EMT in breast
cancer
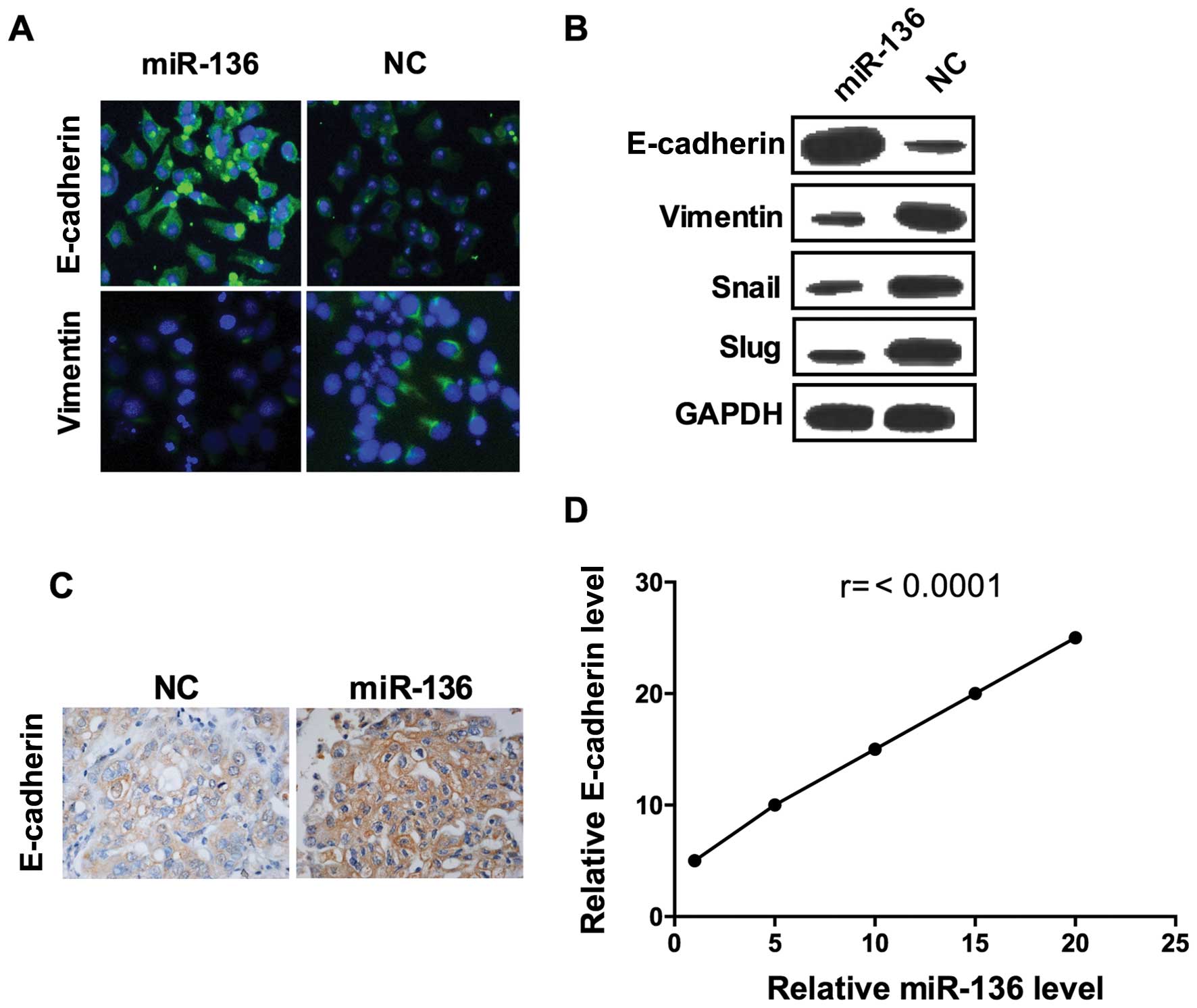
Epithelial-to-mesenchymal transition (EMT) is a
critical step for metastatic dissemination (26) thus, we hypothesized that miR-136 may
affect EMT. As shown, the epithelial marker E-cadherin fluorescence
was visibly enhanced in cells transfected with miR-136 compared to
those transfected with scramble miRNA (Fig. 3A). Consistently, the result of
confocal imaging analysis indicated that mesenchymal marker
vimentin expression was weakened by overexpression of miR-136
(Fig. 3A). Further assessment of a
panel of EMT-related genes by western blot analysis showed that,
miR-136 treatment induced the expression of E-cadherin and
decreased the expression of vimentin, Snail and SLUG, confirmed the
results of immunofluorescence assay (Fig. 3B). To further confirmed these
results, we established a mouse xenograft model by injecting
MDA-MB-231 cells into the mammary gland fat pads. miR-136 mimics
and its negative control miRNA were injected into the tumor every 2
days after day 7 of the graft. In miR-136 overexpressed group,
E-cadherin expression were significant higher than the control
group (Fig. 3C), and the levels of
E-cadherin were correlated with the injection dose of miR-136
mimics (Fig. 3D). These results
elucidated that miR-136 is a suppressor of EMT in the TNBC cell
line.
miR-136 directly regulates RASAL2
expression in breast cancer
Previously we showed that miR-136 may act as tumor
suppressor of TNBC metastasis, then we attempted to uncover the
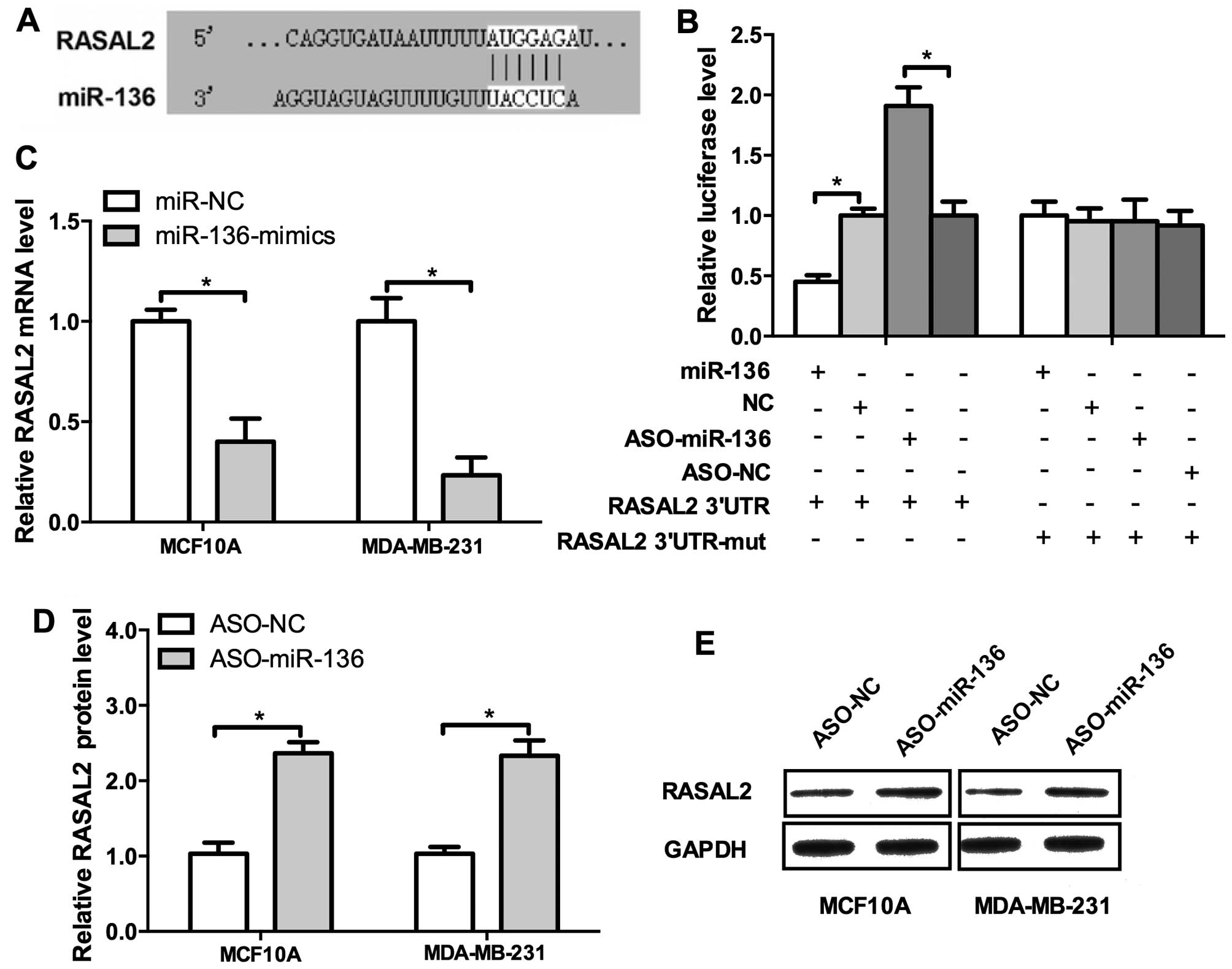
underlying mechanisms. RASAL2 is a newly identified
cancer-promoting gene in TNBC and it drives mesenchymal invasion
and metastasis (14). We
investigated the regulatory sequence of miR-136 in the 3′UTR of
RASAL2 mRNA. Indeed, RASAL2 harbors a binding site of miR-136
(Fig. 4A). The result of luciferase
activity detection revealed miR-136 overexpression repressed,
whereas the ASO elevated the luciferase activities (Fig 4B, left panel). Then we mutated the
binding site of miR-136 in the 3′UTR of RASAL2 mRNA and the
regulatory effect of the mimics or ASO could not be observed
(Fig. 4B, right panel). To further
confirm these results, the miR-136 mimics, ASO and their negative
control were co-transfected into MCF10A and MDA-MB-231 cells.
miR-136 mimics significantly decreased the mRNA level of RASAL2 and
conversely, the ASO increased it (Fig.
4C and D). Western blot analysis confirmed these results
(Fig. 4E). Conclusively, those
results demonstrated that RASAL2 is a direct target of miR-136.
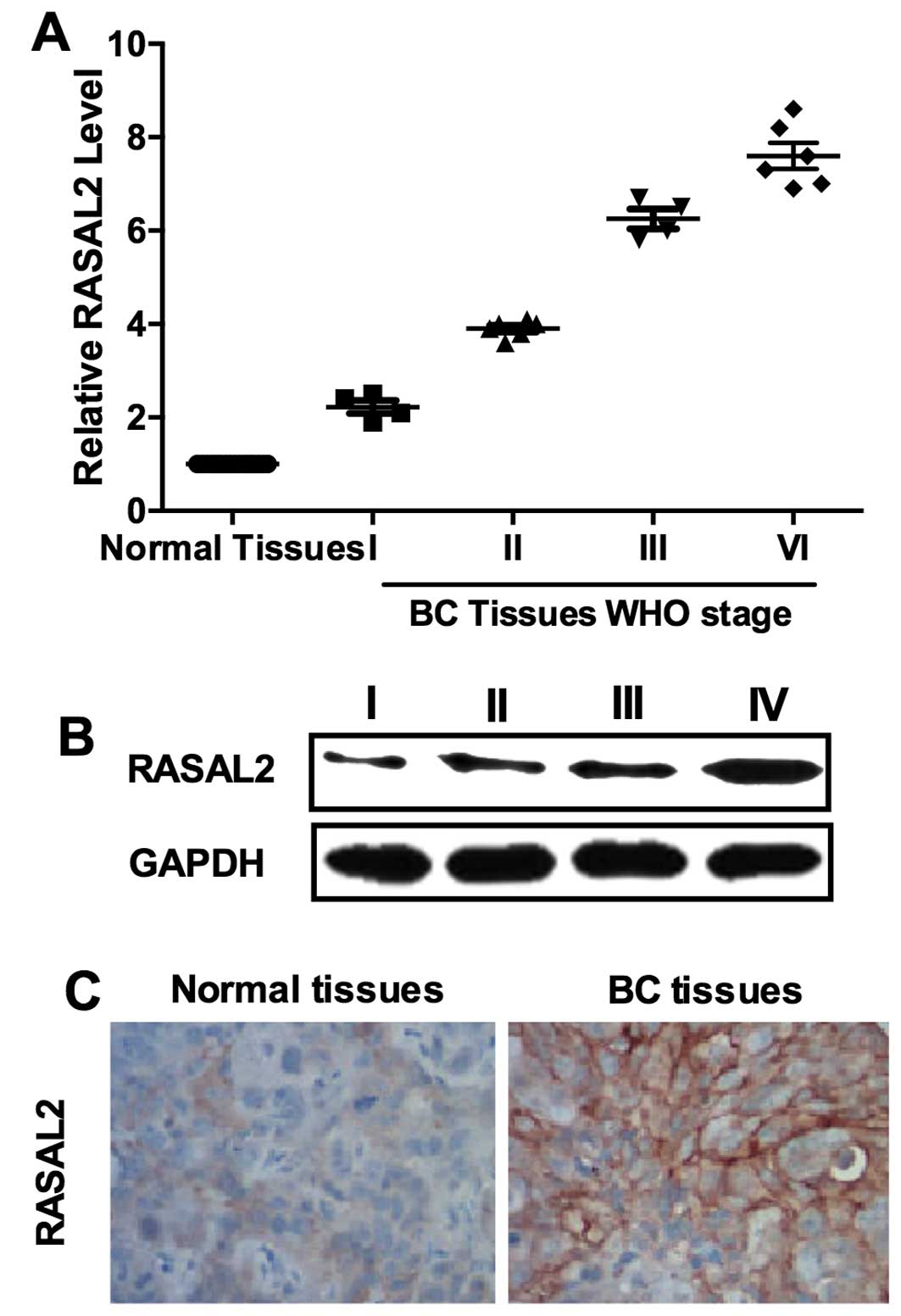
RASAL2 is upregulated in TNBC
Our results showed that RASAL2 was negatively
regulated by miR-136 which functioned as a tumor suppressor in
TNBC. Thus, we inferred that RASAL2 may be a cancer-promoting gene.
We determined the expression of RASAL2 in 20 samples of TNBC
tissues and their normal adjacent tissues (NAT) by qPCR. The
results showed that RASAL2 was unregulated in TNBC tissues and its
levels were correlated with WHO grade 90 (Fig. 5A). Western blot analysis and IHC
results were used as further confirmation (Fig. 5B and C). These results supported
that RASAL2 may function as an oncogene in TNBC.
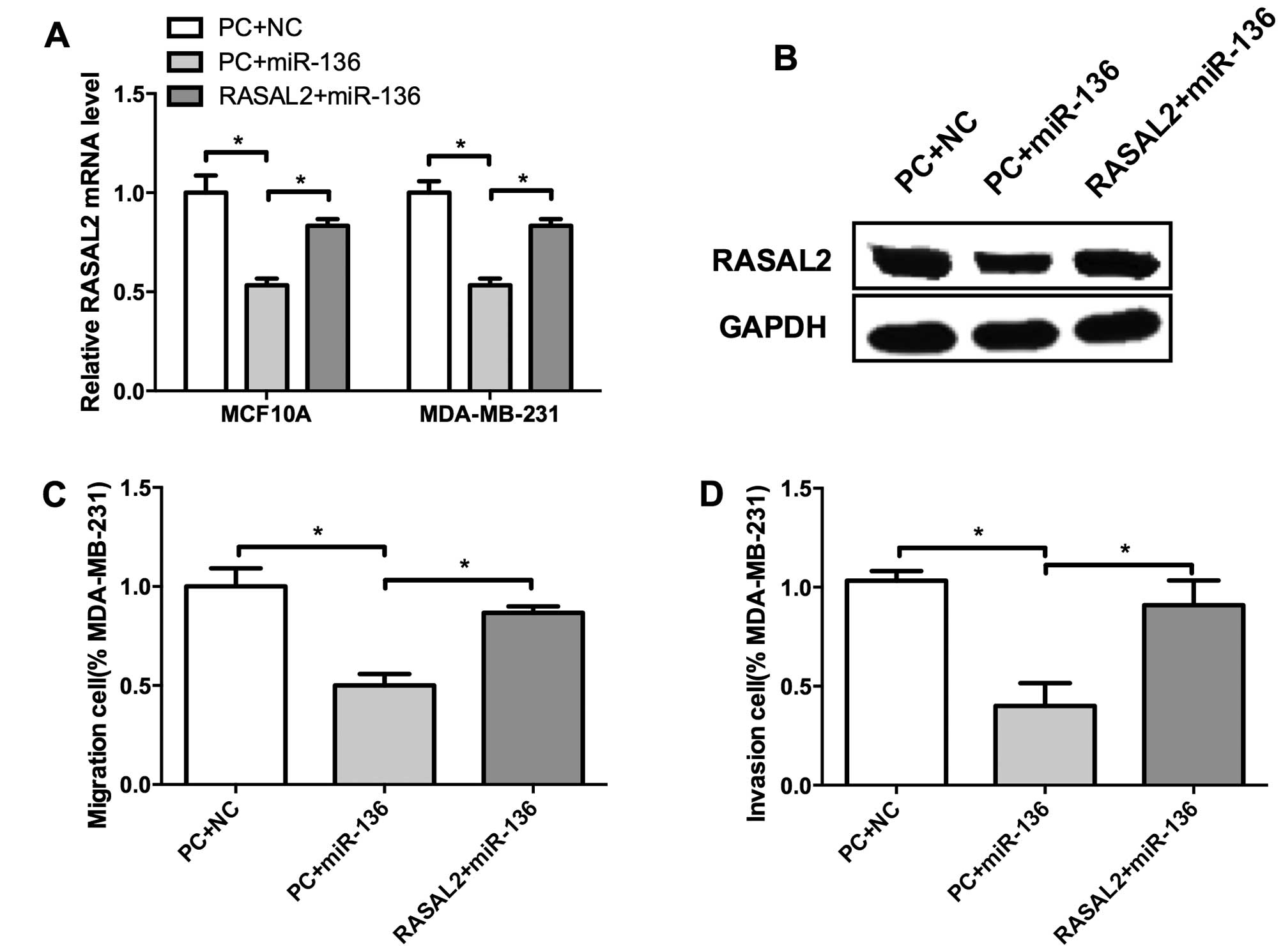
miR-136 suppresses breast cell migration
and invasion through RASAL2
We identified miR-136 as a tumor suppressor of TNBC,
and RASAL2 may served as its target. Whether miR-136 regulate cell
invasion through RASAL2 remains unclear. To validate this
hypothesis, we co-transfected miR-136 mimics and RASAL2 into MCF10A
and MDA-MB-231 cells. Compared with control, miR-136 suppressed
RASAL2 expression both at mRNA and protein levels (Fig. 6A and B). Moreover, RASAL2
restoration significantly elevated the migration and invasion of
MDA-MB-231 cells which were inhibited by miR-136 (Fig. 6C and D), indicating that RASAL2
could rescue miR-136 mediated suppression of migration and invasion
of TNBC cells. These results suggested that RASAL2 may be a
functional target of miR-136 in TNBC cell invasion.
Discussion
Breast cancer is an extremely heterogeneous disease,
comprising a number of different subtypes. TNBC, representing only
10–24% of all breast cancers diagnosed, have been recently
intensely investigated because of their aggressive clinical
behavior. Patients with TNBC are often of younger age, and tend to
develop tumors of larger size, and have an increased risk of
distant metastasis and death within 5 years (27,28).
However, the lack of understanding of the underlying mechanisms of
distant metastasis and early relapse impedes new treatment modality
development for TNBC. In the present study, we identified the
microRNA, miR-136, as an tumor suppressor of TNBC and demonstrated
that it functioned through downregulating RASAL2, a newly
identified cancer-promoting gene in TNBC. This study adds to our
understanding of the underlying mechanism of TNBC invasion and
metastasis.
RASAL2 was first identified as an tumor and
metastasis suppressor in breast cancer (10). However, a more precise analysis
taking the gene subtype into consideration showed that RASAL2
played a pro-oncogenic role in TNBC, rather as a tumor suppressive
RAS-GAP protein in luminal tumors (the most commonly diagnosed
breast cancer subtype) (14,29).
In a Kaplan-Meier meta-analyses consisting of 1,789 patients,
RASAL2 expression level was shown as not prognostic in unselected
patients, but an expression level in the top 30% was significantly
correlated with the poor prognosis in patients with basal tumors,
which overlap largely with TNBC tumors (14). Our results showed that RASAL2 was
markedly upregulated in high-grade TNBC, consistent with other
largescale analyses (12–14). This result may add new evidence to
support RASAL2 as a novel prognostic biomarker of patients with
TNBC.
miR-136 was previously shown to be tightly
associated with tumorigenesis and metastasis. It was reported to
act as a cancer-promoting gene in human NSCLC (24), and may also act as a tumor
suppressor in human glioma (25).
Our results showed that it was downregulated in TNBC and negative
correlated with the WHO grades. It repressed cell migration and
invasion in vitro and restrained the EMT process in a
xenograft mouse model. These results provide new evidence for
miR-136 to be a tumor suppressor specially in the process of
invasion and metastasis of TNBC. miR-136 was recently reported to
be involved in the drug resistance of human epithelial ovarian
cancer and glioma (30,31). Future studies are needed to verify
whether miR-136 regulates sensitivity of chemotherapeutics for
patients diagnosed with TNBC.
In conclusion, our findings demonstrated that
miR-136 was marked downregulated while RASAL2 was upregulated in
TNBC and both were significantly correlated with clinical stage.
Furthermore, our results suggested miR-136 suppressed cell
migration and invasion as well as the EMT process in breast cancer.
Moreover, RASAL2 harbors a binding site of miR-136 and
overexpression of miR-136 significantly decreased the mRNA level of
RASAL2, which indicated miR-136 may directly regulate RASAL2
expression in the development of breast cancer. These results may
validate a pathogenetic role of miR-136 in TNBC and establish a
potential regulatory mechanism involving miR-136/RASAL2/EMT in
TNBC.
Acknowledgments
The present study was supported by the Department of
Pathology, Harbin Medical University.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Autier P, Boniol M, La Vecchia C, Vatten
L, Gavin A, Héry C and Heanue M: Disparities in breast cancer
mortality trends between 30 European countries: Retrospective trend
analysis of WHO mortality database. BMJ. 341:c36202010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kumar P and Aggarwal R: An overview of
triple-negative breast cancer. Arch Gynecol Obstet. 293:247–269.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee J and Gollahon L: Nek2-targeted ASO or
siRNA pretreatment enhances anticancer drug sensitivity in
triple-negative breast cancer cells. Int J Oncol. 42:839–847.
2013.PubMed/NCBI
|
|
6
|
Berrada N, Delaloge S and André F:
Treatment of triple-negative metastatic breast cancer: Toward
individualized targeted treatments or chemosensitization? Ann
Oncol. 21(Suppl 7): vii30–vii35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Downward J: Targeting RAS signalling
pathways in cancer therapy. Nat Rev Cancer. 3:11–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pylayeva-Gupta Y, Grabocka E and Bar-Sagi
D: RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer.
11:761–774. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bos JL, Rehmann H and Wittinghofer A: GEFs
and GAPs: Critical elements in the control of small G proteins.
Cell. 129:865–877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McLaughlin SK, Olsen SN, Dake B, De Raedt
T, Lim E, Bronson RT, Beroukhim R, Polyak K, Brown M, Kuperwasser
C, et al: The RasGAP gene, RASAL2, is a tumor and metastasis
suppressor. Cancer Cell. 24:365–378. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Y, Deng Y, Ji Z, Liu H, Liu Y, Peng H,
Wu J and Fan J: Identification of thyroid carcinoma related genes
with mRMR and shortest path approaches. PLoS One. 9:e940222014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang Y, Zhao M, Xu H, Wang K, Fu Z, Jiang
Y and Yao Z: RASAL2 down-regulation in ovarian cancer promotes
epithelial-mesenchymal transition and metastasis. Oncotarget.
5:6734–6745. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li N and Li S: RASAL2 promotes lung cancer
metastasis through epithelial-mesenchymal transition. Biochem
Biophys Res Commun. 455:358–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng M, Bao Y, Li Z, Li J, Gong M, Lam S,
Wang J, Marzese DM, Donovan N, Tan EY, et al: RASAL2 activates RAC1
to promote triple-negative breast cancer progression. J Clin
Invest. 124:5291–5304. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Yang P and Wang XF:
Microenvironmental regulation of cancer metastasis by miRNAs.
Trends Cell Biol. 24:153–160. 2014. View Article : Google Scholar
|
|
16
|
Pencheva N and Tavazoie SF: Control of
metastatic progression by microRNA regulatory networks. Nat Cell
Biol. 15:546–554. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J, et al: let-7 regulates self
renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Sempere LF, Ouyang H, Memoli VA,
Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, et al:
MicroRNA-31 functions as an oncogenic microRNA in mouse and human
lung cancer cells by repressing specific tumor suppressors. J Clin
Invest. 120:1298–1309. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du
ZW and Zhang JW: Human microRNA clusters: Genomic organization and
expression profile in leukemia cell lines. Biochem Biophys Res
Commun. 349:59–68. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee DY, Jeyapalan Z, Fang L, Yang J, Zhang
Y, Yee AY, Li M, Du WW, Shatseva T and Yang BB: Expression of
versican 3′-untranslated region modulates endogenous microRNA
functions. PLoS One. 5:e135992010. View Article : Google Scholar
|
|
24
|
Shen S, Yue H, Li Y, Qin J, Li K, Liu Y
and Wang J: Upregulation of miR-136 in human non-small cell lung
cancer cells promotes Erk1/2 activation by targeting PPP2R2A.
Tumour Biol. 35:631–640. 2014. View Article : Google Scholar
|
|
25
|
Yang Y, Wu J, Guan H, Cai J, Fang L, Li J
and Li M: MiR-136 promotes apoptosis of glioma cells by targeting
AEG-1 and Bcl-2. FEBS Lett. 586:3608–3612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Podo F, Buydens LM, Degani H, Hilhorst R,
Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J,
Monleon D, et al: FEMME Consortium: Triple-negative breast cancer:
Present challenges and new perspectives. Mol Oncol. 4:209–229.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Carey L, Winer E, Viale G, Cameron D and
Gianni L: Triple-negative breast cancer: Disease entity or title of
convenience? Nat Rev Clin Oncol. 7:683–692. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aysola K, Desai A, Welch C, Xu J, Qin Y,
Reddy V, Matthews R, Owens C, Okoli J, Beech DJ, et al: Triple
negative breast cancer: An overview. Hereditary Genet. 2013(Suppl
2): 20132013.
|
|
30
|
Zhao H, Liu S, Wang G, Wu X, Ding Y, Guo
G, Jiang J and Cui S: Expression of miR-136 is associated with the
primary cisplatin resistance of human epithelial ovarian cancer.
Oncol Rep. 33:591–598. 2015.
|
|
31
|
Wu H, Liu Q, Cai T, Chen YD, Liao F and
Wang ZF: MiR-136 modulates glioma cell sensitivity to temozolomide
by targeting astrocyte elevated gene-1. Diagn Pathol. 9:1732014.
View Article : Google Scholar : PubMed/NCBI
|