1. Introduction
Leukemia is a malignancy of the hematopoietic system
characterized by diffuse replacement of the bone marrow by
neoplastic cells (1). Acute
lymphoid leukemia (ALL) and acute myeloid leukemia (AML) are
oncohematologic diseases in which the process of differentiation
and limited proliferation that characterizes normal hematopoiesis
is altered and replaced by a malignant clonal expansion of immature
hematopoietic cells (blasts) in the bone marrow or peripheral blood
(2–5).
Epidemiological studies indicate that the annual
incidence rates of childhood ALL vary worldwide between one and
four new cases per 100,000 children younger than 15 years, with a
peak incidence at approximately 2–5 years of age (6–8),
whereas AML has been observed with an incidence of 3.7 per 100,000
persons and an age-dependent mortality of 2.7 to nearly 18 per
100,000 persons (9).
Abnormalities in chromosome number as well as
structural rearrangements (translocations) are detected in 60–80%
of patients with ALL, whereas the remaining 20–40% have a normal
karyotype (9–14). In addition to those with a normal
karyotype, t(12;21)(p13;q22);TEL/AML1 (ETV6-RUNX1),
t(9;22)(q34,q11); BCR/ABL (BCR-ABL1), t(4;11) (q21;q23); MLL/AF4
(KMT2A/AFF1), t(1;19)(q23;p13); E2A/PBX1 (TCF3-PBX1), are among the
most common cytogenetic subtypes in ALL (10–12,14),
and have been incorporated in the World Health Organization (WHO)
classification as the criteria for subclassification of acute
leukemia (15).
ALL can be distinguished from AML using morphologic,
immunohistochemical, and immunologic methods (16). The study by Golub et al
showed that the gene expression profiles can discriminate the ALL
from AML (17). However, the
precise genes and pathways that exert critical control over
determination of lineage fate during leukemia development remain
unclear (18).
Hematopoiesis is a highly regulated process of the
differentiation, proliferation hematopoietic stem cells give rise
to all the blood lineages: the myeloid lineage, which comprises
neutrophils, eosinophils, basophils, monocytes, macrophages,
megakaryocytes, platelets and erythrocytes; and the lymphoid
lineage, which includes T and B cells (19,20).
The process is modulated by a series of molecular
events, and there is increasing evidence that miRNAs have important
roles in modulating hematopoietic process by targeting the
expression of transcription factors and genes that are involved in
the regulation of cell proliferation, metabolism, and apoptosis
(21). Aberrant expression of many
different miRNAs has been observed in several cancers, including
hematological malignancies. Furthermore, approximately 50% of
miRNAs are located at fragile sites and genomic regions in the
human genome associated with cancer (22). In this review, we summarized the
association of the miRNA expression with chromosomal translocations
in acute leukemia, with a specific focus on acute lymphoblastic
leukemia.
2. miRNA expression and oncoproteins in
acute lymphoblastic leukemia
TEL/AML1; t(12;21)(p13;q22)
TEL/AML1 t(12;21) (p13;q22) fusion gene, resulting
from 12;21 chromosomal translocation, is believed to be the most
common molecular genetic abnormality in childhood acute
lymphoblastic leukemia (23). The
resulting fusion protein has the AML1 DNA binding domain and the
TEL protein interaction domain and has been shown to maintain
transcription factor properties and bind DNA (24,25).
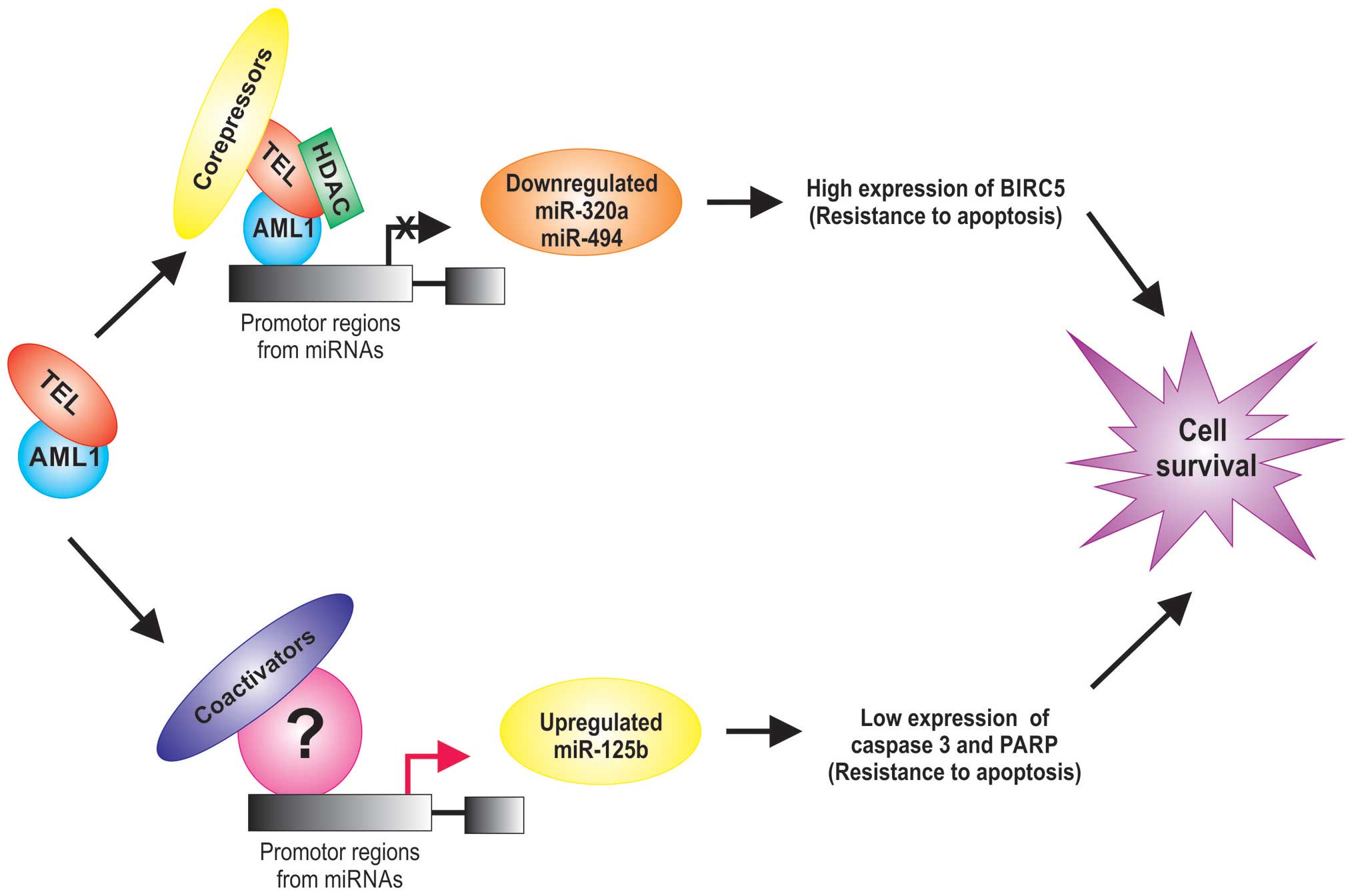
TEL/AML1 act as a transcription factor that reduce the expression
of tumor suppressor and increase antiapoptotic genes (26) and also alters the regulation of
miRNA expression (27,28) (Table
I). For example, the miR-320a and miR-498 tumor suppressor are
significantly lower in TEL/AML1-positive acute lymphoblastic
leukemias (28). Interestingly,
these miRNAs can inhibit the expression of survivin in leukemia
cells, inducing apoptosis, suggests that miR-320a and miR-498 are
potential tumor suppressors or miRNAs that may play a critical role
in the leukemic process (28). The
TEL/AML1 protein is generally a transcriptional repressor due to
its known ability to recruit chromatin repressors such as histone
deacetylases, nuclear receptor corepressors (Fig. 1).
 | Table IExpression of miRNAs in
rearrangement-positive ALL. |
Table I
Expression of miRNAs in
rearrangement-positive ALL.
| miRNAs | Function in
cancera | Expression | Authors/(Ref.) |
|---|
| TEL/AML1-positive
rearrangement |
| miR-26b | OG/TSG | Downregulation | Diakos et al
(28) |
| miR-320a | TSG | | |
| miR-494 | TSG | | |
| miR-213 | TSG | Downregulation | |
| miR-221 | OG/TSG | | |
| miR-99a | OG | Upregulation | Schotte et
al (27) |
| miR-100 | OG/TSG | | |
| miR-125b | OG | | |
| miR-126 | OG/TSG | | |
| miR-383 | OG/TSG | | |
| miR-629 | OG | | |
| miR-125b | OG | Upregulation | Gefen et al
(29) |
| BCR/ABL-positive
rearrangement |
| miR-17 | OG | Upregulation | Scherr et al
(33) |
| miR-18 | OG | | |
| miR-19a | OG | | |
| miR-20a | OG | | |
| miR-93 | OG | Downregulation | Schotte et
al (27) |
| miR-103 | OG/TSG | | |
| miR-148b | TSG | | |
| miR-210 | OG/TSG | | |
| miR-301 | TSG | | |
| miR-331 | TSG | | |
| miR-345 | OG/TSG | | |
| miR-484 | TSG | | |
| miR-1226 | TSG | | |
| MLL/AF4-positive
rearrangement |
| Let-7b | TSG | Downregulation | Mi et al
(18) |
| miR-128a | OG/TSG | Upregulation | |
| miR-128b | OG/TSG | | |
| miR-128a | OG/TSG | Upregulation | Oliveira et
al (47) |
| miR-128b | OG/TSG | | |
| miR-181b | OG | | |
| miR-143 | TSG | Downregulation | Dou et al
(46) |
| mir-196b | OG | Upregulation | Popovic et
al (50) and Li et al
(51) |
| TCF3/PBX1-positive
rearrangement |
| miR-24 | OG/TSG | Downregulation | Schotte et
al (27) |
| miR-126 | OG/TSG | | |
| miR-146a | TSG | | |
| miR-193a | TSG | | |
| miR-365 | TSG | | |
| miR-511 | TSG | | |
| miR-545 | TSG | | |
| miR-181 | OG | Upregulation | Schotte et
al (56) |
| miR-708 | OG | | |
| mir-196b | TSG | Downregulation | |
Other upregulation miRNAs in TEL/AML1-positive acute
lymphoblastic leukemias have also been reported (27–29)
and it has been observed that AML1 can associate with coactivators
to regulate the promoters of its target genes (30). For example, miR-125b is an oncogenic
miRNA by its anti-apoptotic activity which is associated with a
marked inhibition of caspase 3 activation and the cleavage of its
substrate PARP (29), suggesting an
active role for miR-125b in the leukemogenesis (31) and its expression is an independent
event from TEL/AML1 protein (Fig.
1).
BCR/ABL1; t(9;22)(q34,q11)
BCR/ABL1 or Philadelphia (Ph) chromosome is a
product of the t(9;22), which fuses the Abelson kinase gene (ABL1)
from chromosome 9 with the breakpoint cluster region (BCR) from
chromosome 22 that expresses the BCR-ABL1 fusion protein: a
constitutively active tyrosine kinase. The BCR/ABL1 fusion protein
is a hallmark present in a fraction of B progenitor ALL cases that
have a particularly poor prognosis (32).
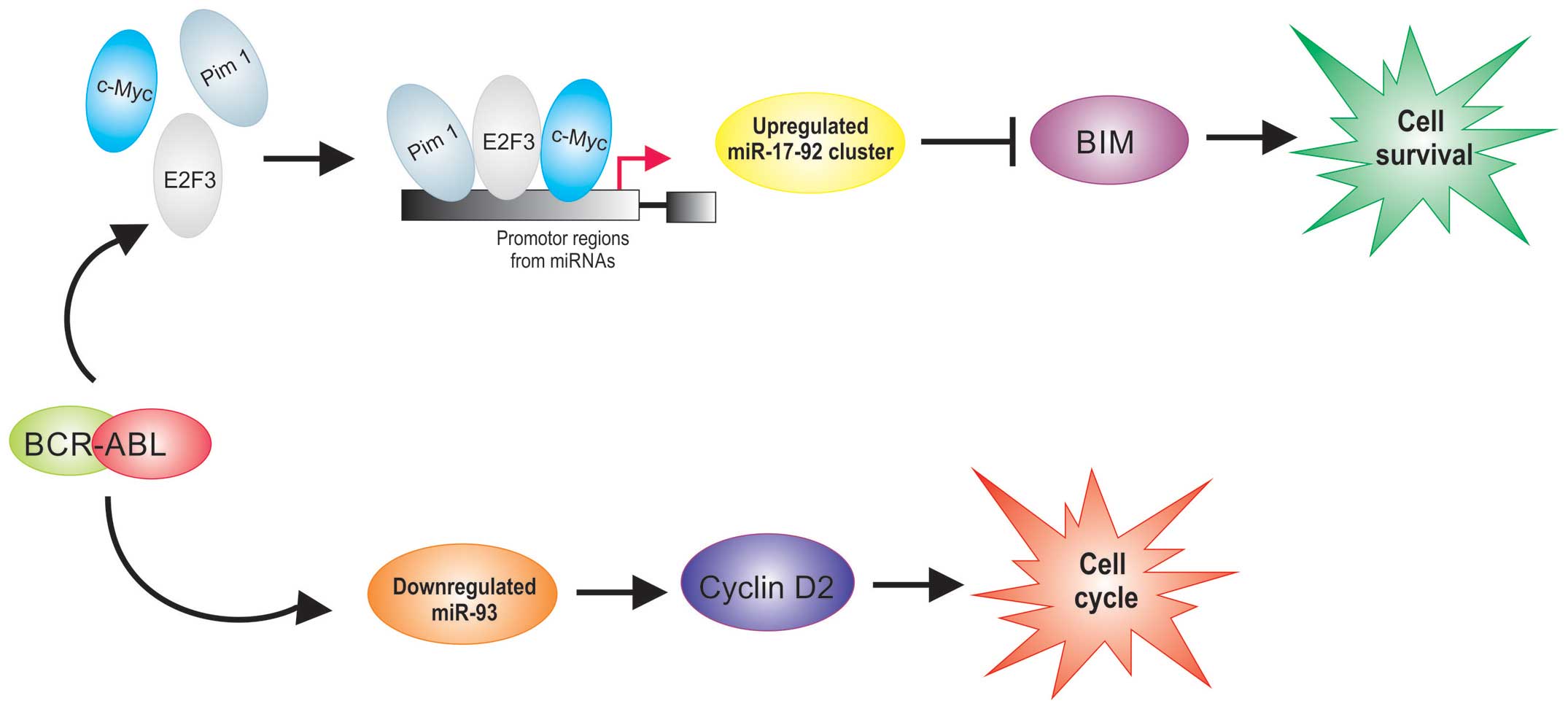
Several miRNAs are overexpressed in
BCR/ABL1-positive ALL (27,33) (Table
I). BCR/ABL has been shown to upregulate Pim-1 (34,35),
E2F3 (36) and c-Myc (35) who regulate the transcriptional
expression of miR-17-92 clusters. Recently, it was reported that
BIM, a proapoptotic member, is a direct target of miR-17-92
clusters (37). It was demonstrated
that BIM is poorly expressed in ALL (38), while the members of the miR-17-92
clusters that have been reported to be upregulated in
BCR/ABL1-positive ALL (33). These
data demonstrate that the upregulation of miR-17-92 cluster
expression observed in BCR/ABL1-positive ALL could have an
important role in survival of cells in leukemia (Fig. 2).
Interestingly, a direct relationship between BCR/ABL
activity and cyclin D2 expression in BCR/ABL-positive cells has
been demonstrated (39,40); these reports suggest the importance
of cyclin D2 in mediating the proliferative signals from BCR/ABL
and show that BCR/ABL regulates cyclin D2 expression at the
transcriptional level. A recent study showed that miR-93 is
downregulated in BCR/ABL-positive patients (27). Interestingly, this miRNA can inhibit
the expression of cyclin D2 which leads to cell cycle progression
(41). Overexpression of cyclins D2
in BCR/ABL-positive hematopoietic cell have been reported (39,40).
The abnormal expression of cyclin D2 may influence the initiation
of tumorigenesis, including leukemogenesis (42), suggesting that downregulation of
miR-93 could be involved to the leukemogenesis at least partly via
upregulation of cyclin D2 expression in BCR/ABL-positive cells
(Fig. 2).
MLL/AF4; t(4;11)(q21;q23)
The t(4;11)(q21;q23) involving the genes MLL and AF4
(alias for AFF1) is detected in 50-70% of infant leukemia. MLL/AF4,
an MLL fusion protein that is associated with infant pro-B acute
lymphoblastic leukemias (43), and
it is usually associated with a poor prognosis (44). Rearrangements of the MLL gene result
in a fusion protein that retain the DNA binding capacities, the
CXXC domain, which binds to non-methylated CpG DNA sites, as well
as their DNA methyltransferase activity (transcriptional
activation/repression domain of MLL), it is possible that the
MLL-fusion proteins directly regulate a subset of genes (45).
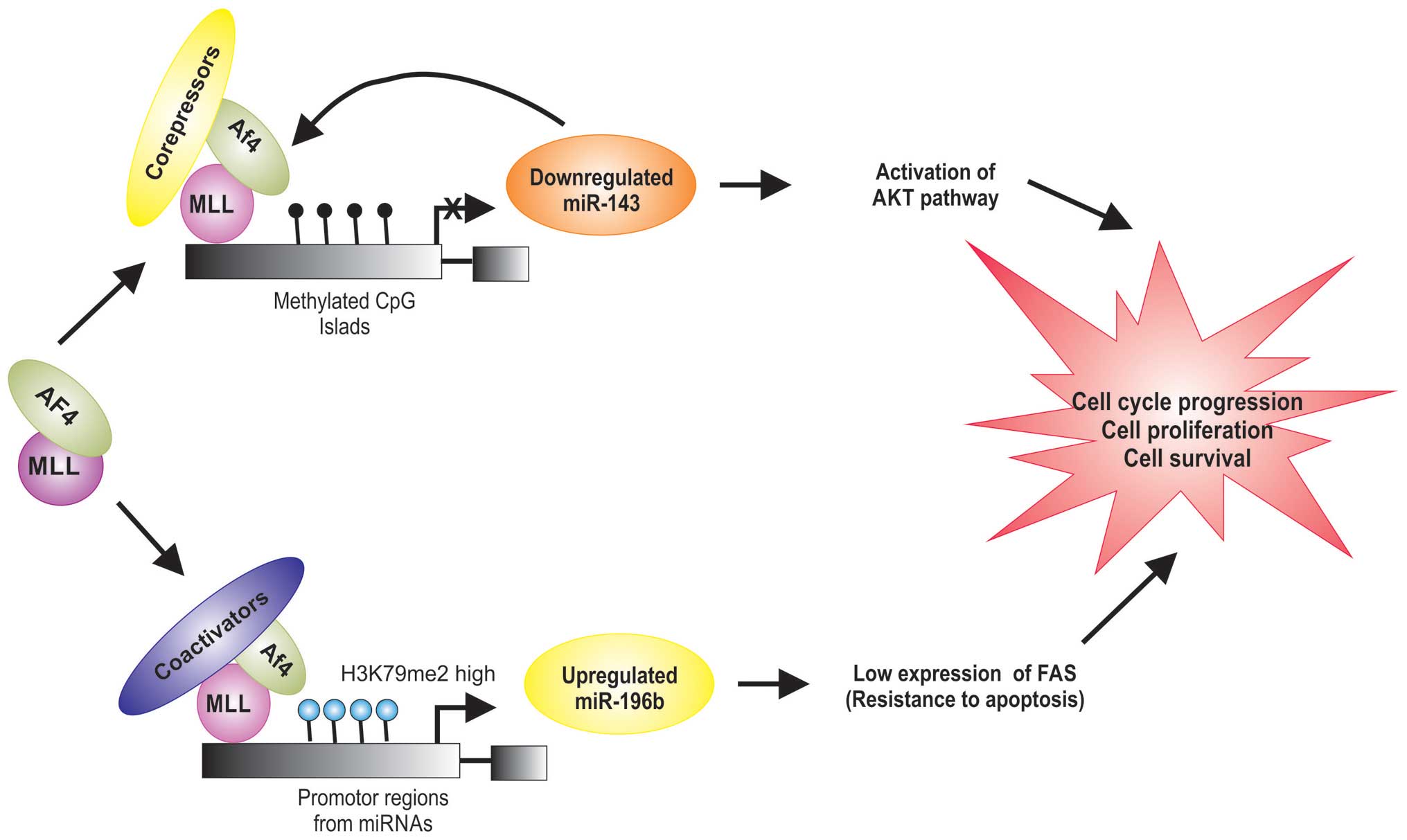
Recent studies indicate that MLL/AF4 regulates the
expression of miRNas (Table I),
which can play a role in leukemogenesis by stimulating
proliferation and inducing a block in differentiation of
hematopoietic cells (46,47). Dou et al demonstrated that
the expression levels of miR-143 are significantly lower in
MLL-AF4-positive cells (46).
PI3K/AKT/mTOR is a survival pathway and plays an important role in
cell proliferation, differentiation and apoptosis. Its abnormal
activation has been found in cases of MLL/AF4-positive ALL
(48). miR-143 has been shown to
decrease the level of Akt at translational level (49). miR-143 is epigenetically repressed
by promoter hypermethylation in MLL/AF4-positive primary blasts and
cell lines, which was directly associated with expression of the
MLL/AF4 oncogene (46).
Additionally, it was shown that MLL/AF4 is a target for miR-143,
and that DNA methylation decreases the expression of miR-143 in
MLL/AF4-positive ALL (46), it is
possible that the MLL/AF4 proteins directly downregulate the miR143
expression by the DNA methyltransferase activity of MLL (Fig. 3).
miR-196b is an oncogene and appears to be required
for MLL/AF4-mediated cell transformation and it was reported that
miR-196b is upregulated in MLL/AF4-positive cell lines (50). MLL/AF4 protein regulate the
transcriptional expression of miR-196b by increasing levels of
K79me2 on the promoter of miR-196b (50,51).
FAS plays a central role in the physiological regulation of
apoptosis (52), and it has been
implicated in the leukemogenesis (51). FAS is direct target of miR-196b and
is negatively regulated at the mRNA and protein levels (51). It was demonstrated that FAS is
poorly expressed in MLL/AF4-positive human leukemic cells, compared
to the normal cells (Table I)
(50,51). Li et al found that the low
expression of FAS leads to inhibition of FAS-mediated apoptosis
(51). These data demonstrate that
the upregulation of miR-196b expression observed in
MLL/AF4-positive cell has an important role in cell survival in
leukemia by targeting FAS mRNA in MLL/AF4-positive cells (Fig. 3).
TCF3/PBX1; t(1;19)(q23;p13)
The t(1;19)(q23;p13) is one of the most frequent
translocations in B-acute lymphoblastic leukemia (B-ALL), and is
observed in both adult and pediatric populations at an overall
frequency of 6% (53). This
translocation is the result from the fusion of TCF3 (transcription
factor 3) found at 19p13 and PBX1 (pre-B cell leukemia homebox 1)
found at 1q23 forming a chimeric gene whose protein product alters
cell differentiation arrest, among other cellular processes
(54). TCF3/PBX1 encodes a
transcription factor bearing the transactivation domain of TCF3 and
the DNA-binding domain of PBX1, which facilitates the activation or
repression of genes (55).
Several miRNAs are downregulated or upregulated in
TCF3/PBX1-positive ALL (27,56)
(Table I). For example, miR-126
plays a pivotal role in restraining cell cycle progression of
hematopoietic stem cell in vitro and in vivo, it was
observed that the downregulation of this miRNA resulted in cell
cycle progression and hematopoietic stem cell expansion (57). miR-126 is significantly lower
expressed in TCF3/PBX1-ALL and was shown to correlate with in
vitro resistance to vincristine and daunorubicin in childhood
ALL (27). This suggests that
miR-126 may have potential in prognosis prediction and therapeutic
application in ALL patients.
3. miRNAs in diagnosis and prognosis of
ALL
MicroRNAs are associated with the regulation of
normal hematopoiesis and their disruption has been related to many
types of cancer, including hematological malignancies. Pediatric
ALL samples showed lower expression levels of miR-100, miR-196b and
let-7e, while miR-128a and miR-181b were overexpressed compared to
normal pediatric samples (47).
miR-451 and miR-373 were downregulated, while an increase in
miR-222, mIR-339 and miR-142-3p was identified in pre-B-ALL cells
compared to control CD19+ (58). It is also possible to identify
between different ALL subtypes (B-ALL and T-ALL) using miRNA
signatures: miR-151 (downregulated in T-ALL), miR-148a and miR-424
(both highly expressed in T-ALL) could all be used to discriminate
between these two leukemic subtypes (59,60).
Furthermore, it is possible to distinguish between B-ALL subtypes
using miRNA expression, as exemplified by miR-629 having high
expression in MLL-AF4, miR-425-5p miR-191 and miR-128 being highly
expressed in E2A-PBX1. BCR-Abl also generates higher expression of
miR-146b and miR-126 (59).
Prognosis can be tested by miRNA expression. miR-708
was upregulated in relapse, whereas both miR-223 and miR-27a were
highly expressed in patients during remission, suggesting that
these dysregulated miRNAs play important roles in controlling
disease (61). It also was reported
that vincristine-resistant ALL patients expressed higher levels of
miR-125b (27). The miR-10a,
miR-134 and miR-214 expression was linked to a favorable outcome in
pediatric ALL, while the expression of miR-33 was associated with
an unfavorable prognosis in T-ALL compared to in normal thymocytes
(27).
4. Conclusions
The discovery of miRNAs and their association with
TEL/AML1, BCR/ABL, MLL/AF4, E2A/PBX1 oncoproteins in acute
lymphoblastic leukemia have provided valuable information on
potential diagnostic and/or prognostic biomarkers, as well as
monitoring the disease progression. Moreover, a potential role of
the microRNAs has been suggested in specific subtypes of acute
lymphoblastic leukemia and in the development of different
phenotypes. Also it is noted that the changes of miRNA expression
levels may play an important role in the genesis and evolution of
acute lymphoblastic leukemia. Thus, the mechanism of miRNA
regulation by TEL/AML1, BCR/ABL, MLL/AF4, E2A/PBX1 oncoproteins is
very complex. Studies are needed to clearly demonstrate the effect
of miRNAs in leukemogenesis and its practical implications.
Acknowledgments
J.O.N and Y.G.G. were recipients of fellowships from
the Programa de Apoyo a los Estudios de Posgrado, Universidad
Nacional Autónoma de Mexico (PAEP-UNAM).
References
|
1
|
Cartwright RA, Alexander FE, McKinney PA
and Ricketts TJ: Leukaemia and lymphoma. An atlas of distribution
within areas of England and Wales 1984–1988. Stat Med. 11:135–136.
1992.
|
|
2
|
Graubert TA and Mardis ER: Genomics of
acute myeloid leukemia. Cancer J. 17:487–491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greaves M: Infection, immune responses and
the aetiology of childhood leukaemia. Nat Rev Cancer. 6:193–203.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greaves MF: Aetiology of acute leukaemia.
Lancet. 349:344–349. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller DR and Miller LP: Acute
lymphoblastic leukemia in children: An update of clinical,
biological, and therapeutic aspects. Crit Rev Oncol Hematol.
10:131–164. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Redaelli A, Laskin BL, Stephens JM,
Botteman MF and Pashos CL: A systematic literature review of the
clinical and epidemiological burden of acute lymphoblastic
leukaemia (ALL). Eur J Cancer Care (Engl). 14:53–62. 2005.
View Article : Google Scholar
|
|
7
|
Linabery AM and Ross JA: Trends in
childhood cancer incidence in the U.S. (1992–2004). Cancer.
112:416–432. 2008. View Article : Google Scholar
|
|
8
|
Howard SC, Metzger ML, Wilimas JA,
Quintana Y, Pui CH, Robison LL and Ribeiro RC: Childhood cancer
epidemiology in low-income countries. Cancer. 112:461–472. 2008.
View Article : Google Scholar
|
|
9
|
Deschler B and Lübbert M: Acute myeloid
leukemia: Epidemiology and etiology. Cancer. 107:2099–2107. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Look AT: Oncogenic transcription factors
in the human acute leukemias. Science. 278:1059–1064. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pui C-H and Jeha S: New therapeutic
strategies for the treatment of acute lymphoblastic leukaemia. Nat
Rev Drug Discov. 6:149–165. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rowley JD: Chromosome translocations:
Dangerous liaisons revisited. Nat Rev Cancer. 1:245–250. 2001.
View Article : Google Scholar
|
|
13
|
Haferlach T, Bacher U, Kern W, Schnittger
S and Haferlach C: Diagnostic pathways in acute leukemias: A
proposal for a multimodal approach. Ann Hematol. 86:311–327. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Armstrong SA and Look AT: Molecular
genetics of acute lymphoblastic leukemia. J Clin Oncol.
23:6306–6315. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steven HS, Swerdlow EC, Harris NL, et al:
International WHO classification of tumours of haematopoietic and
lymphoid tissues. Agency for Research on Cancer; Lyon: pp. 274–288.
2008
|
|
16
|
Löwenberg B, Downing JR and Burnett A:
Acute myeloid leukemia. N Engl J Med. 341:1051–1062. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Golub TR, Slonim DK, Tamayo P, Huard C,
Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri
MA, et al: Molecular classification of cancer: Class discovery and
class prediction by gene expression monitoring. Science.
286:531–537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly
MB, Wang Y, Qian Z, Jin J, Zhang Y, et al: MicroRNA expression
signatures accurately discriminate acute lymphoblastic leukemia
from acute myeloid leukemia. Proc Natl Acad Sci USA.
104:19971–19976. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iwasaki H and Akashi K: Hematopoietic
developmental pathways: On cellular basis. Oncogene. 26:6687–6696.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Doulatov S, Notta F, Laurenti E and Dick
JE: Hematopoiesis: A human perspective. Cell Stem Cell. 10:120–136.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jamil A, Theil KS, Kahwash S, Ruymann FB
and Klopfenstein KJ: TEL/AML-1 fusion gene. its frequency and
prognostic significance in childhood acute lymphoblastic leukemia.
Cancer Genet Cytogenet. 122:73–78. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Golub TR, McLean T, Stegmaier K, Carroll
M, Tomasson M and Gilliland DG: The TEL gene and human leukemia.
Biochim Biophys Acta. 1288:M7–M10. 1996.PubMed/NCBI
|
|
25
|
Lorsbach RB and Downing JR: The role of
the AML1 transcription factor in leukemogenesis. Int J Hematol.
74:258–265. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krug U, Ganser A and Koeffler HP: Tumor
suppressor genes in normal and malignant hematopoiesis. Oncogene.
21:3475–3495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schotte D, De Menezes RX, Akbari Moqadam
F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R and Den
Boer ML: MicroRNA characterize genetic diversity and drug
resistance in pediatric acute lymphoblastic leukemia.
Haematologica. 96:703–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Diakos C, Zhong S, Xiao Y, Zhou M,
Vasconcelos GM, Krapf G, Yeh RF, Zheng S, Kang M, Wiencke JK, et
al: TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and
miRNA-320a. Blood. 116:4885–4893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gefen N, Binder V, Zaliova M, Linka Y,
Morrow M, Novosel A, Edry L, Hertzberg L, Shomron N, Williams O, et
al: Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1
(TEL/AML1) leukemias and confers survival advantage to growth
inhibitory signals independent of p53. Leukemia. 24:89–96. 2010.
View Article : Google Scholar :
|
|
30
|
Zelent A, Greaves M and Enver T: Role of
the TEL-AML1 fusion gene in the molecular pathogenesis of childhood
acute lymphoblastic leukaemia. Oncogene. 23:4275–4283. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bousquet M, Harris MH, Zhou B and Lodish
HF: MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA.
107:21558–21563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Faber J, Gregory RI and Armstrong SA:
Linking miRNA regulation to BCR-ABL expression: The next dimension.
Cancer Cell. 13:467–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scherr M, Elder A, Battmer K, Barzan D,
Bomken S, Ricke-Hoch M, Schröder A, Venturini L, Blair HJ, Vormoor
J, et al: Differential expression of miR-17~92 identifies BCL2 as a
therapeutic target in BCR-ABL-positive B-lineage acute
lymphoblastic leukemia. Leukemia. 28:554–565. 2014. View Article : Google Scholar
|
|
34
|
Nieborowska-Skorska M, Hoser G, Kossev P,
Wasik MA and Skorski T: Complementary functions of the
antiapoptotic protein A1 and serine/threonine kinase pim-1 in the
BCR/ABL-mediated leukemogenesis. Blood. 99:4531–4539. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thomas M, Lange-Grünweller K, Hartmann D,
Golde L, Schlereth J, Streng D, Aigner A, Grünweller A and Hartmann
RK: Analysis of transcriptional regulation of the human miR-17-92
cluster; evidence for involvement of Pim-1. Int J Mol Sci.
14:12273–12296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eiring AM, Neviani P, Santhanam R, Oaks
JJ, Chang JS, Notari M, Willis W, Gambacorti-Passerini C, Volinia
S, Marcucci G, et al: Identification of novel posttranscriptional
targets of the BCR/ABL oncoprotein by ribonomics: Requirement of
E2F3 for BCR/ABL leukemogenesis. Blood. 111:816–828. 2008.
View Article : Google Scholar
|
|
37
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang N, Koh GS, Lim JY, Kham SK, Ariffin
H, Chew FT and Yeoh AE: BIM is a prognostic biomarker for early
prednisolone response in pediatric acute lymphoblastic leukemia.
Exp Hematol. 39:321–329. 329.e1–329.e3. 2011. View Article : Google Scholar
|
|
39
|
Deininger MW, Vieira SA, Parada Y, Banerji
L, Lam EW, Peters G, Mahon FX, Köhler T, Goldman JM and Melo JV:
Direct relation between BCR-ABL tyrosine kinase activity and cyclin
D2 expression in lymphoblasts. Cancer Res. 61:8005–8013.
2001.PubMed/NCBI
|
|
40
|
Parada Y, Banerji L, Glassford J, Lea NC,
Collado M, Rivas C, Lewis JL, Gordon MY, Thomas NS and Lam EW:
BCR-ABL and interleukin 3 promote haematopoietic cell proliferation
and survival through modulation of cyclin D2 and p27Kip1
expression. J Biol Chem. 276:23572–23580. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu X-F, Zou J, Bao Z-J and Dong J: miR-93
suppresses proliferation and colony formation of human colon cancer
stem cells. World J Gastroenterol. 17:4711–4717. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ohtsubo M and Roberts JM: Cyclin-dependent
regulation of G1 in mammalian fibroblasts. Science. 259:1908–1912.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Johansson B, Moorman AV, Haas OA, Watmore
AE, Cheung KL, Swanton S and Secker-Walker LM: Hematologic
malignancies with t(4;11)(q21;q23) - a cytogenetic, morphologic,
immunophenotypic and clinical study of 183 cases. European 11q23
Workshop participants. Leukemia. 12:779–787. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Behm FG, Raimondi SC, Frestedt JL, Liu Q,
Crist WM, Downing JR, Rivera GK, Kersey JH and Pui CH:
Rearrangement of the MLL gene confers a poor prognosis in childhood
acute lymphoblastic leukemia, regardless of presenting age. Blood.
87:2870–2877. 1996.PubMed/NCBI
|
|
45
|
Tamai H and Inokuchi K: 11q23/MLL acute
leukemia: Update of clinical aspects. J Clin Exp Hematop. 50:91–98.
2010. View Article : Google Scholar
|
|
46
|
Dou L, Zheng D, Li J, Li Y, Gao L, Wang L
and Yu L: Methylation-mediated repression of microRNA-143 enhances
MLL-AF4 oncogene expression. Oncogene. 31:507–517. 2012. View Article : Google Scholar
|
|
47
|
de Oliveira JC, Scrideli CA, Brassesco MS,
Morales AG, Pezuk JA, Queiroz RP, Yunes JA, Brandalise SR and Tone
LG: Differential miRNA expression in childhood acute lymphoblastic
leukemia and association with clinical and biological features.
Leuk Res. 36:293–298. 2012. View Article : Google Scholar
|
|
48
|
Urtishak KA, Li-San W, Teachey DT, Sarah
TK, Barrett JS, Chen I-ML, Atlas SR, Harvey RC, Heerema NA, Carroll
AJ, et al: PI3K/AKT/mTOR signaling is a significant druggable
pathway in infant acute lymphoblastic leukemia. Blood.
122:16692013.
|
|
49
|
Noguchi S, Mori T, Hoshino Y, Maruo K,
Yamada N, Kitade Y, Naoe T and Akao Y: MicroRNA-143 functions as a
tumor suppressor in human bladder cancer T24 cells. Cancer Lett.
307:211–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Popovic R, Riesbeck LE, Velu CS, Chaubey
A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et
al: Regulation of mir-196b by MLL and its overexpression by MLL
fusions contributes to immortalization. Blood. 113:3314–3322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Z, Huang H, Chen P, He M, Li Y,
Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, et al: miR-196b
directly targets both HOXA9/MEIS1 oncogenes and FAS tumour
suppressor in MLL-rearranged leukaemia. Nat Commun. 3:6882012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Itoh N, Yonehara S, Ishii A, Yonehara M,
Mizushima S, Sameshima M, Hase A, Seto Y and Nagata S: The
polypeptide encoded by the cDNA for human cell surface antigen Fas
can mediate apoptosis. Cell. 66:233–243. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tirado CA, Shabsovich D, Yeh L, Pullarkat
ST, Yang L, Kallen M and Rao N: A (1;19) translocation involving
TCF3-PBX1 fusion within the context of a hyperdiploid karyotype in
adult B-ALL: A case report and review of the literature. Biomark
Res. 3:42015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Heim S and Mitelman F: Cancer
Cytogenetics. 3. Wiley; Online Library, Hoboken, NJ: 2009
|
|
55
|
Hajingabo LJ, Daakour S, Martin M,
Grausenburger R, Panzer-Grümayer R, Dequiedt F, Simonis N and
Twizere JC: Predicting interactome network perturbations in human
cancer: Application to gene fusions in acute lymphoblastic
leukemia. Mol Biol Cell. 25:3973–3985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Schotte D, Chau JCK, Sylvester G, Liu G,
Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R and
den Boer ML: Identification of new microRNA genes and aberrant
microRNA profiles in childhood acute lymphoblastic leukemia.
Leukemia. 23:313–322. 2009. View Article : Google Scholar
|
|
57
|
Lechman ER, Gentner B, van Galen P,
Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U,
Bachi A, Voisin V, et al: Attenuation of miR-126 activity expands
HSC in vivo without exhaustion. Cell Stem Cell. 11:799–811. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ju X, Li D, Shi Q, Hou H, Sun N and Shen
B: Differential microRNA expression in childhood B-cell precursor
acute lymphoblastic leukemia. Pediatr Hematol Oncol. 26:1–10. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fulci V, Colombo T, Chiaretti S, Messina
M, Citarella F, Tavolaro S, Guarini A, Foà R and Macino G:
Characterization of B- and T-lineage acute lymphoblastic leukemia
by integrated analysis of MicroRNA and mRNA expression profiles.
Genes Chromosomes Cancer. 48:1069–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Saki N, Abroun S, Soleimani M, Hajizamani
S, Shahjahani M, Kast RE and Mortazavi Y: Involvement of microRNA
in T-cell differentiation and malignancy. Int J Hematol Oncol Stem
Cell Res. 9:33–49. 2015.PubMed/NCBI
|
|
61
|
Han B-W, Feng D-D, Li Z-G, Luo XQ, Zhang
H, Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, et al: A set of
miRNAs that involve in the pathways of drug resistance and leukemic
stem-cell differentiation is associated with the risk of relapse
and glucocorticoid response in childhood ALL. Hum Mol Genet.
20:4903–4915. 2011. View Article : Google Scholar : PubMed/NCBI
|