Introduction
Oral squamous cell carcinoma (OSCC) is one of the
leading malignancies worldwide (1,2).
Tongue squamous cell carcinoma (TSCC) is the most common type of
oral cancer and is known for its high rate of proliferation and
nodal metastasis (3). According to
the American Cancer Society, an estimated 13,590 new cases of
tongue cancer were diagnosed in 2013, accounting for approximately
30% of all oral cavity and pharynx cancers, with an estimated 2070
deaths (4). Even with recent
advances in its treatment, the 5-year survival rate remains at ~50%
(5). Although attempts have been
made to identify the molecular pathways that contribute to the
carcinogenesis and progression of TSCC, the mechanisms underlying
TSCC progression are poorly understood.
MicroRNAs (miRNAs) are small, endogenous, non-coding
RNAs involved in post-transcriptional regulation of gene expression
(6) and are involved in cell
growth, development, differentiation, proliferation and apoptosis
(7–10). Recent studies have demonstrated that
miRNAs play a critical role in tumorigenesis (11,12)
and are aberrantly expressed in different cancer types, including
TSCC (13–16).
F-box and WD-40 domain protein 7 (FBXW7) is an E3
ubiquitin ligase that regulates the stability of many proteins,
including cyclin E1, c-Myc, NOTCH1, c-Jun and Mcl-1 (17–19).
Mutations in FBXW7 usually disrupt 3 key residues in the
substrate-binding domain and lead to increased stability of target
proteins. Many of the FBXW7 targets are known oncogenes, thus these
mutations could promote tumorigenesis through multiple pathways.
Based on oncology, clinical and basic research, the reduced
expression or loss of function of FBXW 7 has been frequently found
in a variety of human cancers, with an overall mutation frequency
of 6% (20). Clinically, low
expression of FBXW7 in human solid tumors such as glioma,
colorectal cancer and gastric cancer is associated with a poor
prognosis (21–24).
Upregulation of miR-24 has been observed in a number
of cancers (25–27), including OSCC (28,29).
Our previous study demonstrated that frequent upregulation of
miR-24 occurred in TSCC, and miR-24 induced cell survival and
cisplatin resistance in vitro (29). In the present study, we further
found that miR-24 was associated with a shorter overall survival of
TSCC patients, and that inhibition of miR-24 significantly
suppressed the proliferation, migration and invasion of TSCC cells
in vitro. Moreover, we identified and verified that FBXW7 is
a functional target of miR-24, and miR-24 confers its effects in
part through targeting FBXW7, which in turn regulates its
downstream genes, including c-Myc and c-Jun.
Materials and methods
Patients and TSCC tissues
Eighty-four paired TSCC and corresponding non-tumor
control tissues were obtained from patients through surgical
dissection at the Tianjin Cancer Hospital and Institute. This study
was approved by the Ethics Committee of the Tianjin Cancer Hospital
and Institute, and written informed consent was obtained from all
patients.
Cell lines
Human tongue cancer cell lines (UM1, UM2, Cal27,
SCC1, SCC15 and SCC25) were obtained from the American Type Culture
Collection (ATCC; Manassas, VA, USA). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Invitrogen), supplemented
with 10% fetal bovine serum (FBS); all cell lines were maintained
at 37°C in 5% CO2, respectively.
RNA extraction and quantitative RT-PCR
(qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
used to isolate total RNAs from frozen tissues and TSCC cells
according to the manufacturer's protocol. Expression of miRNAs was
analyzed by mirVana qRT-PCR miRNA detection assay (Ambion) or
TaqMan miRNA assay (Applied Biosystems) according to the
manufacturer's protocol. TaqMan microRNA assays for U6 was used to
normalize the relative abundance of miRNA in the tissues and cell
lines.
Cell transfection
Cell transfection with anti-miR-NC, anti-miR-24,
si-NC or si-FBXW7 was conducted by using Lipofectamine 2000
(Invitrogen) according to the manufacturer's instructions.
Specific miRNA-expression plasmids were created by
PCR amplification of human genomic DNA fragments. The primers were:
miR-24 sense, TGGAAGTTTGAGAGTTGAGCCG and antisense,
ACCTTCAAACTCTCAACTCGGC. The PCR products (pre-miRNAs) were cloned
into the pcDNA3.1/V5-His-TOPO expression vector (Invitrogen) and
confirmed by DNA sequencing. The expression of miRNA was carried
out by transfection of the plasmid into the cells using
Lipofectamine 2000.
Cell proliferation assay
SCC15, SCC25 and Cal27 cells were seeded onto
96-well plates (2×104 cells/well), respectively. The
cells were transfected with anti-miR-NC, anti-miR-24, miR-NC,
miR-24, si-NC, si-FBXW7, empty vector, FBXW7 vector or
cotransfected with the miR-24 and FBXW7 vectors, respectively.
After 48 h, cell proliferation was analyzed using Cell Counting
Kit-8 (CCK-8; Beyotime) according to the manufacturer's
protocol.
Cell migration and invasion assays
SCC15, SCC25 and Cal27 cells were cultured and
transfected with anti-miR-NC, anti-miR-24, miR-NC, miR-24, si-NC,
si-FBXW7, empty vector, FBXW7 vector or co-transfected with miR-24
and FBXW7 vectors, respectively. Cell migration and invasion assays
were performed by using Transwell chambers (Millipore, Billerica,
MA, USA). Filters for the invasion assay were precoated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) in the upper
compartment. Cells (5×105) were seeded in the upper
compartment, and the lower compartment was filled with DMEM
supplemented with 10% FBS. After 24 h of incubation, the migratory
and invasive cells adherent to the undersurface of the filters were
fixed, stained with crystal violet and four random fields were
counted for each group.
Luciferase reporter assays
A FBXW7 3′UTR luciferase reporter was created.
Briefly, complementary 48-mer DNA oligonucleotides containing the
putative binding site of miR-24 were synthesized with flanking
MluI and HindIII restriction enzyme digestion sites.
The pmirGLO Dual-Luciferase miRNA Target expression vector
(Promega, Madison, WI, USA; E1910 expressing both firefly and
Renilla) was employed to construct the luciferase reporter
vector. Double-stranded oligonucleotides corresponding to the
wild-type (wt-3′UTR) or mutant (mu-3′UTR) miR-24 binding site in
the FBXW7 3′UTR were synthesized (Sangon Biotech) and ligated
between the XolI and SbfI restriction sites of the
reporter plasmid pmirGLO (Promega). The sequences of inserted
fragments were confirmed by sequencing.
HEK293T cells were transfected with wild-type
pmir-FBXW7-3′UTR or mutant pmir-FBXW7-3′UTR, and the Renilla
luciferase control vector (pRL-TK; Promega) using Lipofectamine
2000, and then the cells were transfected with miR-NC or miR-24,
respectively. Luciferase activities were measured at 24 h after
transfection by using a dual-luciferase assay kit (E1330; Promega).
Firefly luciferase activities were normalized to Renilla
luciferase activities.
Western blot analysis
To isolate the proteins, cells harvested from 6-well
plates were washed once in phosphate-buffered saline and lysed in
lysis buffer. Protein lysates were separated on 10% Tris-glycine
polyacrylamide gel and then transferred to a polyvinylidene
fluoride (PVDF) membrane (Zhongshan Biotechnology Co. Ltd.,
Beijing, China). Then, the membrane was incubated with monoclonal
antibodies against FBW7, c-Myc and c-Jun, followed by incubation
with HRP (horse-radish peroxidase)-labeled goat-anti-mouse IgG
(Santa Cruz Biotechnology, Inc., TX, USA). β-actin was used as a
protein loading control. The intensity of the protein was
quantified with the Quantity software LANE-1D analyzer (SmartGel;
Sage Creation).
Statistical analysis
Data were analyzed using the Statistical Package for
the Social Sciences (SPSS, Chicago, IL, USA), version 17.0. All
values are presented as mean ± standard deviation (SD). One-way
ANOVA and two-tail Student's t-test were used to analyze the
results. A p-value <0.05 was considered to indicate statistical
significance.
Results
miR-24 is significantly increased in TSCC
tissues and cell lines and upregulation of miR-24 correlates with
poor patient prognosis
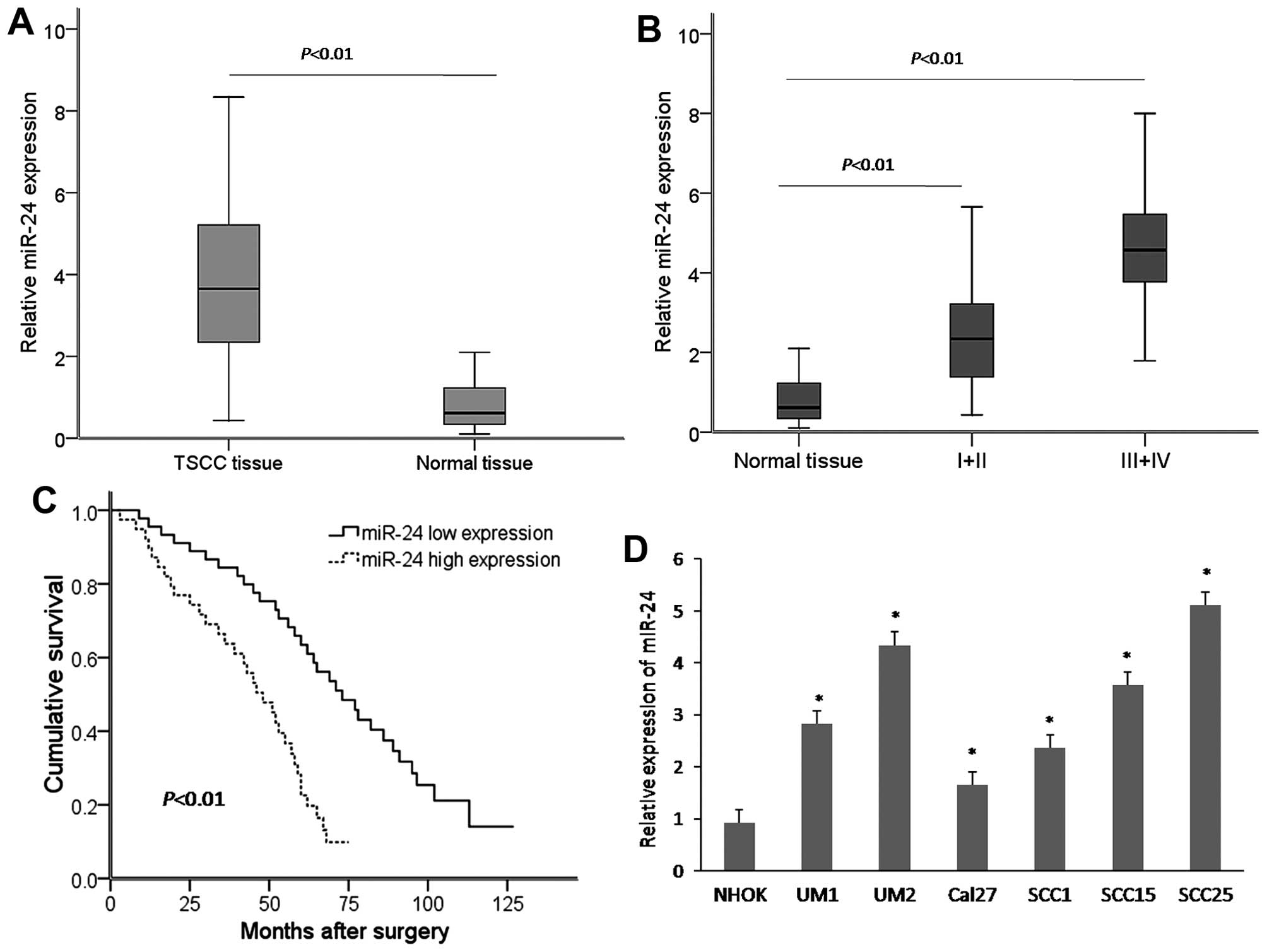
To detect whether there was any difference in the
miR-24 expression level between TSCC tissues and adjacent non-tumor
tissues, a total of 84 paired TSCC tissues and adjacent non-tumor
tissue samples were subjected to qRT-PCR. Compared with the matched
non-tumor tissues, the expression level of miR-24 was significantly
increased in the TSCC tissues (Fig.
1A). Furthermore, the association of miR-24 with
clinicopathologic factors of the TSCC cases was examined. miR-24
overexpression was associated with advanced clinical stage
(Fig. 1B). Kaplan-Meier survival
analysis showed that the overall survival of TSCC patients with
high miR-24 expression was significantly shorter than that with low
miR-24 expression (Fig. 1C). In
additional, miR-24 expression was significantly higher in six TSCC
cell lines (UM1, UM2, Cal27, SCC1, SCC15 and SCC25) compared with
that in the non-tumorigenic cells (NHOK) (Fig. 1D).
miR-24 regulates the proliferation,
migration and invasion of TSCC cells in vitro
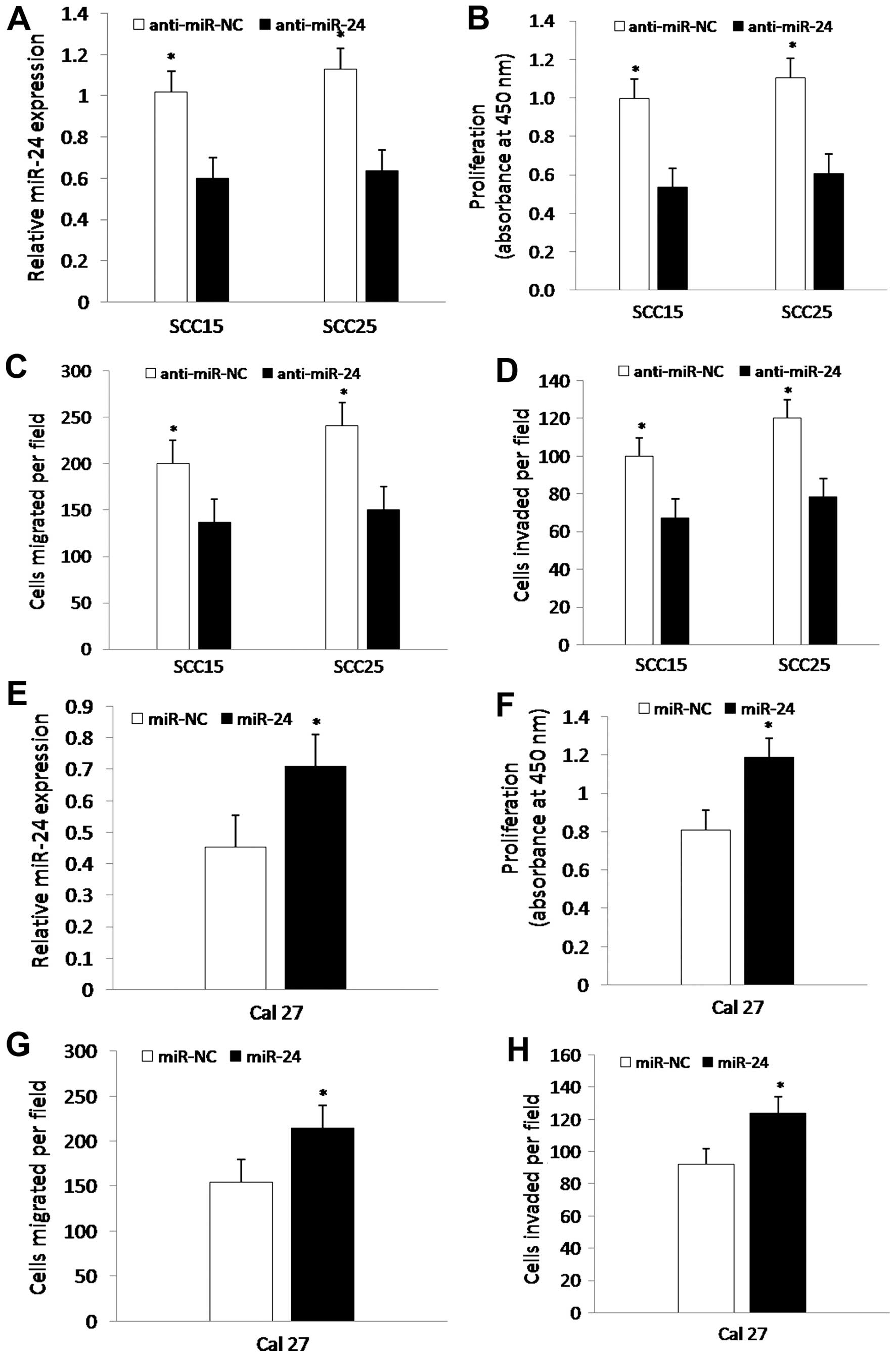
To determine whether miR-24 promotes the
proliferation, migration and invasion potential of TSCC cells, we
carried out experiments using CCK-8, migration and invasion assays.
Briefly, SCC15 and SCC25 cells were transfected with anti-miR-NC or
anti-miR-24 for 24 h, and Cal27 cells were transfected with miR-NC
or miR-24 for 24 h, followed by analysis of the proliferation,
migration and invasion of the cells, respectively. The data showed
that expression of miR-24 in the SCC15 and SCC25 cells transfected
with anti-miR-24 was significantly decreased by qRT-PCR (Fig. 2A), whereas expression of miR-24 was
markedly increased in the Cal27 cells (Fig. 2E). Furthermore, transfection of
anti-miR-24 significantly suppressed the proliferation, migration
and invasion of the SCC15 and SCC25 cells compared to these
parameters in the control group (Fig.
2B–D), whereas miR-24 markedly enhanced the proliferation,
migration and invasion of Cal27 cells (Fig. 2F–H). Together, miR-24 regulates the
proliferation, migration and invasion of TSCC cells.
FBXW7 is a direct target of miR-24 and is
negatively regulated by miR-24
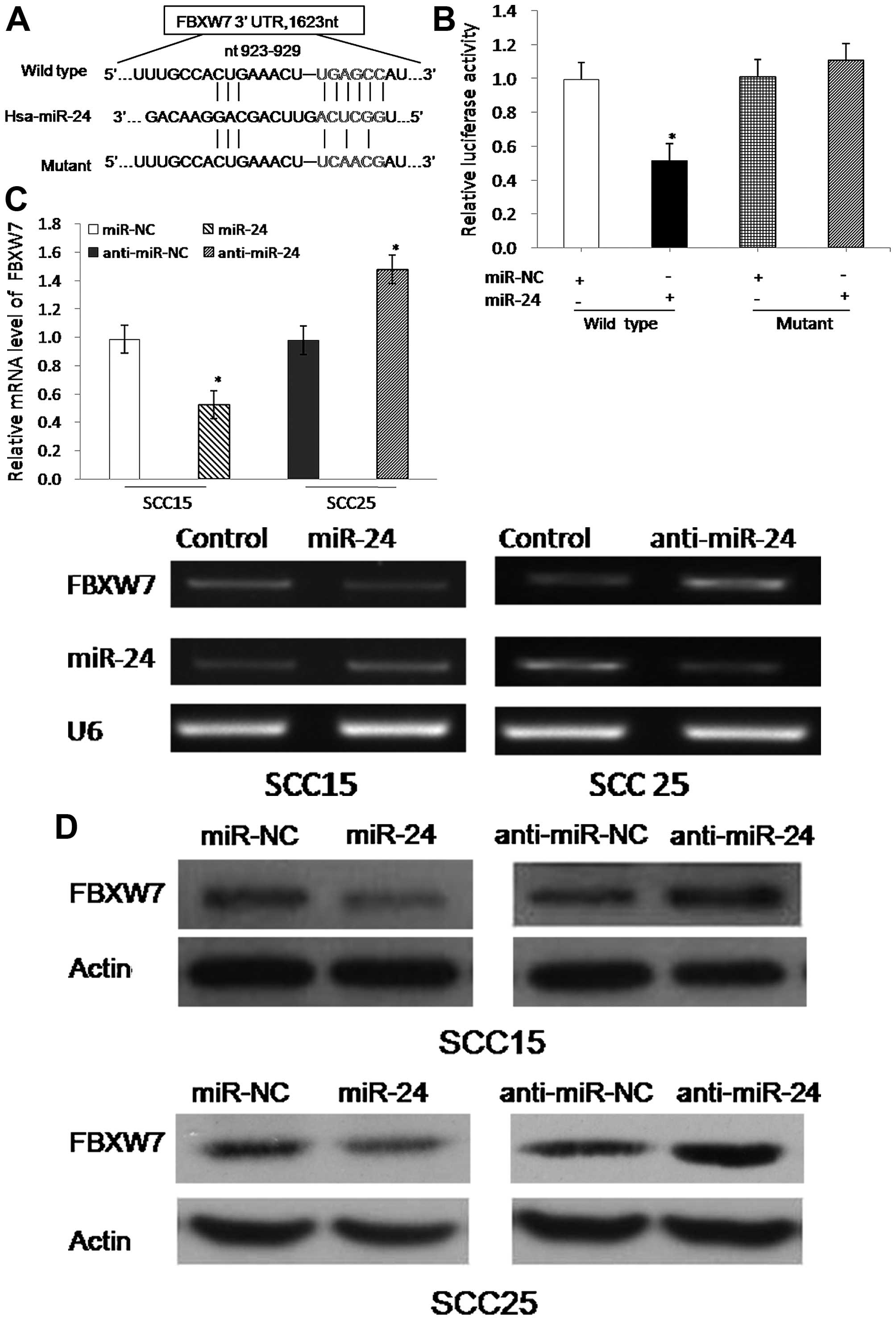
To explore the molecular mechanism of miR-24 in
TSCC, we searched for candidate target genes using bioinformatics
algorithms of TargetScan. We focused on tumor-suppressor gene
FBXW7, which has one potential miR-24 binding site within its
3′UTR.
To identify whether FBXW7 is a target of miR-24, we
constructed vectors containing the wild-type 3′UTR or mutant 3′UTR
of FBXW7 mRNA, which was individually fused directly downstream of
the firefly luciferase gene (Fig.
3A). For the luciferase assays, the wild-type or mutant vector
was co-transfected into HEK293T cells with miR-NC or miR-24.
Luciferase reporter assay showed that miR-24 could inhibit the
luciferase activity of the wild-type 3′UTR of the FBXW7 reporter
vector but not the mutant 3′UTR (Fig.
3B). Furthermore, we confirmed that overexpression of miR-24
could decrease FBXW7 mRNA levels, whereas inhibition of miR-24
increased the FBXW7 mRNA levels (Fig.
3C). Moreover, FBXW7 protein abundance was also negatively
regulated by miR-24 (Fig. 3D).
Together, miR-24 downregulated FBXW7 expression by directly
targeting the 3′UTR of FBXW7.
miR-24 promotes TSCC cell growth and
motility by targeting FBXW7
To investigate the effect of FBXW7 on
tumorigenicity, we designed experiments in which the SCC15 and
SCC25 cells were transfected with si-NC, si-FBXW7, empty vector and
FBXW7 vector. The results showed that the growth and motility of
the SCC15 and SCC25 cells were markedly promoted or suppressed
after transfection with si-FBXW7 or the FBXW7 vector, respectively
(Fig. 4A–C), suggesting that FBXW7
suppresses the proliferation, migration and invasion of TSCC cells.
To further assess whether the effect of miR-24 on tumorigenicity
was via targeting FBXW7, we carried out functional experiments.
Briefly, the SCC15 and SCC25 cells were transfected with miR-NC,
miR-24 or co-transfected with miR-24 and FBXW7. The growth and
motility of the SCC15 and SCC25 cells were significantly enhanced
after transfection with miR-24 compared with the control group,
whereas the restoration of FBXW7 antagonized miR-24 in regards to
the migration, invasion and proliferation (Fig. 4D–F). Furthermore, western blotting
showed that FBXW7 expression was markedly decreased in the TSCC
cells after transfection with miR-24, and was restored when the
TSCC cells were co-transfected with miR-24 and FBXW7, and
subsequently inversely regulated its downstream targets c-Myc and
c-Jun (Fig. 4G). Together, these
findings suggest that miR-24 promotes the proliferation, migration
and invasion, at least partially by targeting FBXW7.
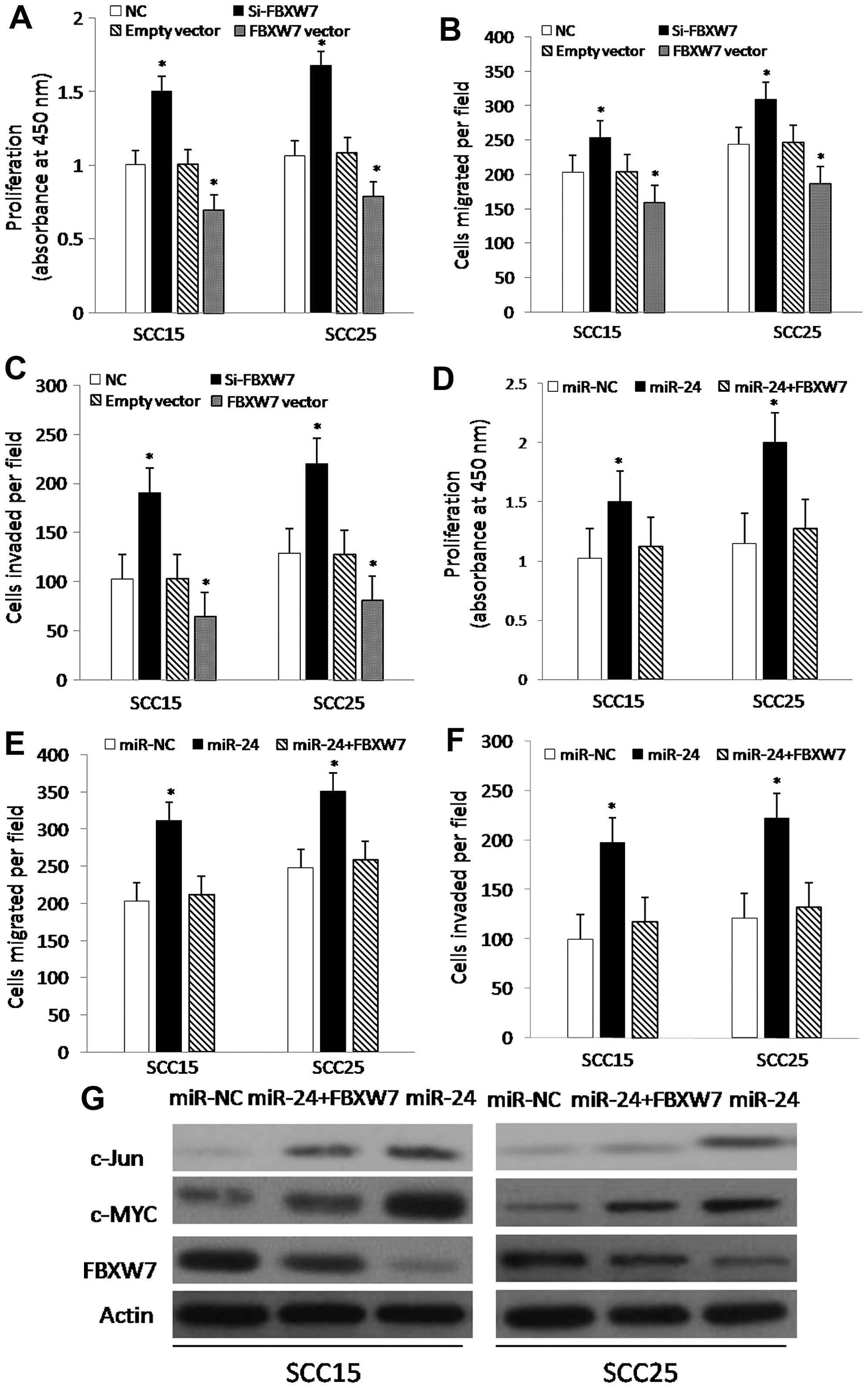 | Figure 4Suppression of FBXW7 promotes TSCC
cell growth and motility and restoration of FBXW7 attenuates cell
growth and motility. (A–C) Cell proliferation, migration and
invasion ability assays of SCC15 and SCC25 cells following
transfection with si-NC, si-FBXW7, empty vector, or FBXW7 vector,
respectively. Histogram reveals the values of absorbance at 450 nm
for proliferation (A). The assays were repeated in duplicates.
(D–F) Cell proliferation, migration and invasion ability assays in
SCC15 and SCC25 cells following transfection with miR-NC, miR-24,
or co-transfection with miR-24 and FBXW7, respectively. Histogram
reveals the values of absorbance at 450 nm for proliferation (D).
The assays were repeated in duplicates. (G) Western blot assays
show the protein expression of FBXW7, c-Myc and c-Jun in the SCC15
and SCC25 cells transfected with miR-NC, miR-24, or co-transfected
with miR-24 and FBXW7, respectively. β-actin served as an internal
control. Western blot analysis confirmed that miR-24 downregulated
the expression of FBXW7 protein and inversely upgulated the
downstream targets c-Myc and c-Jun. The assays were repeated in
duplicates. *P<0.01, compared with the control
group. |
Discussion
The development of TSCC is a multistep process
involving multiple factors. Great efforts have been made to
understand the principles of molecular changes during the
occurrence of this malignancy. To date, accumulating evidence
demonstrates that multiple miRNAs are involved in TSCC development
and progression (13,15,16).
miR-24 has been found to serve as an oncogene in several types of
cancers, including breast carcinoma, glioma, oral carcinoma, acute
myeloid leukemia and squamous cell carcinoma (30–33).
Recently, it was reported that this miR-24 had a close association
with TSCC (28,29), suggesting that it may serve as a
potential biomarker for the diagnosis and prognosis of TSCC.
However, the mechanism of miR-24 in TSCC is not entirely clear.
In our present study, we found that miR-24 was
significantly overexpressed in human TSCC tissues and cell lines
compared with the control groups, and that upregulation of miR-24
was associated with a shorter overall survival of TSCC patients. In
agreement with our findings, previous reports have shown that
miR-24 is overexpressed in many other solid tumors, and is related
to tumor progression and prognosis. It is known that microRNAs
function primarily by negatively regulating the expression of their
target genes. And recent studies have demonstrated some specific
targets of miR-24 in several tumors, such as the sex determining
region Y (SRY)-box 7 (SOX7) (34),
nuclear apoptosis-inducing factor 1 (NAIF1) (35) and dead end 1 (DND1) (14). In addition, our previous study
confirmed that miR-24 induced cell survival and cisplatin
resistance primarily through targeting the PTEN/Akt pathway in TSCC
cell lines (29). We further
confirmed that FBXW7 is a direct target of miR-24. And our
functional analysis showed that miR-24-induced loss of FBXW7
enhanced TSCC cell proliferation, migration and invasion, and that
miR-24 expression responds to alterations in the c-Myc and c-Jun
protein levels, which were regulated by the FBXW7 pathway. These
findings suggest that overexpression of miR-24 promotes the
proliferation, migration and invasion of TSCC cells in
vitro, possibly due to suppression of the function of
FBXW7.
FBXW7 regulates the stability of several substrates,
which are involved in the regulation of apoptosis and cell
proliferation, such as c-Myc, cyclin E, Notch1 and Mcl-1 (17,36–38).
In addition, accumulating evidence shows that FBXW7 plays critical
roles in tumorigenesis. To date, there are a few agents known to
activate FBXW7, such as miR-27, miR-92, miR-25 and miR-223 in
several cancers (39–41). Here, we confirmed that FBXW7 is a
direct target of miR-24. Furthermore, a decrease in the expression
of FBXW7 resulting from the overexpression of miR-24 gave rise to
abnormal accumulation of c-Myc and c-Jun proteins. Our results
suggest that miR-24 may exert its oncogenic function partially
through elevating c-Myc and c-Jun expression. In addition, studies
in mice show that FBXW7 inhibition eliminates CML by increasing Myc
abundance (42). Kurashige
indicates that high expression of miR-223 had a significant adverse
impact on the survival of ESCC patients through repression of the
function of FBXW7 (43). Taking
these results together with our findings, we surmise that
miR-24-induced loss of FBXW7 may be a promising treatment target
for TSCC due to its ability to reduce the levels of several
oncoproteins.
In summary, the present study indicated that
deregulation of miR-24 is a frequent event in TSCC in vitro,
and that miR-24 promoted the growth and motility of TSCC cell
lines, at least partially by targeting FBXW7. We hope that our
investigation can facilitate further exploration of the molecular
mechanisms of miR-24 in TSCC.
Acknowledgments
The present study was partially supported by grants
from the National Natural Science Foundation of China (grant nos.
81402392, 81502322) and the Tianjin Municipal Science and
Technology Project (grant no. 15JCQNJC12800).
References
|
1
|
Marcus B, Arenberg D, Lee J, Kleer C,
Chepeha DB, Schmalbach CE, Islam M, Paul S, Pan Q, Hanash S, et al:
Prognostic factors in oral cavity and oropharyngeal squamous cell
carcinoma. Cancer. 101:2779–2787. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chang KW, Liu CJ, Chu TH, Cheng HW, Hung
PS, Hu WY and Lin SC: Association between high miR-211 microRNA
expression and the poor prognosis of oral carcinoma. J Dent Res.
87:1063–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sano D and Myers JN: Metastasis of
squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev.
26:645–662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao B, Gao K, Li L, Huang Z and Lin L:
miR-184 functions as an oncogenic regulator in hepatocellular
carcinoma (HCC). Biomed Pharmacother. 68:143–148. 2014. View Article : Google Scholar
|
|
7
|
Zhang C: Novel functions for small RNA
molecules. Curr Opin Mol Ther. 11:641–651. 2009.
|
|
8
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eulalio A, Mano M, Dal Ferro M, Zentilin
L, Sinagra G, Zacchigna S and Giacca M: Functional screening
identifies miRNAs inducing cardiac regeneration. Nature.
492:376–381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Swarbrick A, Woods SL, Shaw A,
Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ,
Judson RL, et al: miR-380-5p represses p53 to control cellular
survival and is associated with poor outcome in MYCN-amplified
neuroblastoma. Nat Med. 16:1134–1140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He L, He X, Lowe SW and Hannon GJ:
microRNAs join the p53 network - another piece in the
tumour-suppression puzzle. Nat Rev Cancer. 7:819–822. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu X, Wang A, Heidbreder CE, Jiang L, Yu
J, Kolokythas A, Huang L, Dai Y and Zhou X: MicroRNA-24 targeting
RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS
Lett. 584:4115–4120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia LF, Wei SB, Gong K, Gan YH and Yu GY:
Prognostic implications of micoRNA miR-195 expression in human
tongue squamous cell carcinoma. PLoS One. 8:e566342013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Huang H, Sun L, Yang M, Pan C, Chen
W, Wu D, Lin Z, Zeng C, Yao Y, et al: miR-21 indicates poor
prognosis in tongue squamous cell carcinomas as an apoptosis
inhibitor. Clin Cancer Res. 15:3998–4008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Welcker M and Clurman BE: FBW7 ubiquitin
ligase: A tumour suppressor at the crossroads of cell division,
growth and differentiation. Nat Rev Cancer. 8:83–93. 2008.
View Article : Google Scholar
|
|
18
|
Cheng Y and Li G: Role of the ubiquitin
ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 31:75–87.
2012. View Article : Google Scholar
|
|
19
|
Brandt Y, Mitchell T, Wu Y and Hartley RS:
Developmental downregulation of Xenopus cyclin E is phosphorylation
and nuclear import dependent and is mediated by ubiquitination. Dev
Biol. 355:65–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akhoondi S, Sun D, von der Lehr N,
Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D,
Marth C, et al: FBXW7/hCDC4 is a general tumor suppressor in human
cancer. Cancer Res. 67:9006–9012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hagedorn M, Delugin M, Abraldes I, Allain
N, Belaud-Rotureau MA, Turmo M, Prigent C, Loiseau H, Bikfalvi A
and Javerzat S: FBXW7/hCDC4 controls glioma cell proliferation in
vitro and is a prognostic marker for survival in glioblastoma
patients. Cell Div. 2:92007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bredel M, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended glioma-genesis pathway maps and three novel
MYC-interacting genes in human gliomas. Cancer Res. 65:8679–8689.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iwatsuki M, Mimori K, Ishii H, Yokobori T,
Takatsuno Y, Sato T, Toh H, Onoyama I, Nakayama KI, Baba H, et al:
Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer:
Clinical significance. Int J Cancer. 126:1828–1837. 2010.
|
|
24
|
Yokobori T, Mimori K, Iwatsuki M, Ishii H,
Onoyama I, Fukagawa T, Kuwano H, Nakayama KI and Mori M:
p53-Altered FBXW7 expression determines poor prognosis in gastric
cancer cases. Cancer Res. 69:3788–3794. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu R, Zhang H, Wang X, Zhou L, Li H, Deng
T, Qu Y, Duan J, Bai M, Ge S, et al: The miR-24-Bim pathway
promotes tumor growth and angiogenesis in pancreatic carcinoma.
Oncotarget. 6:43831–43842. 2015.PubMed/NCBI
|
|
28
|
Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS
and Chang KW: miR-24 up-regulation in oral carcinoma: Positive
association from clinical and in vitro analysis. Oral Oncol.
46:204–208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng X, Li J, Peng C, Zhao J, Chi J, Meng
X, Yun X, Li D, Yu Y, Gao M, et al: MicroRNA-24 induces cisplatin
resistance by targeting PTEN in human tongue squamous cell
carcinoma. Oral Oncol. 51:998–1003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du WW, Fang L, Li M, Yang X, Liang Y, Peng
C, Qian W, O'Malley YQ, Askeland RW, Sugg SL, et al: MicroRNA
miR-24 enhances tumor invasion and metastasis by targeting PTPN9
and PTPRF to promote EGF signaling. J Cell Sci. 126:1440–1453.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen L, Zhang A, Li Y, Zhang K, Han L, Du
W, Yan W, Li R, Wang Y, Wang K, et al: miR-24 regulates the
proliferation and invasion of glioma by ST7L via β-catenin/Tcf-4
signaling. Cancer Lett. 329:174–180. 2013. View Article : Google Scholar
|
|
32
|
Zaidi SK, Dowdy CR, van Wijnen AJ, Lian
JB, Raza A, Stein JL, Croce CM and Stein GS: Altered Runx1
subnuclear targeting enhances myeloid cell proliferation and blocks
differentiation by activating a miR-24/MKP-7/MAPK network. Cancer
Res. 69:8249–8255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Papadimitriou E, Vasilaki E, Vorvis C,
Iliopoulos D, Moustakas A, Kardassis D and Stournaras C:
Differential regulation of the two RhoA-specific GEF isoforms
Net1/Net1A by TGF-β and miR-24: Role in epithelial-to-mesenchymal
transition. Oncogene. 31:2862–2875. 2012. View Article : Google Scholar
|
|
34
|
Ma Y, She XG, Ming YZ and Wan QQ: miR-24
promotes the proliferation and invasion of HCC cells by targeting
SOX7. Tumour Biol. 35:10731–10736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao G, Liu L, Zhao T, Jin S, Jiang S, Cao
S, Han J, Xin Y, Dong Q, Liu X, et al: Upregulation of miR-24
promotes cell proliferation by targeting NAIF1 in non-small cell
lung cancer. Tumour Biol. 36:3693–3701. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Koepp DM, Schaefer LK, Ye X, Keyomarsi K,
Chu C, Harper JW and Elledge SJ: Phosphorylation-dependent
ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase.
Science. 294:173–177. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu G, Lyapina S, Das I, Li J, Gurney M,
Pauley A, Chui I, Deshaies RJ and Kitajewski J: SEL-10 is an
inhibitor of notch signaling that targets notch for
ubiquitin-mediated protein degradation. Mol Cell Biol.
21:7403–7415. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for
ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Olive V1, Sabio E, Bennett MJ, De Jong CS,
Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A,
et al: A component of the mir-17-92 polycistronic oncomir promotes
oncogene-dependent apoptosis. Elife. 2:e008222013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang L, Ye X, Liu Y, Wei W and Wang Z:
Aberrant regulation of FBW7 in cancer. Oncotarget. 5:2000–2015.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y
and Sun H: MicroRNA-25 promotes gastric cancer proliferation,
invasion, and migration by directly targeting F-box and WD-40
domain protein 7, FBXW7. Tumour Biol. 36:7831–7840. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Reavie L, Buckley SM, Loizou E, Takeishi
S, Aranda-Orgilles B, Ndiaye-Lobry D, Abdel-Wahab O, Ibrahim S,
Nakayama KI and Aifantis I: Regulation of c-Myc ubiquitination
controls chronic myelogenous leukemia initiation and progression.
Cancer Cell. 23:362–375. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K and
Baba H: Overexpression of microRNA-223 regulates the ubiquitin
ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer.
106:182–188. 2012. View Article : Google Scholar :
|