Introduction
MicroRNAs (miRNAs) are endogenous small RNA
molecules that regulate gene expression by binding to specific
messenger RNAs (mRNAs). miRNA genes are expressed as primary
transcripts (pri-miRNAs) in the nucleus and further processed into
stem-loop precursor miRNAs (pre-miRNAs) by the action of enzymes
Drosha RNase III endonuclease (1,2). The
miRNA precursor is then exported to the cytoplasm by means of the
nuclear export receptor, exportin-5. Once it is in the cytoplasm,
pre-miRNAs are cleaved by Dicer and TRBP, resulting in the
production of ~22-nucleotide double-chain mature miRNAs (2). The mature miRNA subsequently was
loaded onto an AGO protein to form an effecter complex called
RNA-induced silencing complex (RISC), which binds to the
3′-untranslated regions (3'UTRs) of their target mRNAs through
imperfect base pairing, and mediates either endonucleolytic
cleavage or translational repression of target mRNAs (3–6). Since
various abnormally expressed miRNAs were found to influence
tumorigenesis and progression by regulating oncogenes and
tumor-suppressor genes, researchers have tried to identify specific
miRNAs for the diagnosis and treatment of tumors (7–9).
Hepatocellular carcinoma (HCC) is one of the most
fatal cancers in Asia and Africa. Previous studies have shown that
miRNA expression profiles are significantly changed in human HCC
(10–12). Until now several miRNAs have been
discovered associated with poor prognosis and identified as
candidate biomarkers in human HCC, such as miR-26a (13), miR-30d (14), miR-122 (15,16),
and miR-17 family (17). The miR-17
family is composed of the highly conserved miR-106b-25/miR-17-92
cluster. This miRNA cluster is reported to be overexpressed in HCC
clinical samples (17). miR-20a
belongs to the miR-17-92 cluster and is located on chromosome 13.
Although the aberrant expression of miR-20a in HCC has been
observed, data on its function is inconsistent. Li et al
(17) examined the expression of
miR-17 family in 56 pairs of HCC samples and the corresponding
paired non-tumor liver using reverse transcription-real-time PCR
(qRT-PCR). They reported at least 52% of the HCC samples showed a
greater than two-fold increase in the expression for miR-20a; and
miR-17 family (including miR-20a) was necessary for cell
proliferation and anchorage-independent growth. Thus, they
considered miR-20a had oncogenic potential. However, Fan et
al (18) reported that miR-20a
was significantly decreased in HCC samples compared with normal
liver tissue. In addition, patients with lower miR-20a expression
had significantly poorer recurrence-free and overall survival.
Restoration of miR-20a suppressed hepatoma cell proliferation and
induced G1 arrest in cell cycle and apoptosis by directly targeted
myeloid cell leukemia sequence 1 (Mcl-1), so it may be a
tumor-suppressor in HCC. Thus, the expression and function of
miR-20a in HCC have not been fully elucidated, and need to be
clarified.
The information from exploratory analysis using
bioinformatics algorithms (TargetScan, Mirnaviewer and
DIANA-MicroTest) indicated that the 3'UTR of RUNX3 harbored a
potential binding site of miR-20a-5p, which suggested that
miR-20a-5p plays a role in regulating RUNX3 expression. The human
RUNX3 belongs to the RUNX gene family (the runt-related
transcription factor). Clinical and experimental data have shown
that functional inactivation of RUNX3 is frequently observed in
multiple solid tumors and highly associated with tumor progression,
lymph node metastasis and poor prognosis (19–23).
Thus, it has been regarded as a tumor-suppressor. Previous studies
have attributed the loss or reduction of RUNX3 expression to
hemizygous deletion (24–26) and epigenetic silencing (27,28).
To date, several miRNAs were identified as regulators of RUNX3 in
carcinomas, such as miR-532-5p in cutaneous melanoma (29), and miR130b (30) and miR30a (31) in gastric cancer. Nonetheless, the
mechanism underlying the regulation of RUNX3 expression through
miRNAs remains poorly explored.
The aims of the present study were: i) to clarify
the expression and function of miR-20a-5p in HCC; and ii) to
determine whether RUNX3 is a target of miR-20a-5p and evaluate the
role of this mechanism in HCC. Previously, Li et al have
reported the precursors and the mature miRNAs of the miR-106b-25
cluster were highly expressed in HCC tissues and HepG2, and Huh7
hepatoma-derived cells, while only low levels of expression were
observed in primary hepatocytes (17). In another study, Nakanishi et
al, reported that RUNX3 mRNA and protein were undetectable in
eight hepatoma-derived cell lines (HepG2, Hep3B, Huh1, Huh7, JHH1,
JHH2, JHH4 and HLE), in HLF and SK-Hep1 cells, RUNX3 mRNA and
protein were significantly under-expressed (32). Our experimental results showed that
miR-20a was highly expressed and RUNX3 was lower expressed in
SMMC-7721 hepatoma cell, thus, we used SMMC-7721 cells in the
present study to clarify the function of miR-20a and to verify the
hypothesis of RUNX3 as a target of miR-20a-5p in HCC.
Materials and methods
Cell lines
Human hepatoma SMMC-7721 and human embryonic kidney
HEK293T cells were provided by the Institute of Cell and
Biochemistry, Chinese Academy of Sciences (Shanghai, China) and
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS) (both from PAA Laboratories, Pasching, Austria) at 37°C
under a mixture of 95% air and 5% CO2.
Clinical specimens
Formalin-fixed paraffin-embedded (FFPE) tissue
blocks of tumor and corresponding adjacent non-tumorous liver
tissues from 39 cases of HCC patients were obtained from the
Department of Pathology, and 14 cases of fresh frozen clinical
specimens were collected from HCC patients who underwent
hepatectomy in the Department of Surgery of The First Affiliated
Hospital of Xi'an Jiaotong University. No patients received local
or systemic therapies before surgery and both tumor and matched
adjacent non-tumor tissue were histologically confirmed. The
present study was conducted under a protocol approved by the
Hospital Ethics Committee and informed consent was obtained from
each patient.
Reverse transcription quantitative
real-time polymerase chain reaction (qRT-PCR)
Total RNA from cells and frozen tissues was
extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and
from FFPE clinical samples with RecoverAll™ Total Nucleic Acid
Isolation kit (Ambion, Austin, TX, USA) following the
manufacturer's protocol. For detection the expression level of
miR-20a-5p and RUNX3, 500 ng RNA was reverse transcribed using
PrimeScript RT Master Mix (Takara, Otsu, Japan). The primers are
listed in Table I. Subsequently the
cDNA was amplified by SYBR Premix Ex Taq™ II (Takara). U6
snRNA and GAPDH were used as controls for miRNA and mRNA level,
respectively. Relative quantitation was calculated using the
2−ΔΔCt method. All experiments were performed in
triplicates.
 | Table I.Sequences of primers and inhibitor
used in the study. |
Table I.
Sequences of primers and inhibitor
used in the study.
| Name | Sequences |
|---|
| miR-20a-RT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTACCTG |
| miR-20a-5p-F |
ATCCAGTGCGTGTCGTG |
| miR-20a-5p-R |
TGCTTAAAGTGCTTATAGTG |
| U6-RT |
CGCTTCACGAATTTGCGTGTCAT |
| U6-F |
GCTTCGGCAGCACATATACTAAAAT |
| U6-R |
CGCTTCACGAATTTGCGTGTCAT |
|
Anti-miR-20a-5p |
5-CTACCTGCACTATAAGCACTTTA-3 |
| Scrambled |
5′-TGACTGTACTGAACTCGACTG-3′ |
Immunohistochemistry
For immunohistochemical detection of RUNX3
expression in tumor and corresponding non-tumorous tissues from HCC
patients, these tissues were cut at the thickness of 5-µm. The
sections were collected and processed for RUNX3
immunohistochemistry with the avidin-biotin peroxidase complex
(ABC) method following the manufacturer's protocol.
Construction of human miR-20a
precursor expression vector
Human miR-20a precursor (pre-miR-20) was synthesized
by chemical method by Shanghai Sangon Biological Engineering
Technology and Services Co. Ltd. (Shanghai, China). Then,
pre-miR-20 was subcloned between the EcoRI and
HindIII sites of the pcDNA6.2-GW/EmGFP vector (Invitrogen).
The sequences of constructed plasmids were confirmed by DNA
sequencing (Sangon Biotech, Shanghai, China).
Transfection of miRNA or inhibitor
into SMMC-7721 cells
Anti-miR-20a-5p inhibitor and scrambled control
fragment were synthesized by GenePharma Company (Shanghai, China).
The sequence information of anti-miR-20a-5p inhibitor is presented
in Table I. SMMC-7721 cells at
70–80% confluency, were transfected with pre-miR-20a-pcDNA6.2-GW
(0.2–2 µg) or anti-miR-20a-5p inhibitor (16–160 pmol) using
Lipofectamine 2000 (Invitrogen) method following the manufacturer's
protocol. The control vector pcDNA6.2-GW (0.2–2 µg) or scrambled
control RNA (16–160 pmol) were also transfected as negative
controls.
Cell proliferation assay by Cell
Counting Kit-8 (CCK-8) assay
SMMC-7721 cell proliferation was measured by CCK-8
assay. Briefly, cells were seeded into 96-well culture plates at a
density of 6×103 cells/well. Transfection was performed
the next day at the concentration described above. The in
vitro proliferation ability of SMMC-7721 cells were measured
over 24 and 48 h using the CCK-8 kit (Boster, Wuhan, China) assay
according to the manufacturer's instructions. Absorbance was
measured at a wavelength of 450 nm in a microplate
spectrophotometer.
Cell migration assay by wound-healing
test
SMMC-7721 cell migration ability was measured by a
wound-healing assay. Full confluent cells were seeded into 24-well
plates. A cellular area was created by scraping using a pipette
tip. Wound closure was measured at 12, 24, 36 and 48 h
intervals.
Western blot analysis
Cells were lysed in RIPA buffer (Beyotime Biotech,
Jiangsu, China) and protein concentrations were measured by the BCA
assay (Pierce Biotechnology, Inc., Rockford, IL, USA). Total
protein (10 µg) was separated on a 12% SDS-PAGE and transferred
onto polyvinylidene difluoride (PVDF). membranes. The PVDF
membranes were subsequently immune-blotted with the appropriate
primary antibodies including mouse anti-human RUNX3 monoclonal
antibody (Abcam), or mouse anti-human actin monoclonal antibody
(Santa Cruz Biotechnology, Santa Cruz, CA, USA). After extensive
washing, the membranes were incubated with a horseradish
peroxidase-conjugated goat anti-mouse secondary antibody (Pierce
Biotechnology, Inc.). Signals were detected by an ECL kit (Pierce
Biotechnology, Inc.) according to the manufacturer's
instructions.
Luciferase reporter assay
The 3'UTR of RUNX3 (NM-001031680) containing the
predicted miR-20a-5p binding site was constructed. The mutant RUNX3
3'UTR was created by mutating multiple nucleotides complementary to
the miR-20a-5p seed region. The sequences of constructed plasmids
were confirmed by DNA sequencing (Sangon Biotech).
HEK293T cells were cultured into 96-well plates with
50–70% confluency at 24 h before transfection. A mixture of 100 ng
pmiR-RB-Report™ h-RUNX3 wild-type (WT) or mutant (Mut) reporter
plasmid vector along with 50 nM pre-miR-20a-pcDNA6.2-GW or
corresponding control vectors pcDNA6.2-GW were co-transfected. The
luciferase activity was measured 48 h post-transfection using
Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA) with
Renilla (Rluc) luciferase activity as the reporter gene and
firefly luciferase (Luc) as the reference gene. Each assay was
repeated in triplicate.
Statistical analysis
Statistical evaluation was performed using SPSS
software (version 11.05; SPSS, Inc., Chicago, IL, USA). The
statistical analysis between the two groups was conducted with
Student's t-test. Correlations between miR-20a-5p
clinicopathological features were analyzed using Kaplan-Meier and
univariate Cox proportional hazard regression. Moreover, multiple
comparisons between the groups were performed using S-N-K method.
P<0.05 was considered to indicate a statistically significant
result.
Results
miR-20a-5p expression is aberrant in
HCCs
miR-20a is among the most frequently altered miRNAs
in human cancers. To investigate the expression of miR-20a in liver
cancer, we collected the information of miR-20a expression in the
large cohorts of HCC patients available from The Cancer Genome
Atlas (TCGA) database (Liver hepatocellular carcinoma: BCGSC
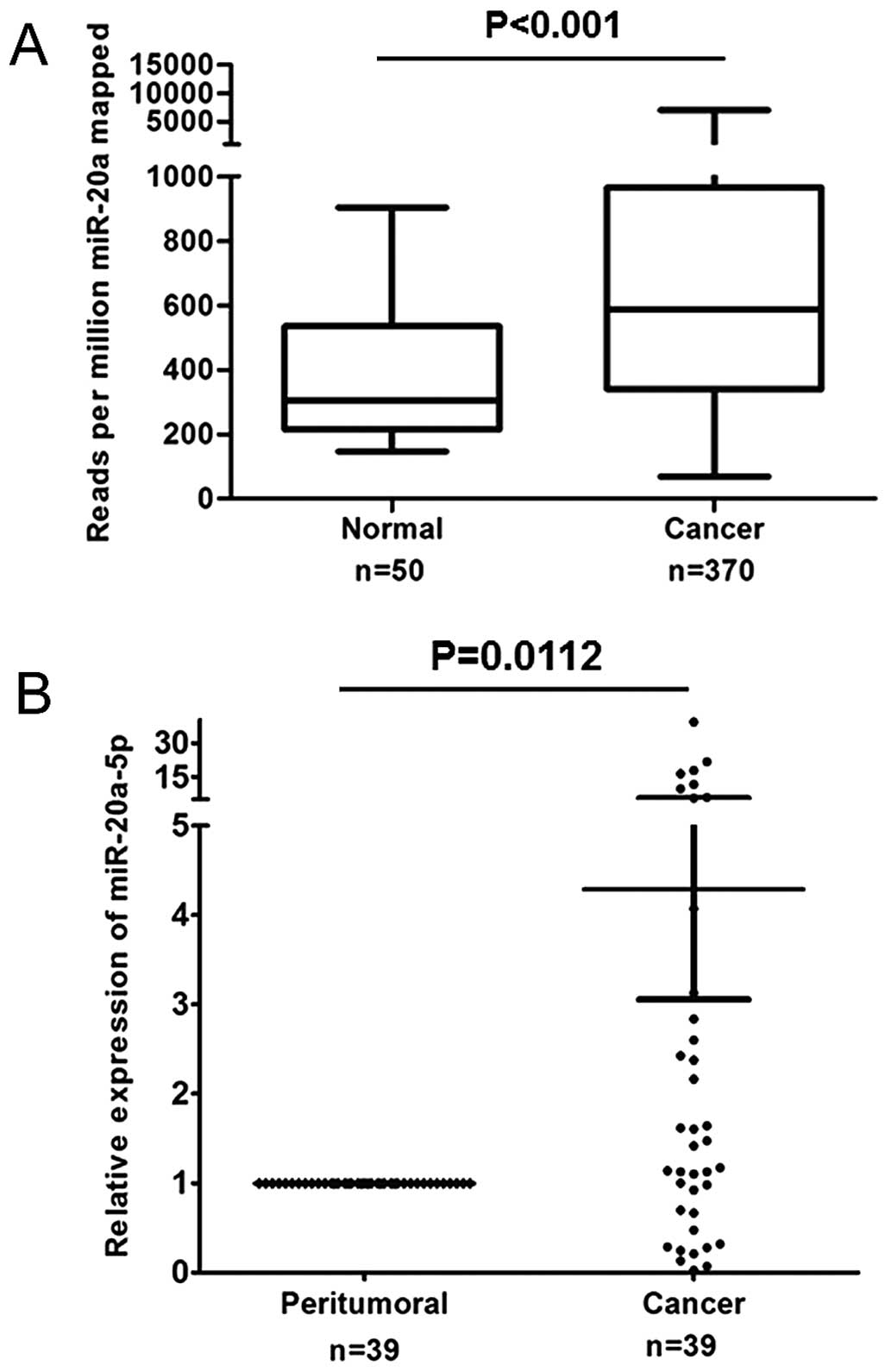
IlluminaHiSeq miRNASeq). The data are presented in Fig. 1A, miR-20a is significantly increased
in HCC tissues compared with normal liver tissues (P<0.001). To
validate the expression of miR-20a in liver cancer, we evaluated
the expression of miR-20a-5p patterns in 39 pairs of HCC and
adjacent non-tumor clinical tissue specimen by qRT-PCR. As shown in
Fig. 1B, an aberrant expression
phenomenon of miR-20a-5p was observed in HCC tissues compared with
the corresponding paired non-tumor tissues. Over 52% of the samples
showed a >1.2-fold upregulation for all members of miR-20a-5p;
28% of the samples showed a >1.2-fold downregulation; and 20% of
the samples showed no significant changes. These results showed
that miR-20a-5p expression was aberrant in human primary HCCs. The
relationships between miR-20a-5p expression and clinicopathological
features were analyzed using Kaplan-Meier and univariate Cox
proportional hazard regression; the data showed that miR-20a-5p
expression had no association with clinicopathological information
(data not shown).
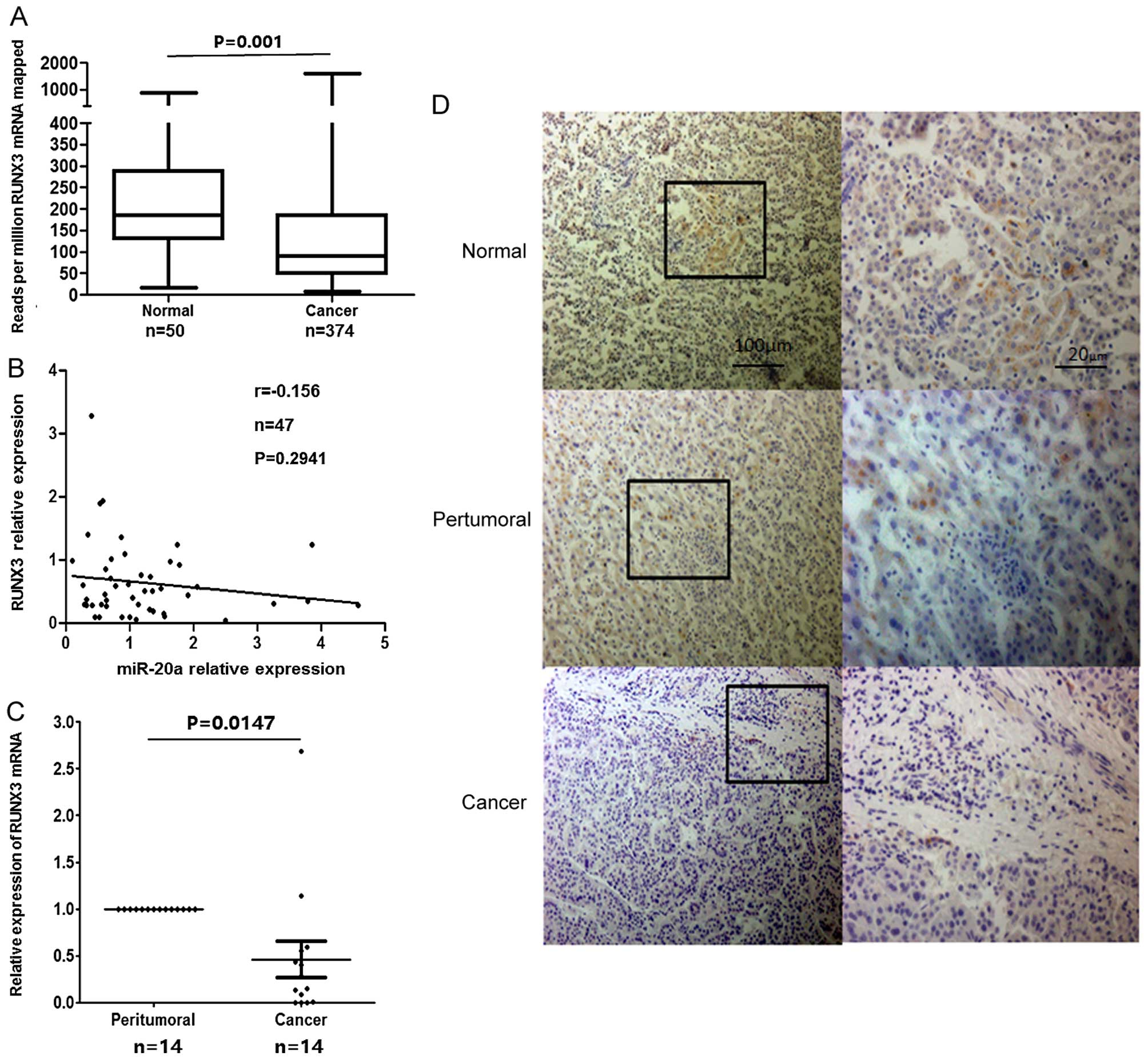
RUNX3 is downregulated in HCC
There are abundant data reported that the expression
of RUNX3 was poor in tumor tissues. We analyzed variation of RUNX3
in HCC using the data from TCGA database (Liver hepatocellular
carcinoma: UNC IlluminaHiSeq RNASeq, UNC IlluminaHiSeq RNASeqV2).
The information showed that RUNX3 is significantly decreased in HCC
tissues compared with no-corresponding normal liver tissues
(P=0.001) (Fig. 2A). Furthermore,
the RUNX3 protein was predicted as a candidate target of miR-20a-5p
using bioinformatics analysis. We assayed the correlations between
miR-20a levels and RUNX3 protein. The information was from 47 pairs
of corresponding data in TCGA datasets. The results showed that the
expression level of miR-20a and RUNX3 mRNA presented a trend of
negative correlation (Fig. 2B;
r=−0.156, P=0.2941).
Next, we examined the mRNA and protein expression of
RUNX3 in clinical specimens of HCC patients using qRT-PCR and IHC
analysis. qRT-PCR analysis from 14 paired specimens showed that
most HCC patients (12/14) exhibited significantly decreased level
of RUNX3 mRNA in hepatoma tissues compared with the paired
non-tumor tissues (Fig. 2C).
Consistently, IHC staining showed that RUNX3 was expressed in
non-tumor tissue and mainly located in the cytoplasm and nucleus,
but it was very rare in hepatoma tissues (Fig. 2D).
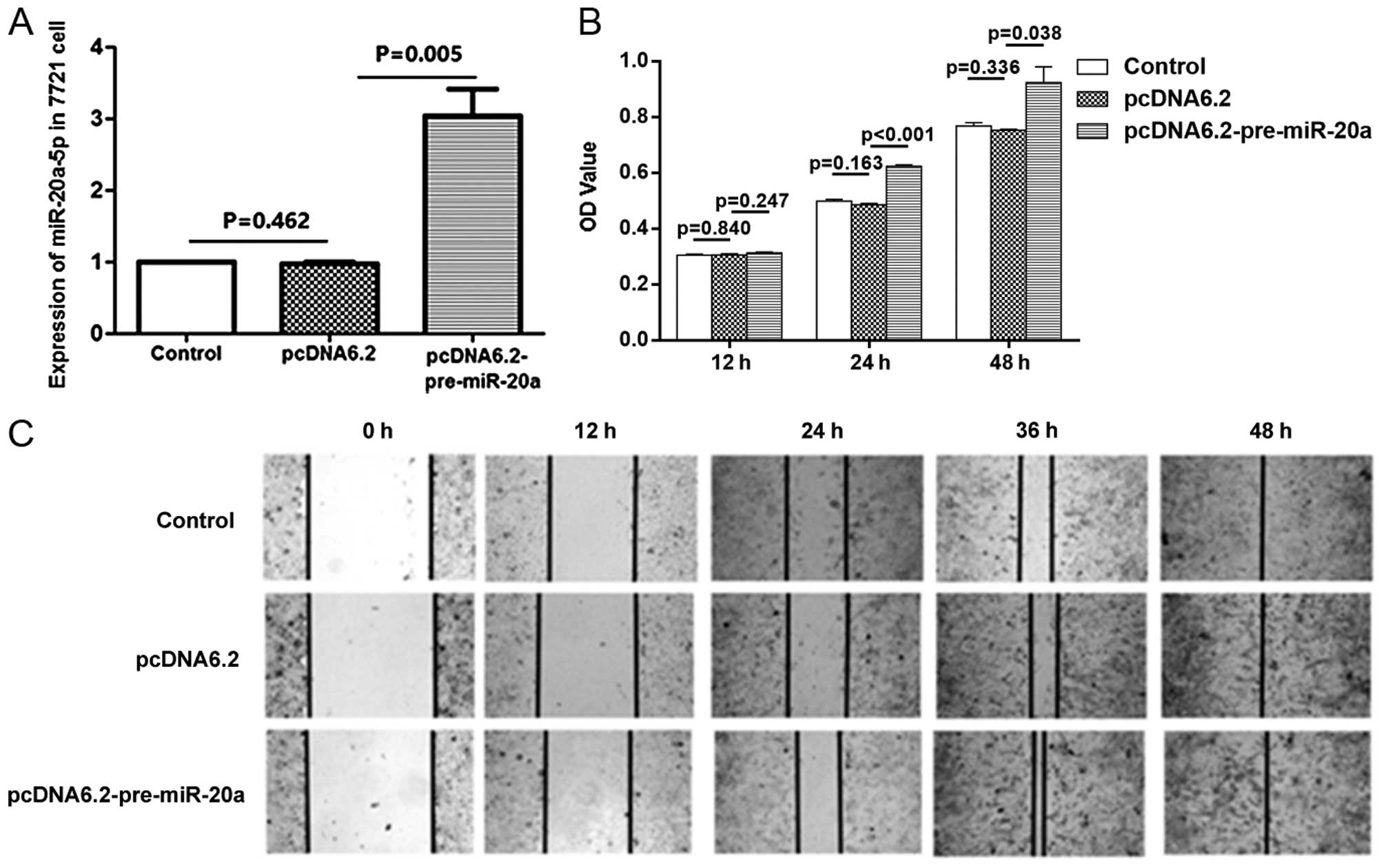
Overexpression of miR-20a promotes the
proliferation and migration of SMMC-7721 hepatoma cancer cells
To investigate the effect of miR-20a on cell
proliferation and migration, we performed both overexpression
studies using plasmid of miR-20a precursor and inhibition studies
using miR-20a-5p-specific antisense oligonucleotide inhibitor
(anti-miR-20a-5p). First, we transfected 2 µg of plasmid into
SMMC-7721 cells. The efficiency of transfection was confirmed by
qRT-PCR. At 48 h post-transfection with miR-20a precursor plasmid
(pre-miR-20a-pcDNA6.2-GW), the expression of miR-20a-5p was
increased ~3-fold (Fig. 3A). The
data from CCK-8 assay showed an increase in proliferation occurred
in cells after transfection with miR-20a precursors plasmid
(Fig. 3B). The wound healing assay
showed that miR-20a overexpression also enhanced cell migration
ability in vitro (Fig. 3C).
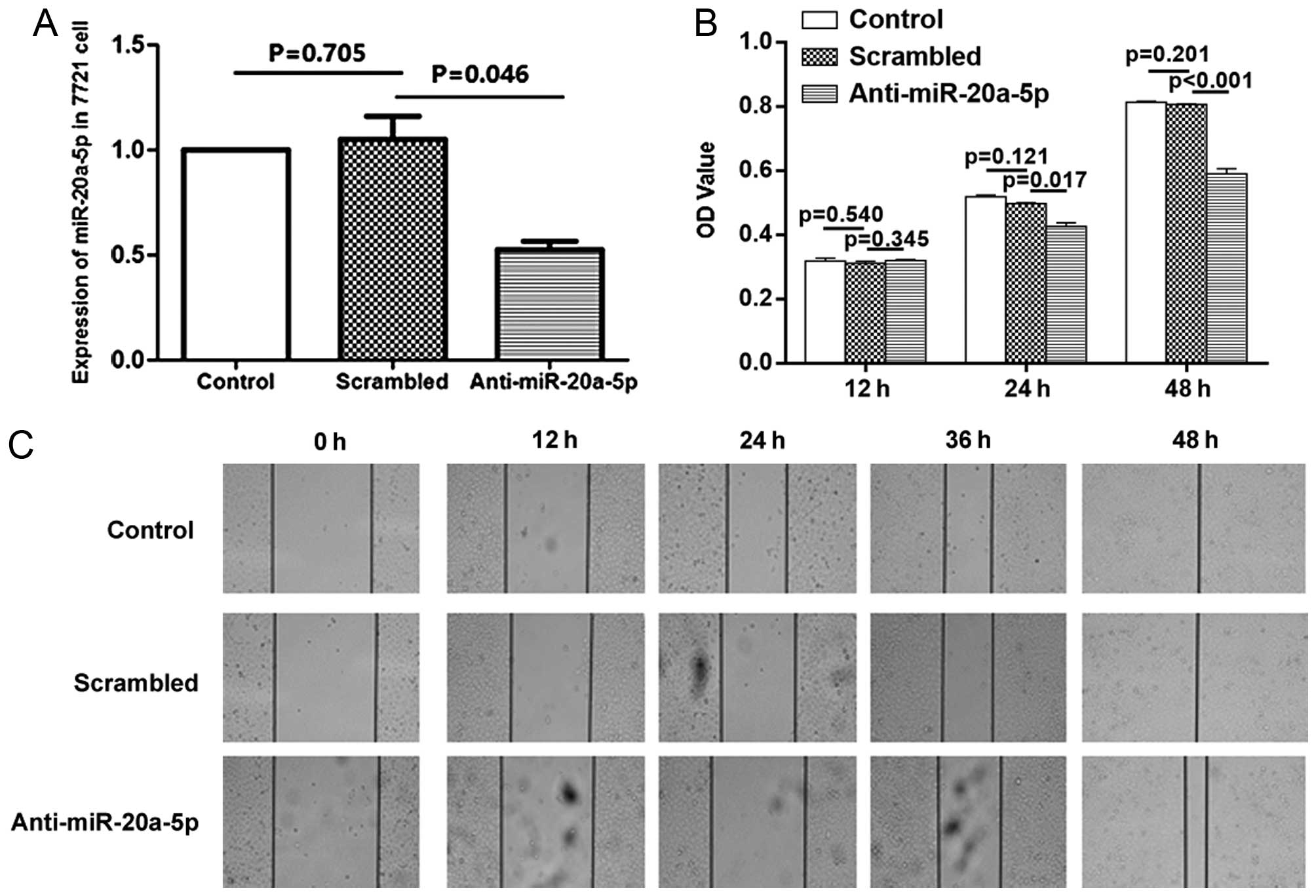
Accordingly, treatment with anti-miR20a-5p inhibitor caused the
opposite effects. Compared with the cells which were treated with
scrambled control oligonucleotides, anti-miR-20a-5p inhibitor
decreased cell proliferation and migration at all-time points
studied (Fig. 4).
When performing analysis on cell migration, and
proliferation behavior of cells affecting migration, in the present
study, we overexpressed or inhibited miR-20a in SMMC-7721 cells,
and we observed the change of proliferation and migration from 12
to 48 h. The data showed an increase in proliferation occurred when
cells were transfected with pre-miR-20a after 24 h, at the time of
12 h cell proliferation did not significantly change. At the same
time (12 h), the migration of cells was influenced significantly by
miR-20a. Therefore, we believe that cell migration detected by
wound healing assay was not due to the cell proliferation at this
time point. These data indicated that HCC cell growth and migration
could be modulated by expression of miR-20a-5p.
miR-20a-5p directly targets the RUNX3
expression
Further analysis was carried out to determine
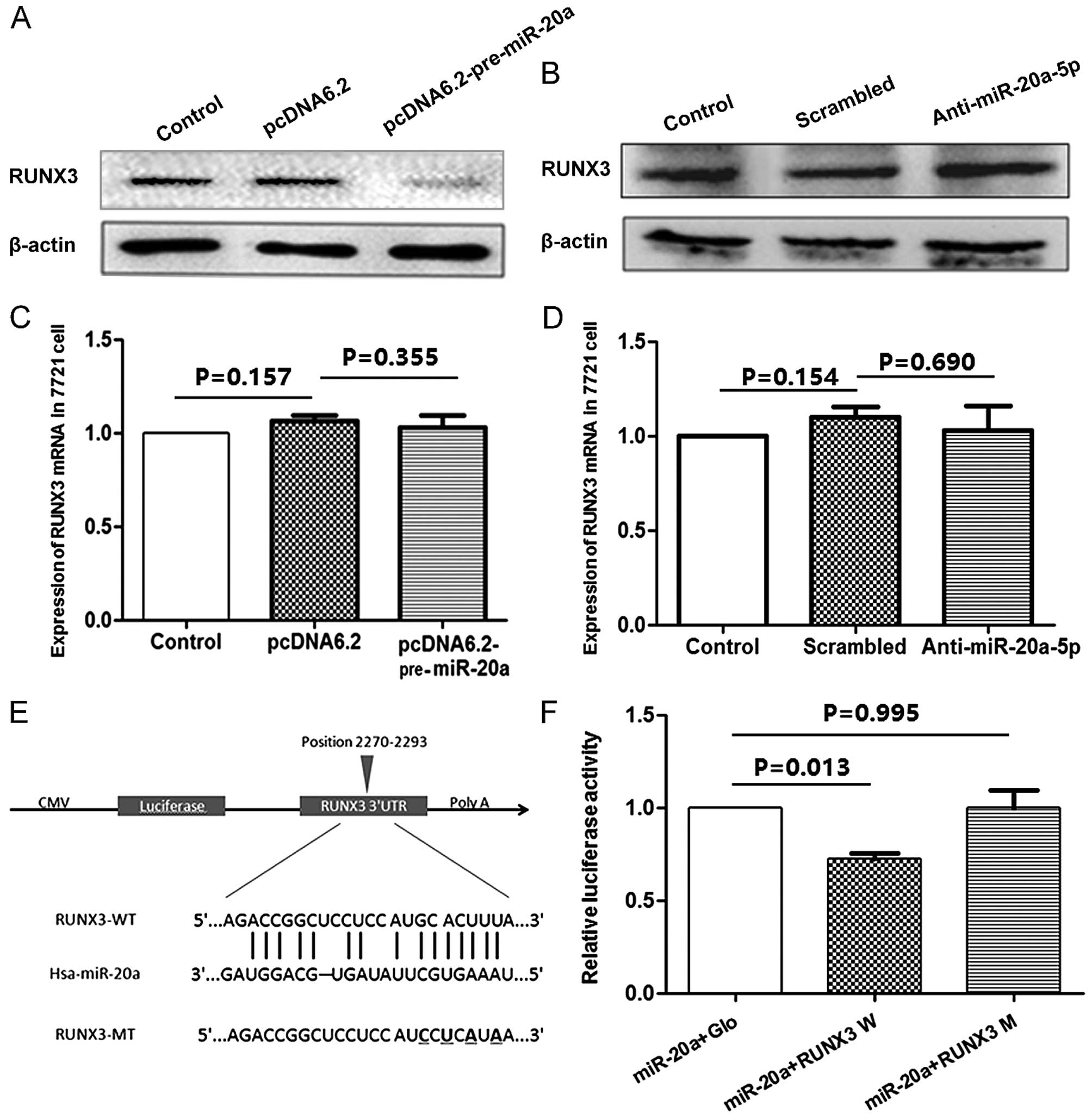
whether miR-20a-5p regulated the expression of RUNX3. The result
from western blotting showed overexpression of miR-20a led to
decreased level of RUNX3 protein (Fig.
5A). Accordingly, transfection of the anti-miR-20a-5p inhibitor
recovered the level of RUNX3 protein (Fig. 5B). However, treatment of SMMC-7721
cells with miR-20a precursor plasmid or anti-miR-20a-5p inhibitors
for 48 h did not affect the level of RUNX3 mRNA as determined by
qRT-PCR (Fig. 5C and D). The data
suggested that miR-20a-5p reduced RUNX3 expression during protein
translation.
To assess whether miR-20a-5p directly alters the
expression of RUNX3, a fragment of the 3'UTR of RUNX3 mRNA
containing the putative miR-20a-5p-binding sequence, cloned into
the pmiRGLO dual-luciferase reporter vector (RUNX3-WT), and a
mutations were generated in the RUNX3 sequences by mutating 4 nt
for the seed region (RUNX3-MT), as indicated in Fig. 5E. Then, HEK293 cells were
co-transfected with miR-20a and RUNX3-WT or RUNX3-MT 3'UTR vector.
It was observed that in the presence of miR-20a-5p, these relative
luciferase activities were significantly reduced (P=0.013), but
miR-20a-5p failed to inhibit the luciferase activity of the
RUNX3-MT reporter vector (Fig. 5F),
which indicated that miR-20a-5p modulated gene expression directly
at the 3'UTR of RUNX3. Thus, these data provide evidence that
miR-20a-5p interacts with the 3'UTR of the RUNX3 transcript and
regulating its translation.
Discussion
miR-20a is among the frequently altered microRNAs
(miRNAs) in HCC, but its expression pattern and role in HCC still
remain controversial. In the present study, we examined miR-20a
expression levels in 39 ormalin-fixed paraffin-embedded (FFPE) and
14 frozen tissues of patients with HCC. Compared with the
corresponding paired non-tumor samples, miR-20a expression was
aberrant, consistent with the observation from Li et al
(17); 52% of the tumor samples
showed a greater increase in the expression for miR-20a in tumor
tissue. Similar results have been described in other members of
miR-17 family in miRNA profiling analysis in HCC (10,11,17,33,34),
although other studies did not observe these changes (12,35),
even opposite results (18). The
difference could be due to the differences in profiling techniques
and control group used. In the studies of Fan et al and
Jiang et al, the change of miR-20a in HCC tumor tissue
compared with the no-paired normal liver specimens was evaluated.
In contrast, in the study of Li et al and ours HCC with
their corresponding paired non-tumor specimens were compared.
Previously, Li et al reported that the precursors and the
mature miRNAs of the miR-17 family were highly expressed in HCC
(17), and we also examined the
expression of the other members, and our result was conformity with
the observation from Li et al (17). Theoretically, the members should
have a similar effect, but differences cannot be ruled out.
Therefore, we may pay attention to these differences in the
follow-up study.
Furthermore, the RUNX3 protein, a tumor-suppressor,
was predicted as a candidate target of miR-20a-5p by bioinformatics
analysis. This assay along with the low expression of RUNX3 in HCC
indicates that RUNX3 may be downregulated by miR-20a-5p. Indeed, we
observed that transfection with miR-20a suppressed RUNX3 protein
expression and enhanced cell proliferation and migration in HCC
cells. Treatment with anti-miR20a-5p caused the opposite effects.
Data from luciferase reporter assay further evidenced RUNX3 was a
direct target of miR-20a-5p.
RUNX3 as a transcription factor, participates in the
aetiology of diverse cancers, and is consider to have a role in
tumor suppression since it is either not expressed or has undefined
expression in the nucleus of many cancers. Nonetheless, in some
cancers, there is evidence that the RUNX3 is expressed in the
cytoplasm (36). In the present
study, using immunohistochemistry we found that the RUNX3 protein
was slightly expressed in HCC with both cytoplasmic and nuclear
patterns. This protein mislocalization has also been found in
various types of human cancers (37,38).
Various researchers believe that, when RUNX3 is expressed in the
cytoplasm, it is thought to be in its inactive state, but this
hypothesis needs to be confirmed by experiments.
Previous studies have demonstrated that losing RUNX3
contributed to tumor progress. RUNX3 suppressed the cancer cell
growth through regulating the expression of Akt and cyclin D1
(38), and the pathways of TGF-β,
Wnt and Notch signaling (39). Loss
of RUNX3 expression was highly associated with increased lymph node
metastasis, reduced cellular differentiation and shorter survival
duration in gastric cancer patients (21). In addition, RUNX3 also slowed down
cancer metastasis through inhibition of epithelial-mesenchymal
transition (EMT) (40). Since these
studies have already demonstrated that RUNX3 is a tumor-suppressor
and inhibit cell proliferation and migration, here we just
postulate that miR-20a-5p influence cell proliferation and
migration through regulating RUNX3 expression, although we cannot
exclude the possibility that other targets may contribute mediating
the action of miR-20a-5p, as various studies have demonstrated that
miR-20a regulates the important tumor suppressor, PTEN, in HCC
(41).
Taken together, our results support the viewpoint
that miR-20-5p has an oncogenic property in HCC; it enhances cancer
cell proliferation and migration through reducing the translation
of RUNX3.
Acknowledgements
The present study was supported by Grants from the
National Natural Science Foundation of China (no. 81101744), the
Fundamental Research Funds for the Central Universities (no.
xjj2014144), and the Funds for Technical Resources sharing Platform
of Shaanxi Province, China (No. 2015FWPT-14).
References
|
1
|
Lee Y, Jeon K, Lee JT, Kim S and Kim VN:
MicroRNA maturation: Stepwise processing and subcellular
localization. EMBO J. 21:4663–4670. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel B and Bartel DP: MicroRNAs: At the
root of plant development? Plant Physiol. 132:709–717. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khvorova A, Reynolds A and Jayasena SD:
Functional siRNAs and miRNAs exhibit strand bias. Cell.
115:209–216. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Doench JG, Petersen CP and Sharp PA:
siRNAs can function as miRNAs. Genes Dev. 17:438–442. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang YS, Dai Y, Yu XF, Bao SY, Yin YB,
Tang M and Hu CX: Microarray analysis of microRNA expression in
hepatocellular carcinoma and non-tumorous tissues without viral
hepatitis. J Gastroenterol Hepatol. 23:87–94. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, Roberts LR and Schmittgen TD: Association
of MicroRNA expression in hepatocellular carcinomas with hepatitis
infection, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang X, Liang L, Zhang XF, Jia HL, Qin Y,
Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, et al: MicroRNA-26a
suppresses tumor growth and metastasis of human hepatocellular
carcinoma by targeting IL-6-Stat3 pathway. Hepatology. 58:158–170.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao J, Liang L, Huang S, Ding J, Tan N,
Zhao Y, Yan M, Ge C, Zhang Z, Chen T, et al: MicroRNA-30d promotes
tumor invasion and metastasis by targeting Galphai2 in
hepatocellular carcinoma. Hepatology. 51:846–856. 2010.PubMed/NCBI
|
|
15
|
Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW,
Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al: MicroRNA-122, a
tumor suppressor microRNA that regulates intrahepatic metastasis of
hepatocellular carcinoma. Hepatology. 49:1571–1582. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bai S, Nasser MW, Wang B, Hsu SH, Datta J,
Kutay H, Yadav A, Nuovo G, Kumar P and Ghoshal K: MicroRNA-122
inhibits tumorigenic properties of hepatocellular carcinoma cells
and sensitizes these cells to sorafenib. J Biol Chem.
284:32015–32027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Tan W, Neo TW, Aung MO, Wasser S,
Lim SG and Tan TM: Role of the miR-106b-25 microRNA cluster in
hepatocellular carcinoma. Cancer Sci. 100:1234–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX
and Zhong L: Decrease expression of microRNA-20a promotes cancer
cell proliferation and predicts poor survival of hepatocellular
carcinoma. J Exp Clin Canc Res. 32:212013. View Article : Google Scholar
|
|
19
|
Fukamachi H and Ito K: Growth regulation
of gastric epithelial cells by Runx3. Oncogene. 23:4330–4335. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng Z, Wei D, Wang L, Tang H, Zhang J, Le
X, Jia Z, Li Q and Xie K: RUNX3 inhibits the expression of vascular
endothelial growth factor and reduces the angiogenesis, growth, and
metastasis of human gastric cancer. Clin Cancer Res. 12:6386–6394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu PI, Hsieh HL, Lee J, Lin LF, Chen HC,
Lu PJ and Hsiao M: Loss of RUNX3 expression correlates with
differentiation, nodal metastasis, and poor prognosis of gastric
cancer. Ann Surg Oncol. 16:1686–1694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chuang LS and Ito Y: RUNX3 is
multifunctional in carcinogenesis of multiple solid tumors.
Oncogene. 29:2605–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramaniam MM, Chan JY, Soong R, Ito K,
Yeoh KG, Wong R, Guenther T, Will O, Chen CL, Kumarasinghe MP, et
al: RUNX3 inactivation in colorectal polyps arising through
different pathways of colonic carcinogenesis. Am J Gastroenterol.
104:426–436. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li QL, Ito K, Sakakura C, Fukamachi H,
Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wada M, Yazumi S, Takaishi S, Hasegawa K,
Sawada M, Tanaka H, Ida H, Sakakura C, Ito K, Ito Y, et al:
Frequent loss of RUNX3 gene expression in human bile duct and
pancreatic cancer cell lines. Oncogene. 23:2401–2407. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yanada M, Yaoi T, Shimada J, Sakakura C,
Nishimura M, Ito K, Terauchi K, Nishiyama K, Itoh K and Fushiki S:
Frequent hemizygous deletion at 1p36 and hypermethylation
downregulate RUNX3 expression in human lung cancer cell lines.
Oncol Rep. 14:817–822. 2005.PubMed/NCBI
|
|
27
|
Nishio M, Sakakura C, Nagata T, Komiyama
S, Miyashita A, Hamada T, Kuryu Y, Ikoma H, Kubota T, Kimura A, et
al: RUNX3 promoter methylation in colorectal cancer: Its
relationship with microsatellite instability and its suitability as
a novel serum tumor marker. Anticancer Res. 30:2673–2682.
2010.PubMed/NCBI
|
|
28
|
Zheng Y, Zhang Y, Huang X and Chen L:
Analysis of the RUNX3 gene methylation in serum DNA from esophagus
squamous cell carcinoma, gastric and colorectal adenocarcinoma
patients. Hepatogastroenterology. 58:2007–2011. 2011.PubMed/NCBI
|
|
29
|
Kitago M, Martinez SR, Nakamura T, Sim MS
and Hoon DS: Regulation of RUNX3 tumor suppressor gene expression
in cutaneous melanoma. Clin Cancer Res. 15:2988–2994. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et
al: Singapore Gastric Cancer Consortium: MicroRNA-130b regulates
the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer.
46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Z, Chen L, Zhang X, Xu X, Xing H,
Zhang Y, Li W, Yu H, Zeng J and Jia J: RUNX3 regulates vimentin
expression via miR-30a during epithelial-mesenchymal transition in
gastric cancer cells. J Cell Mol Med. 18:610–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakanishi Y, Shiraha H, Nishina S, Tanaka
S, Matsubara M, Horiguchi S, Iwamuro M, Takaoka N, Uemura M, Kuwaki
K, et al: Loss of runt-related transcription factor 3 expression
leads hepatocellular carcinoma cells to escape apoptosis. BMC
Cancer. 11:32011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Connolly E, Melegari M, Landgraf P,
Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M,
Tuschl T and Rogler CE: Elevated expression of the miR-17-92
polycistron and miR-21 in hepadnavirus-associated hepatocellular
carcinoma contributes to the malignant phenotype. Am J Pathol.
173:856–864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS,
Kang SG, Kim CH, Lee YJ, Chun JS and Cho MK: Expression of RUNX3 in
skin cancers. Clin Exp Dermatol. 36:769–774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee CWL, Chuang LSH, Kimura S, Lai SK, Ong
CW, Yan B, Salto-Tellez M, Choolani M and Ito Y: RUNX3 functions as
an oncogene in ovarian cancer. Gynecol Oncol. 122:410–417. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin FC, Liu YP, Lai CH, Shan YS, Cheng HC,
Hsu PI, Lee CH, Lee YC, Wang HY, Wang CH, et al: RUNX3-mediated
transcriptional inhibition of Akt suppresses tumorigenesis of human
gastric cancer cells. Oncogene. 31:4302–4316. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Voon DC, Wang H, Koo JK, Nguyen TA, Hor
YT, Chu YS, Ito K, Fukamachi H, Chan SL, Thiery JP, et al: Runx3
protects gastric epithelial cells against epithelial-mesenchymal
transition-induced cellular plasticity and tumorigenicity. Stem
Cells. 30:2088–2099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tanaka S, Shiraha H, Nakanishi Y, Nishina
S, Matsubara M, Horiguchi S, Takaoka N, Iwamuro M, Kataoka J,
Kuwaki K, et al: Runt-related transcription factor 3 reverses
epithelial-mesenchymal transition in hepatocellular carcinoma. Int
J Cancer. 131:2537–2546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Zheng L, Ding Y, Li Q, Wang R,
Liu T, Sun Q, Yang H, Peng S, Wang W, et al: MiR-20a induces cell
radioresistance by activating the PTEN/PI3K/Akt signaling pathway
in hepatocellular carcinoma. Int J Radiat Oncol Biol Phys.
92:1132–1140. 2015. View Article : Google Scholar : PubMed/NCBI
|