Introduction
Hepatocellular carcinoma (HCC) is the sixth most
prevalent human cancer and the second common cause of
cancer-related death worldwide (1).
Although considerable advances have been made in clinical diagnosis
and management of HCC, it is still associated with a high rate of
mortality and poor prognosis (2).
TP53 is a tumor suppressor gene that plays important roles
in cellular stress response and restrains cancer initiation and
progression. TP53 mutations change TP53 protein from a tumor
suppressor into an oncogene (3).
TP53 mutations are among the most frequent genetic
alterations in human cancer, including HCC (4,5).
TP53 mutations primarily occur in the DNA binding domain,
resulting in disruption of tumor suppressor activity due to the
inability to recognize wild-type TP53 consensus sequences.
TP53 mutations also acquire new oncogenic functions through
a gain of function (3,4,6). TP53
protein has been detected immunohistochemically in cancer cells by
virtue of its high accumulation in cell nuclei and is regarded a
highly specific indicator of TP53 gene mutation (7).
Phosphorylation of proteins on serine/threonine
residues that precede proline (pSer/Thr-Pro) is a main signaling
mechanism controlling cell cycle regulation, differentiation and
proliferation. Peptidyl-prolyl isomerase PIN1 can change the
conformation of phosphoproteins and modulate the function and
stability of proteins (8).
Accumulating evidence has demonstrated that PIN1 is overexpressed
in various human cancers and plays a critical role in the
transformation of epithelial cells by activating multiple oncogenic
pathways (9,10). In contrast, several studies have
shown that decreased levels of PIN1 result in cellular
transformation, and restoration of PIN1 can attenuate the growth of
tumor cells (8,11,12).
These contradictory reports regarding the function of PIN1 in
oncogenesis suggest that PIN1 can either function as a conditional
tumor promoter or suppressor. Among the many documented targets of
PIN1-mediated prolyl-isomerization that regulate cell fate, TP53
represents the most relevant one and is frequently deregulated in
cancer (8,10,13). A
recent study has shown that PIN1 conveys oncogenic signals in
concert with mutant TP53 protein to promote aggressiveness in
breast cancer cells, and concomitant high PIN1 expression and
TP53 mutation have been proposed as an independent
prognostic factor of poor clinical outcome in breast cancer
patients (14). Based on these
observations, we hypothesized that PIN1 acts in a different manner
according to the TP53 gene mutation status in HCC.
In the present study, we examined: i) the
relationship between PIN1 and TP53 protein expression in surgical
specimens of human HCC; ii) the relationship between PIN1
expression and TP53 mutation; iii) whether PIN1 silencing by
small interfering RNA (siRNA) differently affects cell growth,
migration, and invasion in HCC cells according to TP53
mutation status and; iv) the association of PIN1 expression and
TP53 gene mutation status with clinical outcome.
Materials and methods
Materials
The present study protocol was approved by the
Institutional Review Board of Chonbuk National University Hospital.
Surgical specimens of 119 formalin-fixed, paraffin-embedded HCC
obtained from the Surgical Pathology Archives of Chonbuk National
University Hospital between 1998 and 2009 were analyzed in the
present study. Patients were 25–74 years in age (mean age, 55.9)
and consisted of 103 males and 16 females. A total of 91 cases were
positive for hepatitis B virus surface antigen, 6 were positive for
anti-hepatitis C virus antibody, 10 were alcohol-related and 12 had
an unknown etiology. Overall survival was calculated from the date
of surgery to the date of death or the final follow-up visit.
Follow-up intervals ranged from 1–194 months. To determine whether
PIN1 expression is associated with TP53 mutation, we
examined PIN1 protein levels in 5 HCC cell lines by western
blotting. The human HCC cell lines HLE, HLF and Huh-7 containing
mutant TP53 (15) were
purchased from the Health Science Research Resources Bank (Osaka,
Japan). The HepG2 cell line (wild-type TP53) was obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). We also used a sarcomatoid HCC cell line, designated as SH-J1
(wild-type TP53) (15). HCC
cell lines were cultured according to the recommendations of the
cell banks.
Immunohistochemical staining and
scoring
Immunohistochemical staining for PIN1 (Santa Cruz
Biotechnology, Santa Cruz, CA, USA) and TP53 (Novocastra,
Newcastle, UK) was performed by a polymer intense detection system
using the Bond-Max Automatic stainer (Leica Bond, Newcastle upon
Tyne, UK) as previously described (16). After deparaffinization, the tissue
sections were heated in a microwave oven in Target Retrieval
Solution (Dako, Glostrup, Denmark) for 12 min. The samples
subjected to immunostaining were rated according to a score
calculated by multiplying the area score by the intensity score of
the staining. The area of staining was scored as follows: 0
(<10% of the cancer cells), 1 (10–29%), 2 (30–59%), or 3 (≥60%).
The intensity of the cell nuclear staining was scored as 0 (none),
1 (weak), 2 (moderate), and 3 (strong). The combined score obtained
by summing the scores was used for further analysis. If the score
was ≥3, the tumor was considered positive; otherwise, the tumor was
considered negative.
TA-cloning and DNA sequencing for TP53
gene
TA-cloning and DNA sequencing of the TP53
gene were performed as previously described (15). The RNeasy Plus Micro kit (Qiagen,
Hilden, Germany) was used according to the manufacturer's protocol
for extraction of total RNA from 10 mg of frozen HCC tissue.
Reverse transcription was performed using avian myeloblastosis
virus reverse transcriptase (CosmoGenetech, Seoul, Korea) with an
oligo(dT) primer supplied by the RT PreMix kit. The primer set for
amplification of a human TP53 cds was designed according to
GenBank NM_000546, using forward primer, 5′-ATGGAGGAGCCGCAGTCAGATC
CTAGCGTCGAG-3′ and reverse primer, 5′-TCAGTCTGAGT
CAGGCCCTTTTCTGTCTTGAA-3′. PCR conditions were 95°C for 45 sec, 60°C
for 45 sec, and 72°C for 90 sec for 35 cycles using LaboPass Pfu
polymerase (CosmoGenetech). PCR products of human TP53 were
purified using a LaboPass PCR purification kit (CosmoGenetech) and
cloned into a pCR2.1 vector (Invitrogen, Carlsbad, CA, USA). We
obtained 5–18 clones for each individual sample and attempted to
sequence as many clones as possible using a BigDye Terminator Cycle
Sequencing Ready Reaction kit with an ABI PRISM 3730xl Genetic
Analyzer (both from Applied Biosystems, Foster City, CA, USA).
Small interfering RNA (siRNA)
transfection
For siRNA transfection, PIN1 siRNA and negative
control siRNA duplexes were synthesized by Bioneer Corporation
(Daejeon, Korea). The PIN1 duplex had the forward and reverse
sequences: 5′-CCAUUUGAAGACGCCUCGU-3′ and 5′-ACGAGGCGU
CUUCAAAUGG-3′, respectively; and the negative control duplex
specific had the forward and reverse sequences: 5′-CCU
ACGCCACCAAUUUCGU-3′ and 5′-ACGAAAUUGGUGGC GUAGG-3′. The specific
PIN1 siRNA or negative control siRNA was transfected using
Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's
instructions. All experiments were performed at 48 h after
transfection.
Enzymatic assay for PIN1
The level of PIN1 activity was determined using the
SensoLyte® Green PIN1 Activity Assay kit
Fluorimetric (AnaSpec, Inc., Fremont, CA, USA), according to
the manufacturer's protocols. Briefly, PIN1 substrate solution was
added to HCC cell extracts and incubated. Then, the fluorescein
signal was read using a Multi-Mode Microplate Reader System
(Perkin-Elmer, Waltham, MA, USA) at excitation and emission
wavelengths of 490 and 520 nm, respectively.
Western blotting
Western blotting was performed as previously
described (17). Briefly, the cells
were collected, and pellets were lysed with PRO-PREP protein
extraction solution (iNtRON Biotechnology, Inc., Seoul, Korea)
containing 1X phosphatase inhibitor cocktails 2 and 3 (Sigma, St.
Louis, MO, USA). The proteins were separated by 10%
SDS-polyacrylamide gel, transferred to a polyvinylidene difluoride
(PVDF) membrane (Thermo Fisher Scientific, Scotts Valley, CA, USA),
and probed with primary antibodies for PIN1 (Santa Cruz
Biotechnology), TP53 (Novocastra) and β-actin (Sigma).
Cell proliferation assay
The cell proliferation ability of PIN1 was
determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium
bromide (MTT) assay (Sigma). Post-transfection, the HepG2 or HLE
cells were re-seeded in 96-well plates at 5 or 3×103
cells/well and incubated for different times (24, 48 and 72 h). The
absorbance of all samples was measured at 560 nm.
In vitro migration and invasion
assays
A 24-Transwell migration assay (Corning Life
Sciences, Acton, MA, USA) was performed to evaluate the migration
ability of the cells. The invasion assay was performed using the
Transwell bioCoat Matrigel Invasion chamber (BD Biosciences). The
cells that migrated to or invaded the lower surface of the filter
were counted in 5 microscopic fields (magnification,
×100)/well.
Statistical analysis
To evaluate the values between the groups, Pearson's
Chi-square test and Student's t-test were used. P-values <0.05
were considered to be statistically significant. All experiments
were repeated a minimum of 3 times, and representative data are
presented. Survival analyses were performed using the Kaplan-Meier
method, and differences in survival between different groups were
determined by the log-rank test.
Results
Relationship between PIN1 and TP53
protein expression
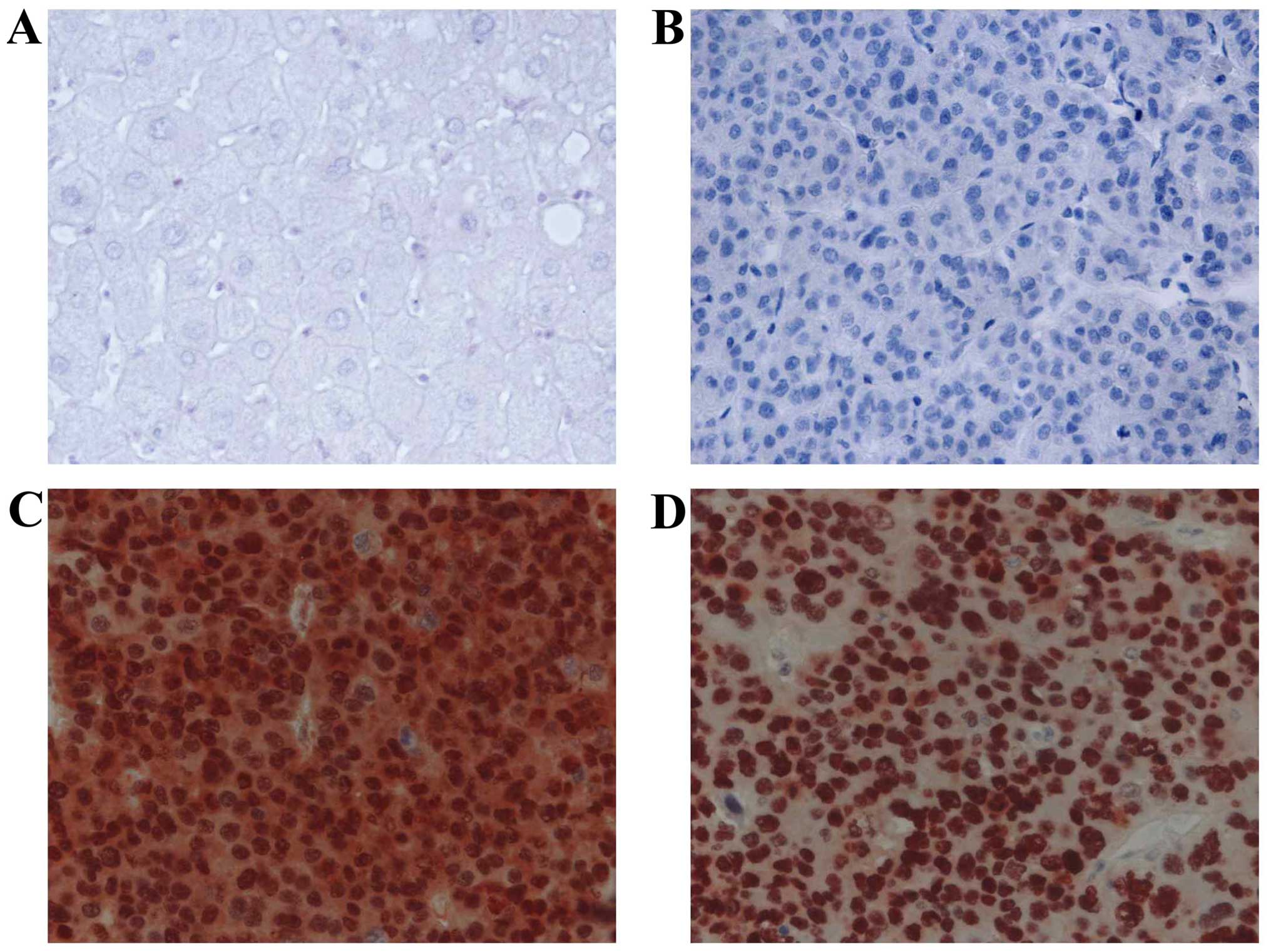
In HCC cells, PIN1 expression was predominantly
localized in the nucleus. Various tumor cells showed both nuclear
and cytoplasmic expression. The expression of TP53 was also mainly
localized in the nuclei of tumor cells. Adjacent benign hepatocytes
showed minimal or no immunoreactivity for PIN1 and TP53 (Fig. 1). Expression of PIN1 and TP53 was
observed in 77 of 119 (64.7%) and 57 of 119 (47.9%) HCC tissues,
respectively. There was a significant correlation between
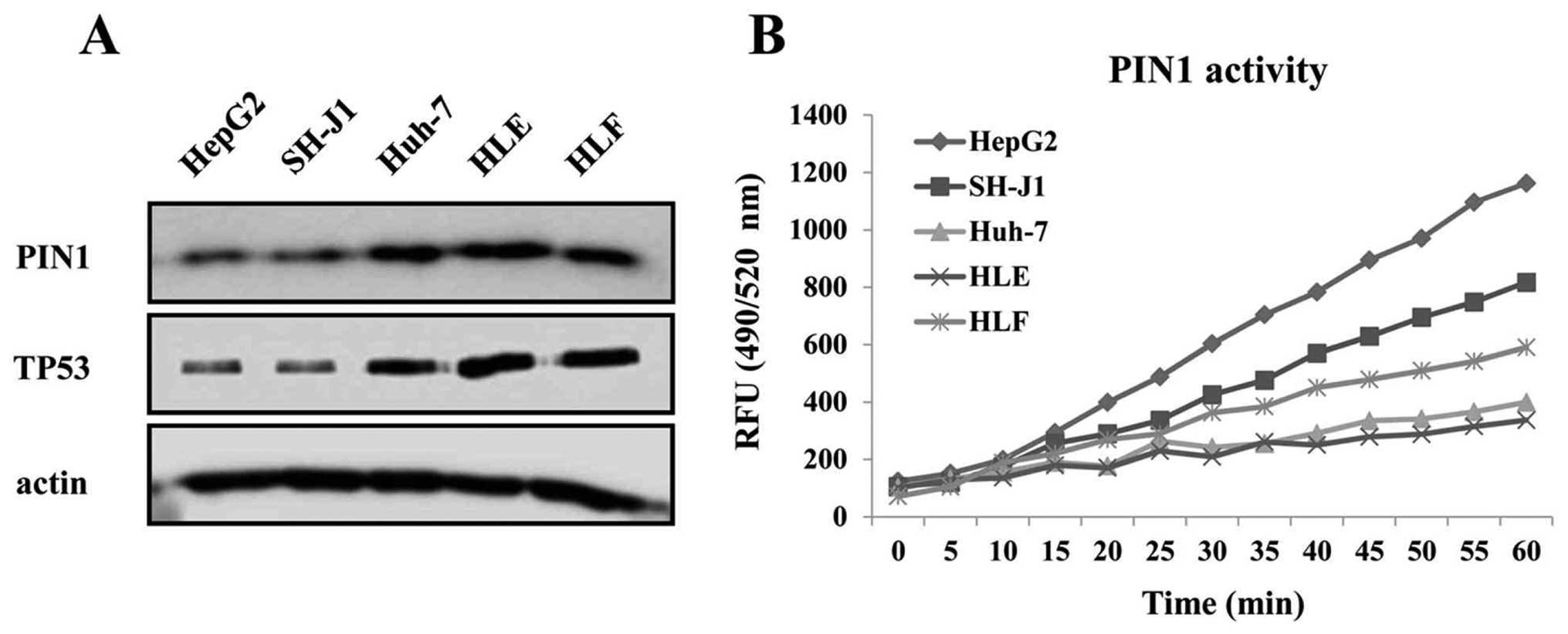
expression of PIN1 and TP53 in HCC tissues (P=0.002) (Table IA). The expression levels of PIN1
and TP53 proteins were higher in the Huh-7, HLE and HLF cell lines
containing mutant TP53 than in the HepG2 and SH-J1 cells
containing wild-type TP53 (Fig.
2A). In contrast, the enzymatic activity of PIN1 was higher in
the HepG2 and SH-J1 cells compared to that of HCC cell lines
containing mutant TP53 (Fig.
2B).
 | Table ICorrelation between PIN1 and
TP53. |
Table I
Correlation between PIN1 and
TP53.
A, Correlation
between PIN1 and TP53 protein expression
|
|---|
| | TP53 expression
| |
|---|
| Positive | Negative | P-value |
|---|
| PIN1
expression | Positive | 45 | 32 | 0.002 |
| Negative | 12 | 30 | |
B, Correlation
between PIN1 expression and TP53 mutation
|
|---|
| | TP53
mutation
| |
|---|
| Mutation | Wild-type | P-value |
|---|
| PIN1
expression | Positive | 19 | 8 | 0.03 |
| Negative | 5 | 9 | |
Relationship between PIN1 expression and
TP53 mutation
Positive immunostaining of TP53 is considered to be
indicative of mutation or overexpression in response to cellular
stress. PIN1 expression correlated with TP53 expression in our
immunohistochemical study. To verify and confirm these
immunohistochemical observations, we examined the relationship
between PIN1 expression and TP53 mutation in HCC tissues
using TP53 DNA sequencing. Forty-three frozen tissues were
selected based on matching with the paraffin-embedded specimens
using the immunohistochemical study. Of the 43 HCC frozen tissues,
26 had a mutation in the TP53 gene. The majority of
mutations were single-nucleotide substitutions (point mutations).
Only 2 HCC tissues showed an insertion mutation. The mutations in
TP53 occurred predominantly in the hot-spot region (exon
5–8) [29 of 64) (45.3%) of TP53 mutations]. Twenty-four HCCs
bearing a TP53 point mutation (missense mutation) were
included in the analysis of the relationship between PIN1
expression and TP53 mutation, since strong diffuse nuclear
staining for TP53 antibody correlates with a missense mutation
(4,6). In agreement with results of
immunohistochemistry, the expression of PIN1 was significantly
correlated with TP53 mutation in HCC tissues (P=0.03)
(Table IB). These results indicate
that there is a good correlation between immunohistochemical
expression and DNA sequencing of TP53 in determining the
relationship between PIN1 expression and TP53 mutation in
HCC tissues.
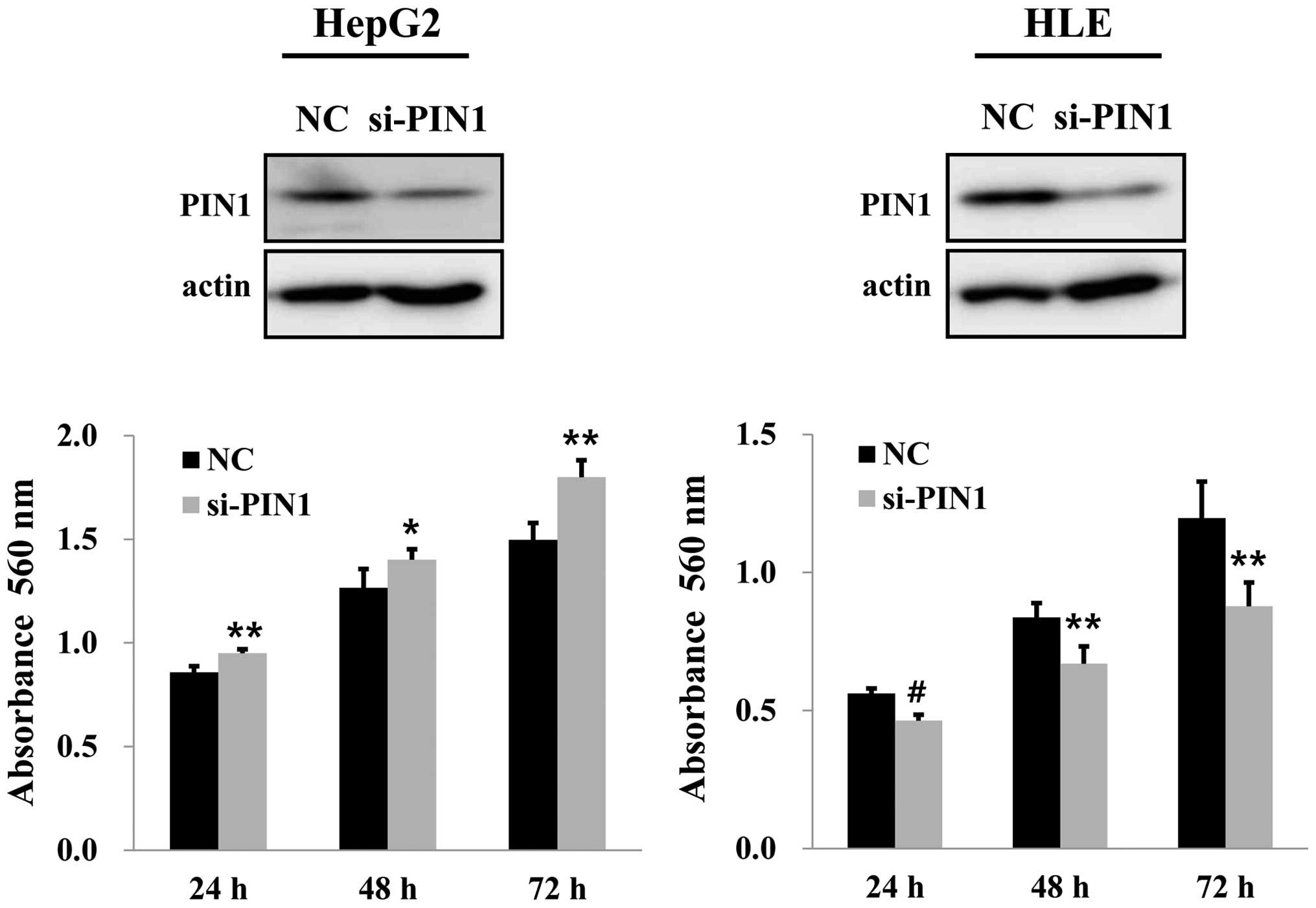
Effects of PIN1 silencing on cell
proliferation, migration and invasion
PIN1 silencing by PIN1 siRNA in HepG2 cells
containing wild-type TP53 increased the cell growth compared
to that of the control. On the contrary, silencing of PIN1 resulted
in a significant decrease in cell growth in HLE cells with mutant
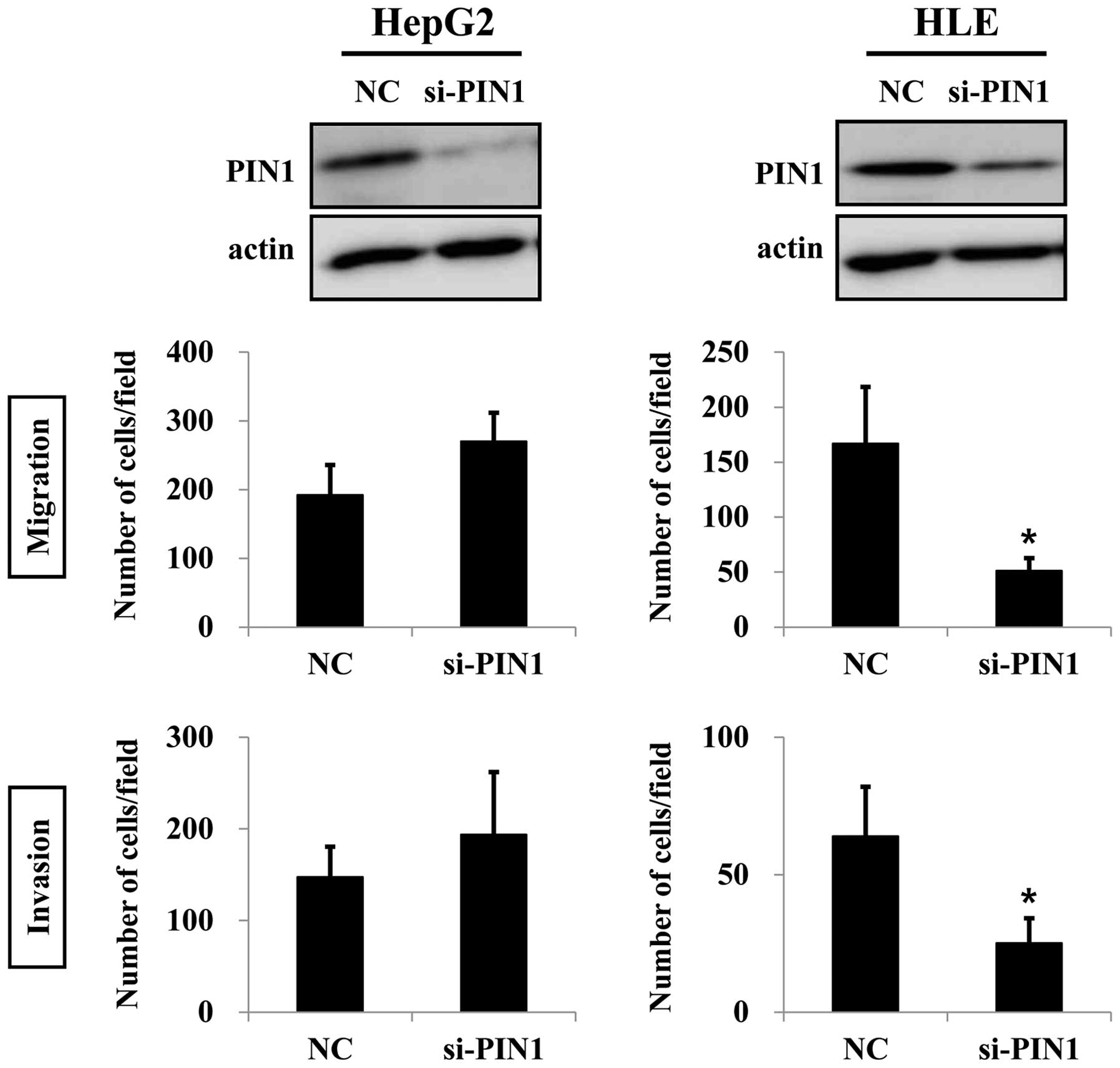
TP53 (Fig. 3). Silencing of
PIN1 in HepG2 cells caused increases in migration and invasion.
However, these results were not statistically significant. On the
contrary, silencing of PIN1 significantly suppressed migration and
invasion of HLE cells (Fig. 4).
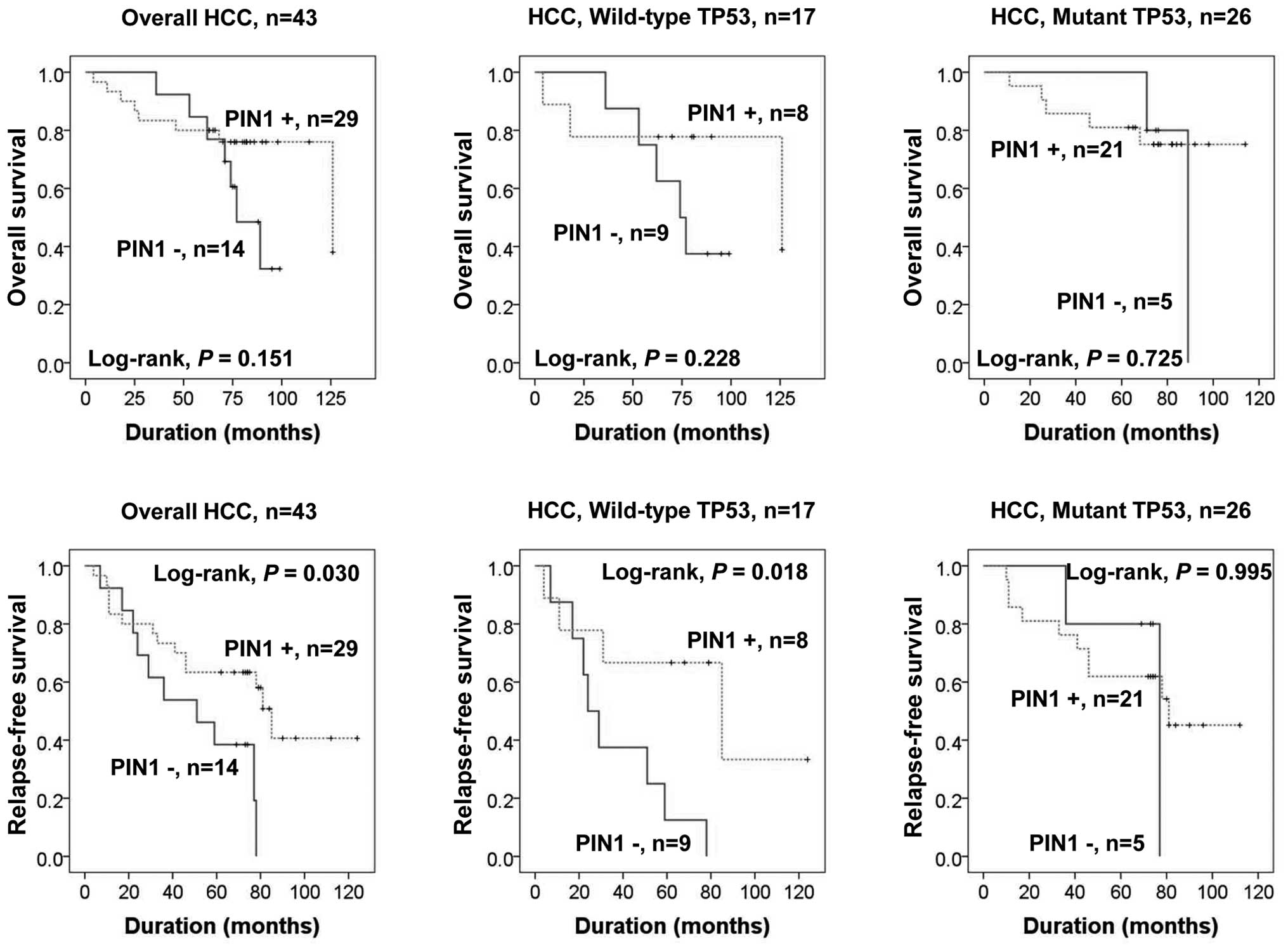
Clinical relevance of PIN1 expression and
TP53 mutation
We next examined the association of PIN1 expression
and TP53 gene mutation status with clinical outcome in 43
TP53 DNA-sequenced HCC patients. The mean overall and
relapse-free survival durations of patients were 71.2 and 65.6
months, respectively, for PIN1-positive HCC and 73.5 and 48.4
months for PIN1-negative HCC. PIN1 expression was associated with
favorable relapse-free survival (RFS) independent of TP53
mutation status (P=0.03). When we determined the RFS using a
putative combination of PIN1 expression with TP53 mutation
status, we found that RFS was significantly longer in patients with
HCCs bearing wild-type TP53 and PIN1 expression compared to
that in PIN1-negative HCC patients, according to Kaplan-Meier
survival analysis (P=0.018) (Fig.
5). However, we did not find a significant effect on overall
survival (OS) or RFS in patients with HCCs bearing mutant
TP53 according to PIN1 expression status.
Discussion
Frequent high expression of PIN1 in various human
cancers strongly implicates PIN1 in carcinogenesis of many
different cancer types, including HCC (8–10,18–21).
PIN1 is critically involved in hepatocarcinogenesis via
accumulation of β-catenin and cyclin D (19), or interaction with HBx in hepatitis
B virus-related HCC (20). PIN1
also facilitates NF-κB activation and promotes tumor progression in
HCC (21). However, it remains
controversial whether PIN1 acts as a tumor promoter (10,14,18–20) or
tumor suppressor (11,12,22).
TP53 is one of the more than 50 critical regulatory proteins
catalyzed by PIN1 (8,13). TP53 mutation is also one of
the most common genetic alterations in HCC and leads to the
accumulation of mutant TP53 protein that endows oncogenic
activities (5,7,23). It
has been proposed that the PIN1/mutant TP53 axis promotes
aggressiveness in breast cancer (14). Accordingly, the role of PIN1
expression in conjugation with TP53 mutation status in human
malignancy needs to be investigated. However, there have been no
studies on the relationship between PIN1 expression with respect to
TP53 gene status and its roles in HCC.
The present study is the first to demonstrate the
following: i) there is a significant correlation between
immunohistochemical expression of PIN1 and TP53 protein in HCC; ii)
expression of PIN1 is strongly associated with TP53 mutation
in HCC; iii) PIN1 and TP53 expression in TP53 mutant HCC
cell lines is higher compared to that in TP53 wild-type HCC
cell lines; in contrast, the enzymatic activity of PIN1 is higher
in HCC cells with wild-type TP53; iv) PIN1 silencing
effectively reduces tumor cell proliferation, but also cell
migration and invasion capacity in HLE cells containing mutant
TP53 gene. In contrast to PIN1 silencing in HLE cells, PIN1
silencing in HepG2 cells containing functional wild-type
TP53 yields different results: PIN1 silencing enhances tumor
cell proliferation, migration and invasion; and v) patients bearing
wild-type TP53 with PIN1 expression show favorable
relapse-free survival. These findings strongly suggest that a
functional interaction between PIN1 and TP53 produces different
biologic outputs in tumor cell growth, migration and invasion
depending on TP53 gene mutation status.
TP53 is a tumor suppressor that initiates cell cycle
arrest, apoptosis, and senescence in response to cellular stress
(3,4,6). The
isomerization of TP53 protein by PIN1 results in an alteration of
protein structure/or function, which is often coupled to the
stabilization of the TP53 protein (8–10). We
found a correlation between expression of PIN1 and TP53 protein or
TP53 mutation in HCC. These observations are consistent with
those of previous studies, which have reported a correlation
between high expression level of PIN1 and high levels of TP53
protein in non-small cell lung and esophageal cancer (24,25).
Since PIN1 has been shown to stabilize wild-type TP53 (8–10),
these observations suggest a possible role of PIN1 in stabilization
of mutant TP53. In the present study, transfection with PIN1 siRNA
in HepG2 cells containing wild-type TP53 caused an increase
in cell growth; these findings are consistent with the physiologic
role of PIN1, which induces cell cycle arrest and growth inhibition
through interaction with wild-type TP53. This observation suggests
that PIN1 functions as a tumor suppressor in conditions of intact
TP53 signaling. In contrast, silencing of PIN1 resulted in a
significant decrease in cell growth in HLE cells. This observation
is in agreement with results from previous studies showing that
RNAi-mediated PIN1 suppression inhibits HCC cell proliferation
using PLC/PRF/5 and Huh-7 cell lines, which have a mutant
TP53 gene (18,21). It is noteworthy, that the
underexpression of PIN1 is frequently seen in human cancers where
the TP53 mutation is rare, including kidney and skin cancer
(9,12,22).
PIN1 negatively influences the growth of human clear cell renal
cell carcinoma (ccRCC), and this suppressive ability may be
dependent on the presence of functional TP53 (12). Collectively, these observations
support the notion that PIN1 can either function as a tumor
promoter or suppressor depending on the genetic context. In
particular, interaction of PIN1 with mutant TP53 promotes its
oncogenic activities in TP53 mutant HCC. However, PIN1 may
have a tumor inhibitory role in the presence of functional
wild-type TP53.
Several studies have indicated that, in addition to
the role of PIN1 in oncogenesis, its expression is also associated
with cancer cell migration and invasion. Downregulation of PIN1
expression significantly reduces tumor cell migration and invasion
in various types of cancers occurring in the breast (14), lung (26) and prostate (27), where the TP53 mutation is
frequent. Consistent with these observations, we found that PIN1
silencing effectively suppressed the migration and invasion in HLE
cells containing mutant TP53 gene. Similarly, Girardini
et al have demonstrated that RNAi-mediated knockdown of
either mutant TP53 or PIN1 significantly attenuated cell migration
and invasion of MDA-MB-231 breast cancer cells containing mutant
TP53 (14). Notably, in
contrast to PIN1 silencing in HLE cells, PIN1 silencing in HepG2
cells containing wild-type TP53 gene yielded slightly
increased migration and invasion. These findings suggest that an
interaction between PIN1 and functional TP53 results in cellular
effects opposite to those occurring in the presence of mutant TP53.
High expression of PIN1 is correlated with poor prognosis in
several types of cancers, including HCC (21,24–27).
However, previous studies did not perform TP53 gene mutation
analysis, or details of the TP53 gene status were not
provided. The biological behavior of PIN1 according to the context
of TP53 gene in HCC remains unclear. We found that RFS was
significantly longer in patients with HCC wild-type TP53
with PIN1 expression. Similar to our result, Lill et al have
demonstrated that high expression of PIN1 is a good prognostic
factor in patients with Merkel cell carcinoma, which shows
infrequent TP53 mutation (22,28).
Girardini et al have reported that OS is significantly
decreased in patients with breast cancer expressing high levels of
PIN1 and mutant TP53 compared to that of patients with low
PIN1 expression and mutant or wild-type TP53 (14). However, PIN1 expression in
combination with TP53 mutation was not found to be a poor
prognostic factor in the present study. This discrepancy in the
clinical relevance of PIN1 may be partly explained by differences
in tumor type and an insufficient number of patients in the present
study. Additional investigations with a larger population of HCC
patients with simultaneous assessment of the TP53 gene and
other cellular partner genes of PIN1 are necessary to determine the
combination of PIN1 expression and mutant TP53 gene that
serves as a prognostic and predictive tool.
In conclusion, our findings strongly suggest that
the PIN1 can exert a conditional tumor promoter or suppressor role
depending on the TP53 gene mutation status in HCC. Our
results also support the need to evaluate the status of the
TP53 gene in the development of therapeutic approaches for
targeting PIN1 in HCC patients.
Acknowledgments
The present study was supported by the National
Research Foundation of Korea (NRF) grant funded by the Korean
Government (MSIP) (no. 2008-0062279), and fund of Biomedical
Research Institute, Chonbuk National University Hospital.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Graf D, Vallböhmer D, Knoefel WT, Kröpil
P, Antoch G, Sagir A and Häussinger D: Multimodal treatment of
hepatocellular carcinoma. Eur J Intern Med. 25:430–437. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rivlin N, Brosh R, Oren M and Rotter V:
Mutations in the p53 tumor suppressor gene: Important milestones at
the various steps of tumorigenesis. Genes Cancer. 2:466–474. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li
W, Wang F and Wu E: Alterations of TP53 are associated with a poor
outcome for patients with hepatocellular carcinoma: Evidence from a
systematic review and meta-analysis. Eur J Cancer. 48:2328–2338.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim MP, Zhang Y and Lozano G: Mutant p53:
Multiple mechanisms define biologic activity in cancer. Front
Oncol. 5:2492015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dowell SP, Wilson PO, Derias NW, Lane DP
and Hall PA: Clinical utility of the immunocytochemical detection
of p53 protein in cytological specimens. Cancer Res. 54:2914–2918.
1994.PubMed/NCBI
|
|
8
|
Yeh ES and Means AR: PIN1, the cell cycle
and cancer. Nat Rev Cancer. 7:381–388. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bao L, Kimzey A, Sauter G, Sowadski JM, Lu
KP and Wang DG: Prevalent overexpression of prolyl isomerase Pin1
in human cancers. Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Finn G and Lu KP: Phosphorylation-specific
prolyl isomerase Pin1 as a new diagnostic and therapeutic target
for cancer. Curr Cancer Drug Targets. 8:223–229. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yeh ES, Lew BO and Means AR: The loss of
PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts
to genomic instability. J Biol Chem. 281:241–251. 2006. View Article : Google Scholar
|
|
12
|
Teng BL, Hacker KE, Chen S, Means AR and
Rathmell WK: Tumor suppressive activity of prolyl isomerase Pin1 in
renal cell carcinoma. Mol Oncol. 5:465–474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mantovani F, Zannini A, Rustighi A and Del
Sal G: Interaction of p53 with prolyl isomerases: Healthy and
unhealthy relationships. Biochim Biophys Acta. 1850:2048–2060.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Girardini JE, Napoli M, Piazza S, Rustighi
A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson
A, et al: A Pin1/mutant p53 axis promotes aggressiveness in breast
cancer. Cancer Cell. 20:79–91. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choi HN, Bae JS, Jamiyandorj U, Noh SJ,
Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG and Moon WS: Expression
and role of SIRT1 in hepatocellular carcinoma. Oncol Rep.
26:503–510. 2011.PubMed/NCBI
|
|
16
|
Sung JJ, Noh SJ, Bae JS, Park HS, Jang KY,
Chung MJ and Moon WS: Immunohistochemical expression and clinical
significance of suggested stem cell markers in hepatocellular
carcinoma. J Pathol Transl Med. 50:52–57. 2016. View Article : Google Scholar :
|
|
17
|
Bae JS, Noh SJ, Kim KM, Jang KY, Chung MJ,
Kim DG and Moon WS: Serum response factor induces epithelial to
mesenchymal transition with resistance to sorafenib in
hepatocellular carcinoma. Int J Oncol. 44:129–136. 2014.
|
|
18
|
Pang RW, Lee TK, Man K, Poon RT, Fan ST,
Kwong YL and Tse E: PIN1 expression contributes to hepatic
carcinogenesis. J Pathol. 210:19–25. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pang R, Yuen J, Yuen MF, Lai CL, Lee TK,
Man K, Poon RT, Fan ST, Wong CM, Ng IO, et al: PIN1 overexpression
and beta-catenin gene mutations are distinct oncogenic events in
human hepatocellular carcinoma. Oncogene. 23:4182–4186. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pang R, Lee TK, Poon RT, Fan ST, Wong KB,
Kwong YL and Tse E: Pin1 interacts with a specific serine-proline
motif of hepatitis B virus X-protein to enhance
hepatocarcinogenesis. Gastroenterology. 132:1088–1103. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shinoda K, Kuboki S, Shimizu H, Ohtsuka M,
Kato A, Yoshitomi H, Furukawa K and Miyazaki M: Pin1 facilitates
NF-κB activation and promotes tumour progression in human
hepatocellular carcinoma. Br J Cancer. 113:1323–1331. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lill C, Schneider S, Pammer J, Loewe R,
Gedlicka W, Houben R, Heiduschka G, Brunner M and Thurnher D:
Significant correlation of peptidyl-prolyl isomerase overexpression
in Merkel cell carcinoma with overall survival of patients. Head
Neck. 33:1294–1300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He J, Zhou F, Shao K, Hang J, Wang H,
Rayburn E, Xiao ZX, Lee SW, Xue Q, Feng XL, et al: Overexpression
of Pin1 in non-small cell lung cancer (NSCLC) and its correlation
with lymph node metastases. Lung Cancer. 56:51–58. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jin H, Jiang J, Sun L, Zheng F, Wu C, Peng
L, Zhao Y and Wu X: The prolyl isomerase Pin1 is overexpressed in
human esophageal cancer. Oncol Lett. 2:1191–1196. 2011.
|
|
26
|
Tan X, Zhou F, Wan J, Hang J, Chen Z, Li
B, Zhang C, Shao K, Jiang P, Shi S, et al: Pin1 expression
contributes to lung cancer: Prognosis and carcinogenesis. Cancer
Biol Ther. 9:111–119. 2010. View Article : Google Scholar
|
|
27
|
Matsuura I, Chiang KN, Lai CY, He D, Wang
G, Ramkumar R, Uchida T, Ryo A, Lu K and Liu F: Pin1 promotes
transforming growth factor-beta-induced migration and invasion. J
Biol Chem. 285:1754–1764. 2010. View Article : Google Scholar
|
|
28
|
Lill C, Schneider S, Item CB, Loewe R,
Houben R, Halbauer D, Heiduschka G, Brunner M and Thurnher D: P53
mutation is a rare event in Merkel cell carcinoma of the head and
neck. Eur Arch Otorhinolaryngol. 268:1639–1646. 2011. View Article : Google Scholar : PubMed/NCBI
|