Introduction
Pancreatic cancer (PC), a highly malignant digestive
system tumor, is the fourth major cause of cancer-related deaths
worldwide (1). Pancreatic ductal
adenocarcinoma (PDAC) is the most aggressive PC, and accounts for
>80% of PC cases. Despite continuous progress in diagnosis and
treatment in recent decades, PDAC remains a great clinical
challenge due to its dismal prognosis (2–7).
Currently, the key obstacle to progress is the lack of accurate and
specific targets for the early diagnosis of PDAC (8–10).
Therefore, the identification of novel biomarkers and development
of new therapeutic approaches are of great value for PDAC.
The oncoprotein DEK was initially discovered as a
fusion protein with CAN/NUP214 nucleoporin due to the (6;9)
(p23;q64) translocation in a subset of acute myeloid leukemia (AML)
(11,12). Now, it is emerging as a class of DNA
topology modulators encoded by a gene located at chromosome 6p22.3
(13). The functions of DEK involve
DNA supercoiling, mRNA splicing, DNA damage repair, transcriptional
control and cell viability in cell progression and metabolism
(14–17). As an architectural chromatin
protein, DEK has been detected in numerous human malignancies
including glioblastoma (18), AML
(19), bladder cancer (20) and hepatocellular carcinoma (21). Khodadoust et al showed that
the level of DEK expression can distinguish benign nevi from
malignant melanomas, indicating that this protein may be highly
useful for differentiating diagnoses (12).
Our previous study found that DEK was significantly
expressed in patients with colorectal cancer, and this
overexpression was associated with poor prognostic factors
(22). We also revealed that the
level of DEK expression was significantly increased in various
solid tumors, such as breast and gastric cancer using
immunohistochemical (IHC) staining (23,24).
However, to date, the detailed role of DEK overexpression in PDAC
remains unclear. Therefore, we identified the clinical features
correlated with DEK overexpression and the potential prognostic
value of DEK in PDAC. The results revealed a significant increase
in DEK expression in PDAC tissues compared to levels in the normal
pancreas tissues. These findings suggest that DEK overexpression
may be an independent reliable biomarker for poor prognosis in
patients with PDAC.
Materials and methods
Ethics statement
The present study complied with the Helsinki
Declaration and was approved by the Human Ethics Committee and the
Research Ethics Committee of Yanbian University Medical College.
Patients were informed that the resected specimens were stored by
the hospital and potentially used for scientific research, and that
their privacy may be maintained. Follow-up survival data were
retrospectively collected through medical-record analyses.
Clinical samples
A total of 139 samples of pancreas tissues,
including 87 PDAC and 52 adjacent normal pancreas tissues, were
collected from the Tumor Tissue Bank of Yanbian University Medical
College. All tissues were routinely fixed in 10% buffered formalin
and embedded in paraffin blocks. The institutional Review Board of
Yanbian University Medical College approved the study protocol. The
pathological parameters, including gender, age, tumor location,
tumor size, grading, clinical tumor-node-metastasis (TNM) stage,
perineural invasion status, lymph node metastasis and survival
data, were carefully reviewed for all 87 PDACs. The male to female
ratio was 48:39, and 52 cases were <50 years and 35 cases were
≥50 years (median age of 59 years). Tumors were located in the head
of the pancreas in 59 cases, and in the body and tail of the
pancreas in 28 cases. Of the 87 PDACs, 48 cases had tumor size
<3 cm and 39 cases had tumor size ≥3 cm (mean size of 3.36 cm).
In regards to the grading of PDAC, 25 cases were grade 1, 34 cases
were grade 2, and 28 cases were grade 3. Concerning the clinical
TNM stage, 53 cases were stage I–II and 34 cases were TNM stage
III–IV. Clinicopathological classification and staging were
assessed according to the staging system established by the
American Joint Committee on Cancer (AJCC). In addition, 42 cases
had perineural invasion, and 45 cases had no perineural invasion;
42 cases had lymph node (LN) metastasis, and 45 cases had no LN
metastasis. The normal pancreases were obtained from the resection
margins of radical specimens of PDAC.
A total of 87 patients with PDAC had received
surgical treatment, but not adjuvant chemotherapy at the time of
data collection. The survival information of the patients was
successfully collected during 30 months or until death.
Immunofluorescence (IF) staining
analysis
Human PC cell line PANC-1 was obtained from the Cell
Bank of the Chinese Academy of Medical Science (Shanghai, China).
The cells were grown and cultured in Dulbeccos modified Eagles
medium (DMEM) (Gibco, Gaithersburg, MD, USA) supplemented with 10%
fetal bovine serum and 1% penicillin/streptomycin in humidified 5%
CO2 at 37°C.
PANC-1 cells were grown on coverslips to 70–80%
confluency, and fixed with 4% paraformaldehyde for 10 min and
permeabilized with 0.5% Triton X-100 for 10 min at room
temperature. Subsequently, after blocking with 3% albumin bovine V
(A8020; Solarbio, Beijing, China) for 1 h, the cells were gently
washed with phosphate-buffered saline (PBS). A primary antibody
against DEK (1:50; 610948; BD Biosciences, Franklin Lanes, NJ, USA)
was incubated with the cells at 4°C overnight, followed by
incubation with Alexa Fluor® 568 goat anti-mouse IgG (H
+ L) (1:1,000; A11004; Invitrogen, Carlsbad, CA, USA) for 1 h.
Then, the cells were washed with PBS and counterstained with
4′,6-diamidino-2-phenylindole (DAPI) (C1006; Beyotime, Beijing,
China). The coverslips were mounted with Antifade Mounting Medium
(P0126; Beyotime). Finally, IF signals were visualized and recorded
using a Leica SP5 II confocal microscope.
Immunoenzyme staining analysis
Immunoenzyme staining was performed using the
standard streptavidin-peroxidase (SP) method. Briefly, all tissue
sections were deparaffinized, rehydrated and incubated with 3%
H2O2 in methanol for 15 min at room
temperature. Subsequently, the antigen was retrieved in 0.01 M
sodium citrate buffer (pH 6.0). The slides were incubated with a
primary antibody against DEK (1:50; 610948; BD Biosciences) at 4°C
overnight. After incubation with biotinylated secondary antibody at
room temperature for 30 min, the slides were covered with SP
complex at room temperature for 30 min. Immunostaining was
developed using 3,3′-diaminobenzidine and counterstaining with
Mayer's hematoxylin. Mouse IgG isotope was used as the control and
the result was negative. Furthermore, the positive tissue sections
were processed as negative controls by omitting the primary
antibody.
Two pathologists (Y. Yang and F. Bi) independently
evaluated all tissue specimens without knowledge of the clinical
data. In case of discrepancies, a final score was established by
reassessment on a double-headed microscope. The scoring system for
the interpretation criteria was previously described (22). Briefly, staining intensity of the
tissue sections was scored as ‘−’ for no staining, ‘+’ was defined
as weak staining, and ‘++’ was considered as intense staining,
respectively. The staining area was scored as follows: ‘−’
(negative, no or <5% positive cells), ‘+’ (5–50% positive
cells), ‘++’ (>50% positive cells). For the double scoring
system together, ‘++’ scored samples were considered as DEK
overexpression, and ‘−’ or ‘+’ scored samples were considered as
DEK low-expression.
Statistical analysis
Statistical analyses were conducted using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). DEK mRNA expression data
were obtained from GEO database. Correlations between DEK protein
expression and clinicopathological features were evaluated by
Chi-squared (χ2) and Fisher's exact tests. The survival
curves were performed using the Kaplan-Meier method, and
significant differences were assessed by log-rank tests.
Multivariate survival analysis was performed on all significant
characteristics measured by univariate survival analysis with the
Cox proportional hazard regression model. A P-value <0.05 was
considered to indicate a statistically significant result.
Results
DEK expression in PDAC
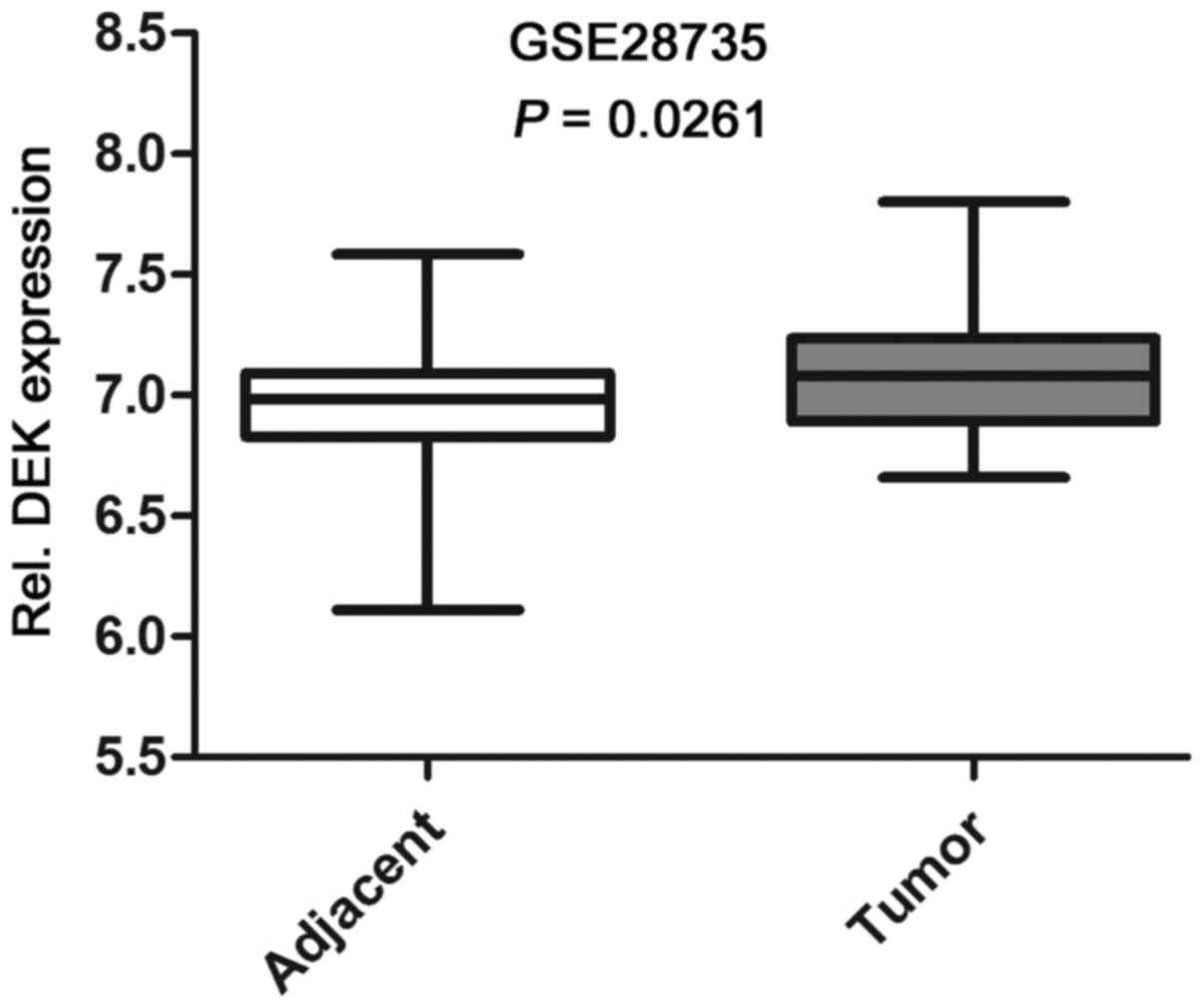
Based on the data from Gene Expression Omnibus
(GEO), we found that the expression level of DEK mRNA in PDAC
tissues was significantly higher than that in the adjacent
non-tumor tissues (Fig. 1). To
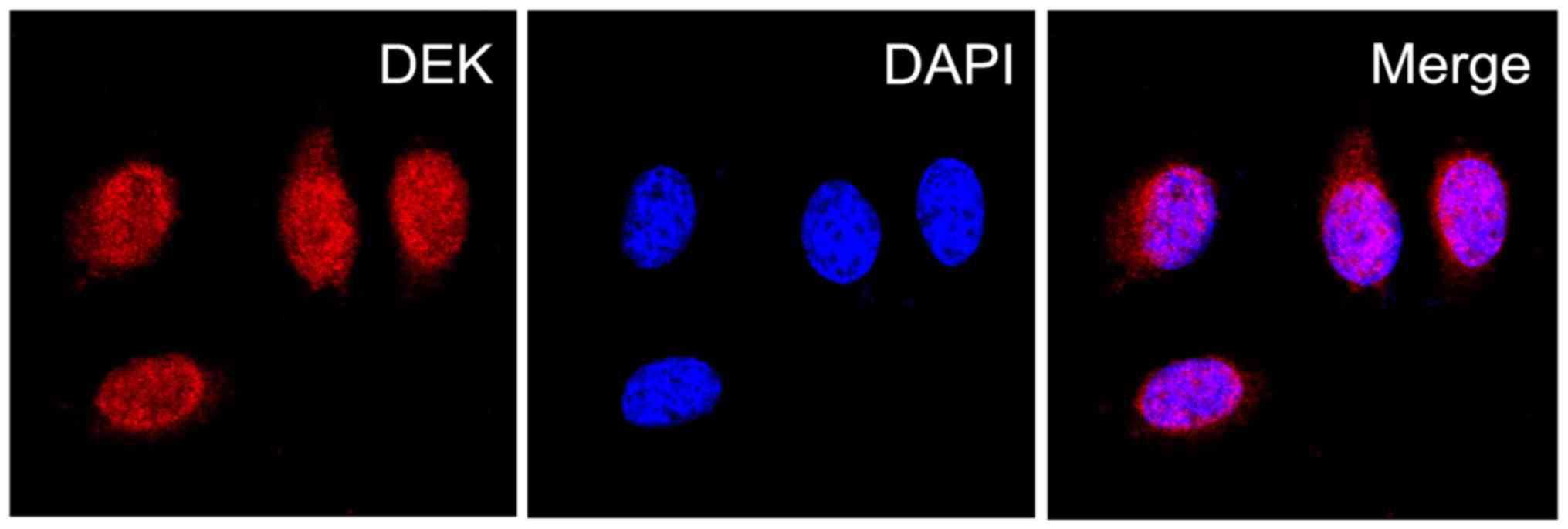
explore the role of DEK protein in PDAC, we then determined the
localization of DEK protein expression in PDAC PANC-1 cells via IF
staining, and assessed the expression levels of DEK protein in PDAC
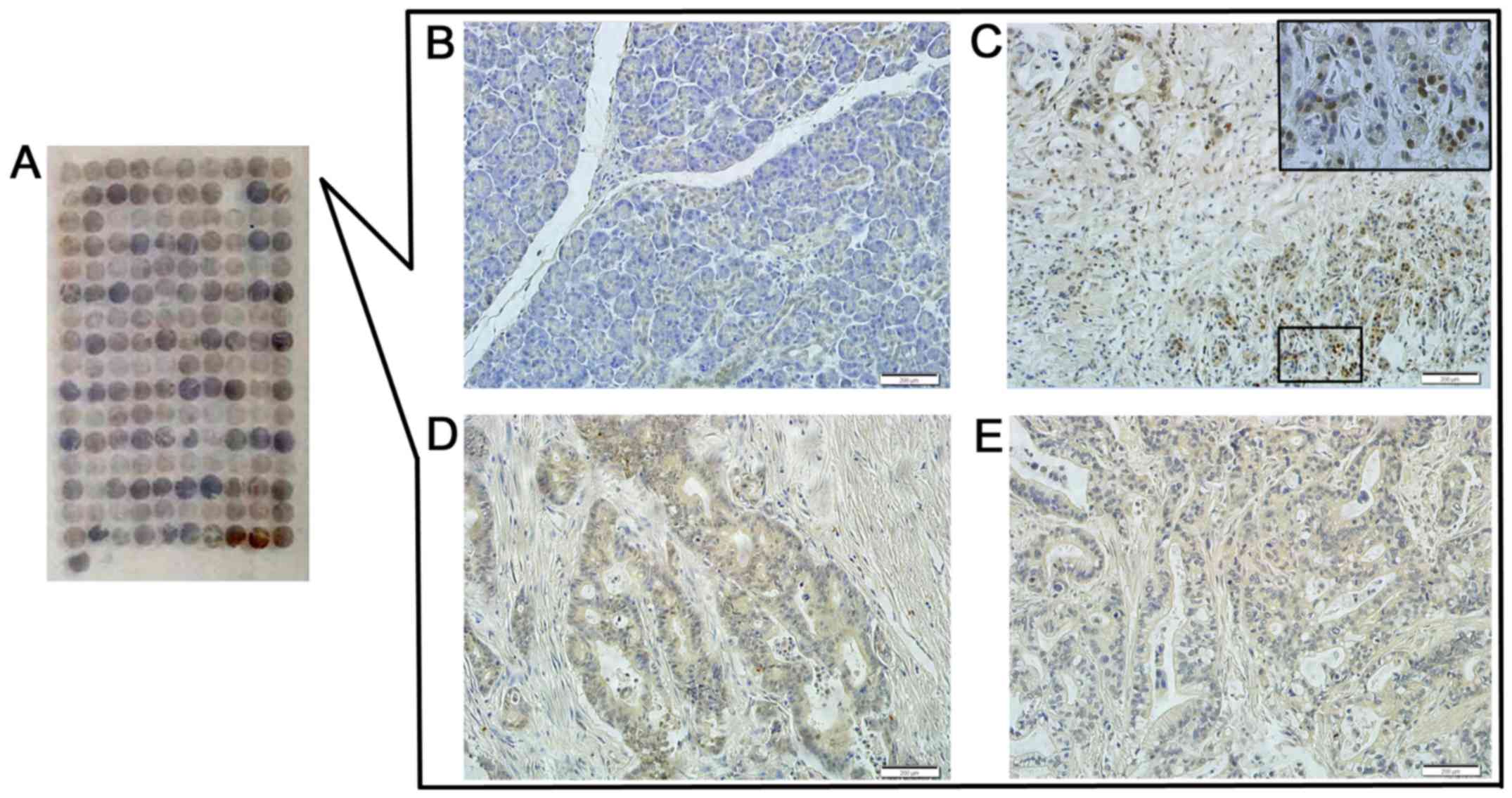
and the normal pancreas tissues via immunoenzyme staining. The DEK
protein showed a strictly nuclear staining pattern in PDAC
(Figs. 2 and 3). Simultaneously, the positive rate of
the DEK protein was 52.9% (46/87) in PDAC, which was significantly
higher than in the adjacent normal pancreatic tissues (7.7%, 4/52)
(P<0.01). Similarly, the strong positive rate of the DEK protein
was also higher in PDAC (13.8%, 12/87) compared with the adjacent
normal pancreatic tissues (0%, 0/52) (P<0.01) (Table I).
 | Table I.DEK protein expression in the PDAC
cases. |
Table I.
DEK protein expression in the PDAC
cases.
|
|
| Negative cases | Positive cases |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Diagnosis | No. of cases | − | + | ++ | Positive rate
(%) | Strongly positive
rate (%) |
|---|
| PDACs | 87 | 41 | 34 | 12 | 52.9a | 13.8a |
| Normal pancreas | 52 | 48 | 4 | 0 | 7.7 | 0 |
Correlations between DEK protein
overexpression and clinical features of PDAC
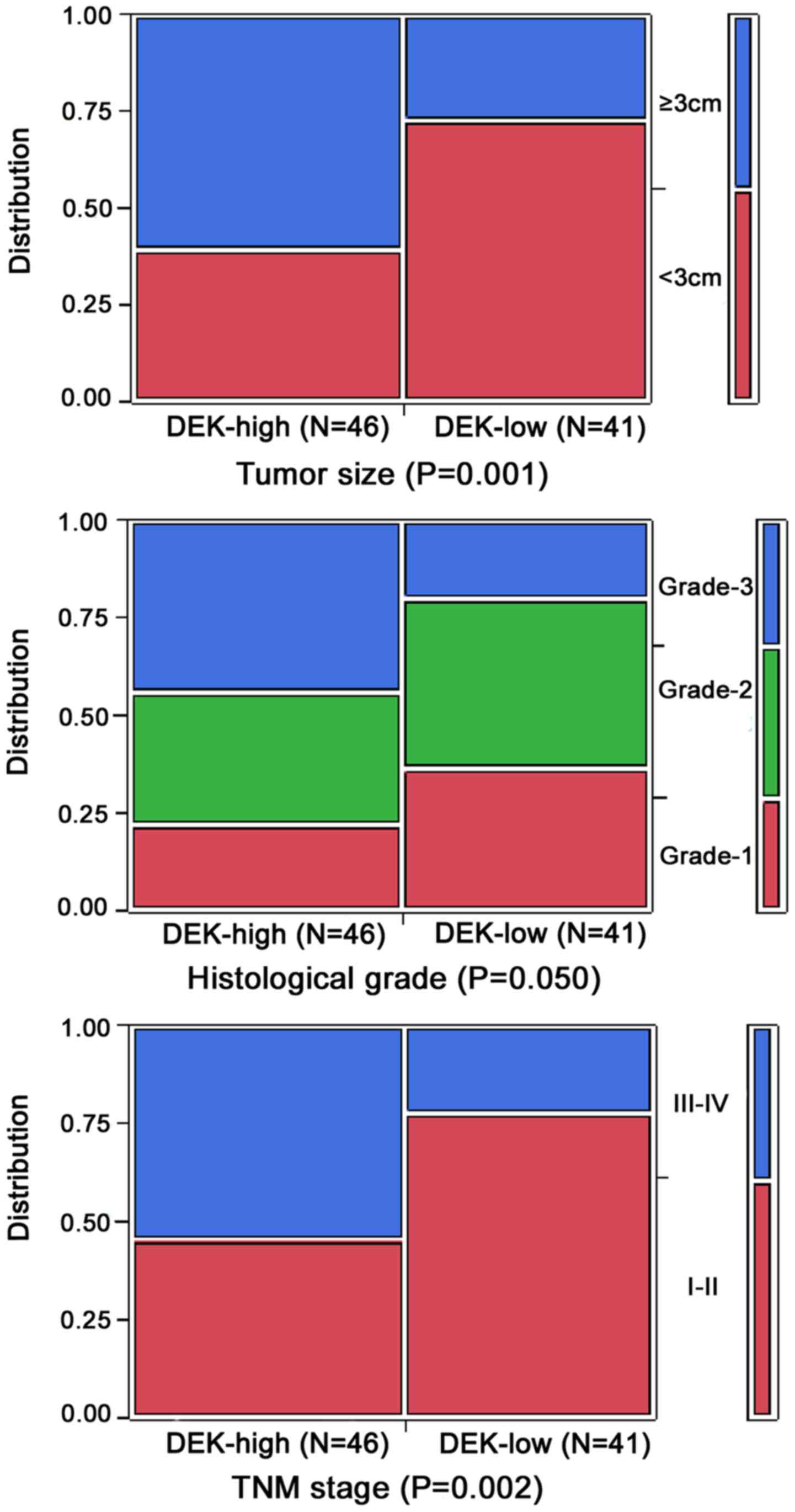
To evaluate the role of the DEK protein in PDAC
progression, we analyzed the correlation between DEK overexpression
and clinicopathological features of the PDAC patients. Generally,
DEK overexpression was significantly correlated with tumor size,
TNM stage and grade of PDAC, but not related to gender, age, tumor
location, perineural invasion status and lymph node metastasis of
patients with PDAC (P>0.05).
The positive rate of the DEK protein was
significantly higher in PDAC cases with ≥3 cm tumor size (71.8%,
28/39) than in patients with <3 cm tumor size (37.5%, 18/48)
(P<0.01). For TNM clinical stage, the positive rate of DEK
protein in the advanced stage (III–IV) PDAC cases was 73.5%
(25/34), but only 39.6% (21/53) in the early stage (I–II) cases
(P<0.05). Moreover, the positive rate of DEK was significantly
higher in grade 3 (71.4%, 20/28) than in grade 2 (47.1%, 16/34) and
grade 1 cases (40.0%, 10/25) (P<0.05) (Table II and Fig. 4).
 | Table II.Correlation of DEK protein expression
and the clinicopathological features of PDAC. |
Table II.
Correlation of DEK protein expression
and the clinicopathological features of PDAC.
| Variables | No. of cases | DEK-positive cases
(%) | χ2 | P-value |
|---|
| Gender |
|
| 0.355 | 0.551 |
|
Male | 48 | 24 (50.0) |
|
|
|
Female | 39 | 22 (56.4) |
|
|
| Age (years) |
|
| 1.194 | 0.275 |
|
<50 | 52 | 25 (48.1) |
|
|
|
≥50 | 35 | 21 (60.0) |
|
|
| Location |
|
| 0.464 | 0.496 |
|
Head | 59 | 32 (54.2) |
|
|
| Body
and tail | 28 | 13 (46.4) |
|
|
| Tumor size
(cm) |
|
| 10.156 | 0.001a |
|
<3 | 48 | 18 (37.5) |
|
|
| ≥3 | 39 | 28 (71.8) |
|
|
| Histological
grade |
|
| 5.993 | 0.050a |
| Grade
1 | 25 | 10 (40.0) |
|
|
| Grade
2 | 34 | 16 (47.1) |
|
|
| Grade
3 | 28 | 20 (71.4) |
|
|
| TNM stage |
|
| 9.557 | 0.002a |
| Stage
I–II | 53 | 21 (39.6) |
|
|
| Stage
III–IV | 34 | 25 (73.5) |
|
|
| Perineural
invasion |
|
| 0.116 | 0.733 |
|
Absent | 45 | 23 (51.1) |
|
|
|
Presence | 42 | 23 (54.8) |
|
|
| LN metastasis |
|
| 0.900 | 0.343 |
|
Negative | 45 | 26 (57.8) |
|
|
|
Positive | 42 | 20 (47.6) |
|
|
DEK overexpression is an independent
prognostic biomarker of PDAC
To evaluate the role of DEK overexpression in PDAC
progression, we analyzed the prognostic factors and overall
survival (OS) in 87 PDAC cases using the Cox proportional hazards
model. Univariate analysis showed that tumor size (P=0.034),
histological grade (P=0.000), TNM stage (P=0.000), perineural
invasion status (P=0.034), LN metastasis (P=0.004) and the level of
DEK expression (P=0.000) were associated with OS in patients with
PDAC (Table III), indicating DEK
overexpression may be a valuable prognostic factor for PDAC.
Therefore, further multivariate analysis was performed for all of
the significant variables examined in the univariate analysis.
These data suggest that DEK overexpression [hazard ratio (HR),
2.023; 95% confidence interval (CI), 1.287–3.274; P=0.003],
histological grade (HR, 1.801; 95% CI, 1.266–2.563; P=0.001) and
TNM stage (HR, 3.396; 95% CI, 2.018–5.713; P=0.000) proved to be
independent prognostic factors in prognosis of PDAC. To further
substantiate the importance of DEK overexpression in PDAC
progression, we analyzed the association between DEK expression and
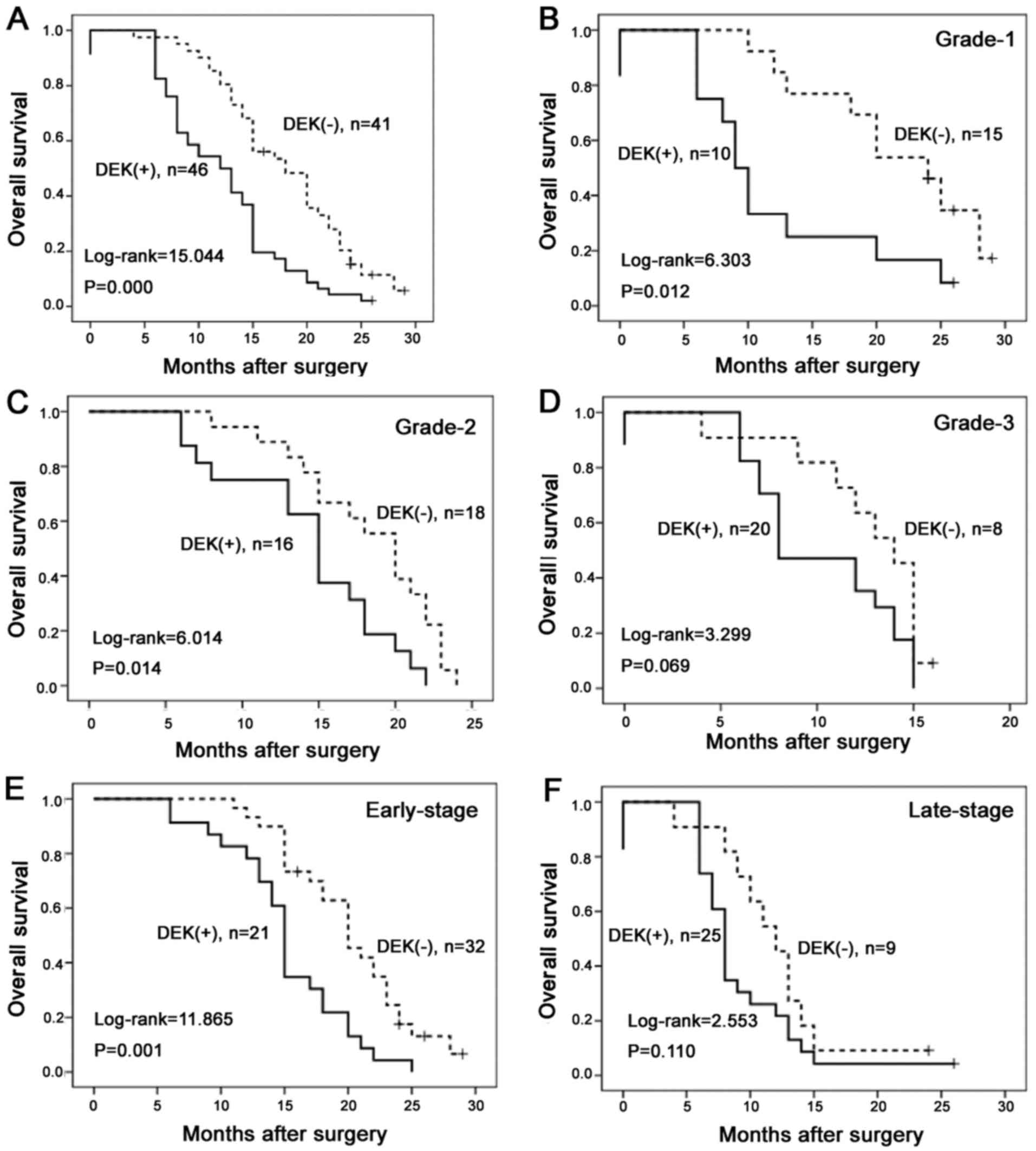
OS of 87 PDAC cases using the Kaplan-Meier method. OS rates were
significantly higher in PDAC cases with DEK low-expression than in
those with DEK overexpression (Fig.
5A). Combination analysis showed that DEK overexpression
influenced OS rates of PDAC in grade 1 and 2, and early-stage
(I–II) groups (log-rank=6.303, 6.014 and 11.865, respectively;
P=0.012, 0.014 and 0.001, respectively) (Fig. 5B, C and E). However, in the groups
of patients with grade 3 and late-stage tumors (III–IV), the OS
rate was not correlated with DEK expression status (log-rank=3.299
and 2.553, respectively; P=0.069 and 0.110, respectively) (Fig. 5D and F).
 | Table III.Univariate and multivariate survival
analyses of the clinicopathological features in 87 PDAC cases. |
Table III.
Univariate and multivariate survival
analyses of the clinicopathological features in 87 PDAC cases.
|
|
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|
|
|---|
|
Characteristics | B | SE | Wald | HR | Lower | Upper | P-value |
|---|
| Univariate survival
analyses |
|
|
|
|
|
|
|
|
Gender | 0.278 | 0.229 | 1.477 | 1.320 | 0.843 | 2.066 | 0.224 |
|
Age | 0.278 | 0.230 | 1.457 | 1.320 | 0.841 | 2.071 | 0.227 |
|
Location | 0.190 | 0.237 | 0.643 | 0.827 | 0.520 | 1.315 | 0.423 |
| Tumor
size | 0.479 | 0.226 | 4.508 | 1.614 | 1.038 | 2.512 | 0.034a |
|
Histological grade | 0.734 | 0.182 | 16.261 | 2.083 | 1.458 | 2.976 | 0.000b |
| TNM
stage | 1.173 | 0.241 | 23.629 | 3.233 | 2.014 | 5.189 | 0.000b |
|
Perineural invasion | 0.475 | 0.223 | 4.514 | 1.608 | 1.037 | 2.491 | 0.034a |
| LN
metastasis | 0.641 | 0.225 | 8.098 | 1.899 | 1.221 | 2.954 | 0.004b |
|
DEK | 0.824 | 0.232 | 12.639 | 2.280 | 1.447 | 3.591 | 0.000b |
| Multivariate
survival analyses |
|
|
|
|
|
|
|
| Tumor
size | 0.476 | 0.244 | 3.799 | 1.609 | 0.997 | 2.596 | 0.051 |
|
Histological grade | 0.588 | 0.180 | 10.697 | 1.801 | 1.266 | 2.563 | 0.001b |
| TNM
stage | 1.222 | 0.265 | 21.213 | 3.396 | 2.018 | 5.713 | 0.000b |
|
Perineural invasion | 0.428 | 0.255 | 2.816 | 1.534 | 0.931 | 2.528 | 0.093 |
| LN
metastasis | 0.259 | 0.267 | 0.941 | 1.296 | 0.768 | 2.187 | 0.332 |
|
DEK | 0.719 | 0.238 | 9.110 | 2.023 | 1.287 | 3.274 | 0.003b |
Discussion
Pancreatic ductal adenocarcinoma (PDAC), a frequent
and challenging tumor, is a deadly disease with a dismal prognosis.
The characteristics of PDAC include an aggressive rate of tumor
growth and high incidence of metastasis (25). Currently, the most patients are in
an advanced or metastatic condition at the time of diagnosis, and
only ~15% of cases can be surgically removed (3). Zhou et al reported that the
median survival time of patients with PDAC was only 13.4 months
after curative resection (26).
Therefore, the identification of a sensitive and reliable biomarker
for the early detection of PDAC is greatly needed. In the recent
study, we evaluated the clinicopathological value of DEK
overexpression in patients with PDAC.
DEK, a transcription factor, is a conserved
non-histone nucleoprotein without known paralogs (27,28).
The human DEK gene is an important proto-oncogene that is involved
in a variety of tumor-associated transcriptional and
post-translational modifications (29). Numerous studies have shown that DEK
functions as a positive supporting transcriptional factor to induce
the expression of target genes. Sawatsubashi et al showed
that DEK was correlated with numerous transcriptionally active
areas of chromatin and the nuclear ecdysone receptor, exerting its
functions as a transcriptional activator in Drosophila
(30). Sandén et al found
that DEK preferentially bound to regions of euchromatin near the
transcription start sites of highly expressed genes in lymphoma
cells and was involved with common transcriptional regulators
including SP1 and RNA polymerase II (31). Vinnedge et al showed that DEK
drove the expression of Wnt ligands, enhancing β-catenin
transcriptional activity in breast cancer cells (32,33).
Adams et al reported that DEK can activate transcription via
interaction with IRAK1 in head and neck cancer (34). These findings indicate that DEK
potentially plays important roles in the progression of tumor
cells.
Recently, Datta et al validated that the
level of DEK expression was markedly higher in bladder cancer than
normal counterparts using western blotting, suggesting that DEK may
be a biomarker for the detection of bladder cancer (35). In the present study, our principal
aim was to determine whether DEK overexpression is a biomarker for
the prognostic evaluation of PDAC. In the present study, we
assessed DEK mRNA expression in PDAC clinical samples using
microarray data from GEO, and performed Immunoenzyme staining of
DEK in 87 PDAC tissues and 52 adjacent normal pancreas tissues. We
found that the expression levels of DEK mRNA in tumor tissues were
significantly higher than that in the adjacent non-tumor tissues.
Simultaneously, the DEK protein gave a primarily nuclear staining
pattern based on immunoenzyme staining, which was consistent with
Kappes et al and our IF staining results for PANC-1
pancreatic cancer (PC) cells (29).
In the present study, using immunoenzyme staining of the DEK
protein, we found that the DEK protein was highly expressed in PDAC
tissues, while the staining was weak positive or negative in normal
tissues. These findings demonstrated that DEK may play an important
role in the progression and aggressiveness of PDAC.
Despite the significant association between DEK
overexpression and numerous types of cancers, studies of DEK
expression-based outcome in patients are limited. Liu et al
demonstrated a significant association between DEK overexpression
and poor survival of non-small cell lung carcinoma patients
(36). Shibata et al showed
that DEK overexpression was associated with tumor initiation
activity and a poor prognosis in high grade neuroendocrine
carcinoma of the lung (13). Our
previous study reported that DEK overexpression was not only
strongly associated with breast cancer, but that the expression was
also higher in high grade breast cancers, as well as advanced stage
tumors (23). In the present study,
we also found that DEK overexpression was significantly correlated
with tumor size (P=0.001), histological grade (P=0.050) and TNM
stage (P=0.002). Unfortunately, high histological grade and
advanced TNM stage indicate poor outcomes and recurrence in
patients with PDAC. Therefore, DEK protein may be a novel biomarker
related to progression and aggressiveness of PDAC.
In regards to survival rates, we found that the
level of DEK expression was strongly correlated with the survival
rates in patients with PDAC. Additionally, univariate survival
analysis showed that tumor size, histological grade, TNM stage,
perineural invasion status and LN metastasis were all associated
with OS rates in patients with PDAC. Multivariate survival analysis
revealed that DEK overexpression was an independent prognostic
factor along with histological grade and TNM stage. Furthermore,
combination analysis showed that DEK overexpression influenced OS
rates of PDAC in grade 1 and 2, and early-stage groups. However, in
the groups of patients with grade 3 and late-stage tumors, the OS
rate was not correlated with DEK expression status. Apparently,
these findings indicated that DEK may be a potentially predictive
biomarker of poor prognosis, particularly in patients with low
histological grade and early-stage PDAC.
In conclusion, DEK plays an important role in the
progression of PDAC. Its overexpression may be associated with PDAC
progression, and may be used as a biomarker for prognostic
evaluation and as a therapeutic target in PDAC. Further studies are
required to confirm this hypothesis using molecular biology
experiments.
Acknowledgements
The present study was supported by grants from the
Special Research Project of the ‘973 Plan’ (2014CB560708), the
National Natural Science Funds of China (no. 61371067), and the
International Cooperation Project of Science and Technology
Department of Jilin Province (no. 20150414030GH).
References
|
1
|
Urayama S: Pancreatic cancer early
detection: Expanding higher-risk group with clinical and
metabolomics parameters. World J Gastroenterol. 21:1707–1717. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vernejoul F, Faure P, Benali N, Calise D,
Tiraby G, Pradayrol L, Susini C and Buscail L: Antitumor effect of
in vivo somatostatin receptor subtype 2 gene transfer in primary
and metastatic pancreatic cancer models. Cancer Res. 62:6124–6131.
2002.PubMed/NCBI
|
|
3
|
Recaldini C, Carrafiello G, Bertolotti E,
Angeretti MG and Fugazzola C: Contrast-enhanced ultrasonograpic
findings in pancreatic tumors. Int J Med Sci. 5:203–208. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stathis A and Moore MJ: Advanced
pancreatic carcinoma: Current treatment and future challenges. Nat
Rev Clin Oncol. 7:163–172. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee SH, Kim H, Hwang JH, Lee HS, Cho JY,
Yoon YS and Han HS: Breast cancer resistance protein expression is
associated with early recurrence and decreased survival in
resectable pancreatic cancer patients. Pathol Int. 62:167–175.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu K, Ji B, Zhang W, Liu S, Wang Y and
Liu Y: Comparison of iodine-125 seed implantation and
pancreaticoduodenectomy in the treatment of pancreatic cancer. Int
J Med Sci. 11:893–896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamashita K, Miyamoto A, Hama N, Asaoka T,
Maeda S, Omiya H, Takami K, Doki Y, Mori M and Nakamori S: Survival
impact of pulmonary metastasis as recurrence of pancreatic ductal
adenocarcinoma. Dig Surg. 32:464–471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van den Broeck A, Vankelecom H, Van
Eijsden R, Govaere O and Topal B: Molecular markers associated with
outcome and metastasis in human pancreatic cancer. J Exp Clin
Cancer Res. 31:682012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Niccolai E, Cappello P, Taddei A, Ricci F,
D'Elios MM, Benagiano M, Bechi P, Bencini L, Ringressi MN, Coratti
A, et al: Peripheral ENO1-specific T cells mirror the intratumoral
immune response and their presence is a potential prognostic factor
for pancreatic adenocarcinoma. Int J Oncol. 49:393–401.
2016.PubMed/NCBI
|
|
10
|
Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H,
Wang W, Huang X, Yuan Z and Ai K: miR-212 promotes pancreatic
cancer cell growth and invasion by targeting the hedgehog signaling
pathway receptor patched-1. J Exp Clin Cancer Res. 33:542014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
von Lindern M, Fornerod M, van Baal S,
Jaegle M, de Wit T, Buijs A and Grosveld G: The translocation
(6;9), associated with a specific subtype of acute myeloid
leukemia, results in the fusion of two genes, dek and can, and the
expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell
Biol. 12:1687–1697. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khodadoust MS, Verhaegen M, Kappes F,
Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz
DM and Soengas MS: Melanoma proliferation and chemoresistance
controlled by the DEK oncogene. Cancer Res. 69:6405–6413. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shibata T, Kokubu A, Miyamoto M, Hosoda F,
Gotoh M, Tsuta K, Asamura H, Matsuno Y, Kondo T, Imoto I, et al:
DEK oncoprotein regulates transcriptional modifiers and sustains
tumor initiation activity in high-grade neuroendocrine carcinoma of
the lung. Oncogene. 29:4671–4681. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sammons M, Wan SS, Vogel NL, Mientjes EJ,
Grosveld G and Ashburner BP: Negative regulation of the RelA/p65
transactivation function by the product of the DEK proto-oncogene.
J Biol Chem. 281:26802–26812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gamble MJ and Fisher RP: SET and PARP1
remove DEK from chromatin to permit access by the transcription
machinery. Nat Struct Mol Biol. 14:548–555. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wise-Draper TM, Morreale RJ, Morris TA,
Mintz-Cole RA, Hoskins EE, Balsitis SJ, Husseinzadeh N, Witte DP,
Wikenheiser- Brokamp KA, Lambert PF, et al: DEK proto-oncogene
expression interferes with the normal epithelial differentiation
program. Am J Pathol. 174:71–81. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kappes F, Fahrer J, Khodadoust MS, Tabbert
A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Bürkle A,
Markovitz DM and Ferrando-May E: DEK is a poly(ADP-ribose) acceptor
in apoptosis and mediates resistance to genotoxic stress. Mol Cell
Biol. 28:3245–3257. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kroes RA, Jastrow A, McLone MG, Yamamoto
H, Colley P, Kersey DS, Yong VW, Mkrdichian E, Cerullo L, Leestma
J, et al: The identification of novel therapeutic targets for the
treatment of malignant brain tumors. Cancer Lett. 156:191–198.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
von Lindern M, van Baal S, Wiegant J, Raap
A, Hagemeijer A and Grosveld G: can, a putative oncogene associated
with myeloid leukemogenesis, may be activated by fusion of its 3
half to different genes: Characterization of the set gene. Mol Cell
Biol. 12:3346–3355. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sanchez-Carbayo M, Socci ND, Lozano JJ, Li
W, Charytonowicz E, Belbin TJ, Prystowsky MB, Ortiz AR, Childs G
and Cordon-Cardo C: Gene discovery in bladder cancer progression
using cDNA microarrays. Am J Pathol. 163:505–516. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kondoh N, Wakatsuki T, Ryo A, Hada A,
Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita
M, et al: Identification and characterization of genes associated
with human hepatocellular carcinogenesis. Cancer Res. 59:4990–4996.
1999.PubMed/NCBI
|
|
22
|
Lin L, Piao J, Gao W, Piao Y, Jin G, Ma Y,
Li J and Lin Z: DEK over expression as an independent biomarker for
poor prognosis in colorectal cancer. BMC Cancer. 13:3662013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu S, Wang X, Sun F, Kong J, Li Z and Lin
Z: DEK overexpression is correlated with the clinical features of
breast cancer. Pathol Int. 62:176–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Piao J, Shang Y, Liu S, Piao Y, Cui X, Li
Y and Lin Z: High expression of DEK predicts poor prognosis of
gastric adenocarcinoma. Diagn Pathol. 9:672014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chatterjee D, Katz MH, Rashid A, Wang H,
Iuga AC, Varadhachary GR, Wolff RA, Lee JE, Pisters PW, Crane CH,
et al: Perineural and intraneural invasion in posttherapy
pancreaticoduodenectomy specimens predicts poor prognosis in
patients with pancreatic ductal adenocarcinoma. Am J Surg Pathol.
36:409–417. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou HY, Wang Y, Zhang J, Ruan CP, Wang
WJ, Sun YP and Hu ZQ: Retrograde vs conventional dissection
technique in pancreaticoduodenectomy: A pilot study. JAMA Surg.
149:604–607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vinnedge LM Privette, Kappes F, Nassar N
and Wells SI: Stacking the DEK: From chromatin topology to cancer
stem cells. Cell Cycle. 12:51–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pease NA, Wise-Draper T and Vinnedge L
Privette: Dissecting the potential interplay of DEK functions in
inflammation and cancer. J Oncol. 2015:1065172015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kappes F, Damoc C, Knippers R, Przybylski
M, Pinna LA and Gruss C: Phosphorylation by protein kinase CK2
changes the DNA binding properties of the human chromatin protein
DEK. Mol Cell Biol. 24:6011–6020. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sawatsubashi S, Murata T, Lim J, Fujiki R,
Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, et al: A
histone chaperone, DEK, transcriptionally coactivates a nuclear
receptor. Genes Dev. 24:159–170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sandén C, Järvstråt L, Lennartsson A,
Brattås PL, Nilsson B and Gullberg U: The DEK oncoprotein binds to
highly and ubiquitously expressed genes with a dual role in their
transcriptional regulation. Mol Cancer. 13:2152014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vinnedge LM Privette, McClaine R, Wagh PK,
Wikenheiser-Brokamp KA, Waltz SE and Wells SI: The human DEK
oncogene stimulates β-catenin signaling, invasion and mammosphere
formation in breast cancer. Oncogene. 30:2741–2752. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vinnedge LM Privette, Benight NM, Wagh PK,
Pease NA, Nashu MA, Serrano-Lopez J, Adams AK, Cancelas JA, Waltz
SE and Wells SI: The DEK oncogene promotes cellular proliferation
through paracrine Wnt signaling in Ron receptor-positive breast
cancers. Oncogene. 34:2325–2336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Adams AK, Bolanos LC, Dexheimer PJ, Karns
RA, Aronow BJ, Komurov K, Jegga AG, Casper KA, Patil YJ, Wilson KM,
et al: IRAK1 is a novel DEK transcriptional target and is essential
for head and neck cancer cell survival. Oncotarget. 6:43395–43407.
2015.PubMed/NCBI
|
|
35
|
Datta A, Adelson ME, Mogilevkin Y,
Mordechai E, Sidi AA and Trama JP: Oncoprotein DEK as a tissue and
urinary biomarker for bladder cancer. BMC Cancer. 11:2342011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Qi D, Qi J, Mao Z, Li X, Zhang J,
Li J and Gao W: Significance of DEK overexpression for the
prognostic evaluation of non-small cell lung carcinoma. Oncol Rep.
35:155–162. 2016.PubMed/NCBI
|