Introduction
Multiple myeloma (MM) is one of the most common
hematological cancers characterized by high proliferation of plasma
cells expressing the surface marker CD138 in the bone marrow
(1,2). Despite the advancement in treatment
options over the past decades, MM remains an intractable disease
with a poor survival rate and a relatively high incidence rate in
recent years (3,4). The underlying mechanism of the
pathogenesis of MM remains poorly understood. Therefore, it is of
great importance to develop novel and effective therapies for
MM.
In recent years, microRNAs (miRNAs) have been taken
as promising therapeutic targets for cancer treatments, including
MM (5). miRNAs are a type of small,
non-coding regulatory RNA molecules (20–25 nucleotides) that
post-transcriptionally and negatively modulate target gene
expression by targeting the 3-untranslated region (UTR) of target
gene (6,7). Therefore, miRNAs regulate a variety of
cellular processes, including cell proliferation, apoptosis,
differentiation, migration and invasion (8). Increasing evidence has reported that
numerous miRNAs are frequently dysregulated in MM serving as
oncogenes or tumor suppressors in regulating MM cell proliferation,
apoptosis, cell cycle, drug resistance, migration and invasion
(9–15). In addition, the dysregulated miRNAs
also represent potential biomarkers for MM diagnosis and prognosis
(16–18). However, the precise mechanism of
miRNAs in MM remains largely unknown.
Hedgehog (HH) signaling pathway is a highly
conserved pathway from Drosophila to vertebrates that
regulate embryonic development and adult tissue homeostasis
(19). Three HH ligands exist
including desert hedgehog, Indian hedgehog and sonic hedgehog in
mammals (20,21). In the state of inactivation, the HH
receptor patched1 (Ptch1) inhibits smoothened (SMO), which is a
seven-transmembrane protein essential for HH activation (22). When HH ligands bind to Ptch1, SMO is
released and activated, which then initiates the transcriptional
program of HH signaling including Ptch1 and glioma-associated
oncogene homolog 1 (Gli1) (20,23).
Although HH signaling pathway is essential for embryonic
development, tissue repair, and homeostasis, the abnormal
activation of HH signaling pathway can lead to tumorigenesis
(22,24–27).
The abnormal activation of HH signaling pathway has been found in
solid tumors (prostate, pancreatic and lung cancers) (28–30)
and hematologic malignancies (MM and B-cell lymphoma) (31–33).
HH signaling pathway regulates cell proliferation, apoptosis,
survival, drug resistance, colony growth and self-renewal of MM
cells (34–36). Therefore, strategies inhibiting HH
signaling may be a promising and effective anticancer
intervention.
A recent study has suggested that miR-1271 functions
as a tumor suppressor gene in various cancer types (37–40).
However, whether miR-1271 plays a potential role in MM is unknown.
In this study, we investigated the potential role and underlying
mechanism of miR-1271 in MM. We found that miR-1271 was
significantly decreased in MM samples and MM cell lines.
Overexpression of miR-1271 inhibited proliferation and promoted
apoptosis of MM cells. Bioinformatics algorithms analysis showed
that SMO was a predicted target gene of miR-1271 that was confirmed
by a Dual-luciferase reporter assay. The overexpression of miR-1271
inhibited SMO expression as well as HH signaling in MM cells. The
restoration of SMO expression significantly abrogated the effects
of miR-1271. The present study suggests that miR-1271 functions as
a tumor suppressor of MM, indicating a potential molecular
candidate for MM treatment.
Materials and methods
Cell cultures
Bone marrow aspirates were collected from 10 MM
patients and 10 healthy donors in Xi'an Jiaotong University Medical
College Red Cross Hospital. The primary MM cells were isolated from
bone marrow aspirates as previously described (41). Briefly, mononuclear cells were
separated from bone marrow aspirates by Ficoll-Hypaque density
gradient centrifugation (Amersham, Little Chalfont, UK). Then, the
cells were suspended in ice-cold phosphate-buffered saline (PBS)
and incubated with microbeads labeled with a mouse anti-human CD138
monoclonal antibody (Miltenyi Biotech, Auburn, CA, USA). The
CD138+ cells were sorted on a BD fluorescence-activated
cell sorting FACSAria flow cytometer (BD Biosciences, San Jose, CA,
USA) in accordance with the manufacturers instruction. The
collection and use of clinical samples were approved by the
Institutional Human Experiment and Ethics Committee of Xi'an
Jiaotong University Medical College Red Cross Hospital with written
informed consents from all MM patients and healthy donors. The
human MM cell lines including NCI-H929, U266 and MM1.R were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The KMS-12-BM MM cell line was purchased from
Beijing Abace Biology, Co., Ltd. (Beijing, China). The primary MM
cells, human MM cell lines, and normal plasma cells (nPCs) were
cultured in RPMI-1640 (Gibco, Rockville, MD, USA) in supplement
with 10% fetal bovine serum (FBS; Gibco) and 1%
penicillin/streptomycin mix (Sigma-Aldrich, St. Louis, MO, USA) in
a humidified atmosphere containing 5% CO2 at 37°C.
Real-time quantitative PCR (RT-qPCR)
analysis
Total RNA from tissues or cells was extracted using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA
was synthesized using M-MLV reverse transcriptase (BioTeke,
Beijing, China) for mRNA expression analysis or miScript reverse
transcription kit (Qiagen, Dusseldorf, Germany) for miRNA
expression analysis. PCR amplification was performed on an Applied
Biosystems AB 7500 Real-Time PCR system (Applied Biosystems, In.,
Carlsbad, CA, USA) using Power SYBR-Green PCR Master Mix (Applied
Biosystems). The primers used were as follows: miR-1271 forward,
5-CAGCACTTGGCACCTAGCA-3 and reverse, 5-TATGGTTGTTCTCCTCTCTGTCTC-3;
U6 forward, 5-CGCTTCGGCAGCACATATACTAA-3 and reverse,
5-TATGGAACGCTTCACGAATTTGC-3; SMO forward,
5-TGCTCATCGTGGGAGGCTACTT-3 and reverse, 5-ATCTTGCTGGCAGCCTTCTCAC-3;
Gli1 forward, 5-TATGGACCTGGCTTTGGA-3 and and reverse,
5-CCTATGTGAAGCCCTATTT GC-3; Ptch1, CTCTGGAGCAGATTTCCAAGG; Ptch1
forward, 5-CTCTGGAGCAGATTTCCAAGG-3 and reverse,
5-TGCCGCAGTTCTTTTGAATG-3 and GAPDH forward,
5-CCATGTTCGTCATGGGTGTG-3 and reverse, 5-GGTGCTAAGCAGTTGGTGGTG-3. U6
was used as the internal control of miR-1271 and GAPDH was used as
the internal control of mRNA. The relative gene expression was
determined by 2−ΔΔCt method, normalized against GAPDH or
U6, and then compared with control.
Cell transfection
The miR-1271 mimics, miR-1271 inhibitor, and their
scrambled controls (Scr) were purchased from Shanghai GenePharma
(Shanghai, China). Cells were transfected with miR-1271 mimics or
miR-1271 inhibitor at a final concentration of 50 nM using
Lipofectamine 2000 (Invitrogen). The open reading frame of SMO
without 3-UTR was inserted into pcDNA3.1 (BioVector, Beijing,
China). For overexpression of SMO, the pcDNA/SMO constructs were
transfected into cells using Lipofectamine 2000 (Invitrogen).
Colony formation assay
Cells were transfected with miR-1271 mimics or
miR-1271 inhibitor for 48 h. Then, the cells (200 cells/well) were
seeded in 6-well plates and grown in a medium containing 0.3% noble
agar for 2 weeks at 37°C. After fixing with 100% methanol for 30
min, the cells were stained with 0.1% crystal violet
(Sigma-Aldrich). The number of colonies was observed and counted
under a microscope (Olympus Corp., Tokyo, Japan).
3-(4,5-Dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell proliferation was assessed by MTT assay.
Briefly, cells were seeded into a 96-well plate at a density of
5×103 cells/well and transfected with miR-1271 mimics or
miR-1271 inhibitor for 24, 48 and 72 h. After replacement with
fresh medium, 20 µl MTT (0.5 mg/ml in PBS; Sigma) was added to each
well and incubated for 4 h at 37°C. Then, the supernatant was
discarded and 200 µl dimethyl sulfoxide (Sigma-Aldrich) was added
to each well. After incubation for 15 min, the absorbance at 490 nm
was measured with a microplate reader (Thermo Fisher Scientific,
Rockford, IL, USA).
Cell apoptosis assay
Cell apoptosis was determined by the terminal
deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and
caspase-3 activity assay. For TUNEL assay, briefly, cells were
fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton
X-100 followed by incubation with TUNEL reaction mixtures (Roche
Diagnostics, Indianapolis, IN, USA) for 1 h at 37°C. The apoptotic
cells were observed under a microscope (Olympus), and five fields
per slide were randomly chosen to quantitatively calculate the
apoptotic cells. For caspase-3 activity assay, cells were lysed and
the protein concentration was measured. A total of 100 µg of
protein with 50 µl of reaction buffer was treated with 5 µl
DEVD-pNA substrate (4 mM; Roche Diagnostics) for 2 h at 37°C. The
absorbance at 405 nm was measured with a microplate reader (Thermo
Fisher Scientific).
Dual-luciferase reporter assay
The 3-UTR of SMO containing the target sequence of
miR-1271 was inserted into pmirGLO vector (Promega, Madison, WI,
USA) to obtain pmirGLO-SMO 3-UTR, and the 3-UTR of SMO containing
mutant miR-1271 target sites was inserted into pmirGLO vector
(Promega) to obtain pmirGLO-mutant SMO 3-UTR. NCI-H929 cells were
co-transfected with pmirGLO-SMO 3-UTR or pmirGLO-mutant SMO 3-UTR
and miR-1271 mimics, miR-1271 inhibitor or Scr controls. At 4 h
after incubation, the luciferase activity was detected by a
Dual-luciferase reporter assay system (Promega).
Western blot analysis
Total proteins were extracted, separated and
transferred onto a nitrocellulose membrane (Bio-Rad Laboratories,
Hercules, CA, USA). The membrane was blocked by 5% non-fat milk and
incubated with primary antibodies at 4°C overnight. After three
washes with Tris-buffered saline containing 0.1% Tween-20, the
membrane was incubated with horseradish peroxidase conjugated
secondary antibodies (1:2,000; Bosis, Beijing, China) for 1 h at
37°C. Then, the protein was detected using enhanced
chemiluminescence (Millipore, Boston, MA, USA). The primary
antibodies (anti-SMO and anti-GAPDH) used in this study were
purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The
intensities of protein bands were quantified by Image-Pro Plus 6.0
software (Media Cybernetics, Inc., Rockville, MD, USA). The
relative protein expression was normalized against GAPDH and then
compared with the control.
Data analysis
All the data were reported as means ± standard
deviation. Statistical analyses were analyzed by the Students
t-test for two group comparison or one-way analysis of variance
followed by Bonferroni post hoc for multiple group comparison
(>2) using SPSS version 11.5 (SPSS, Inc., Chicago, IL, USA). At
P<0.05, the difference was considered statistically
significant.
Results
Expression of miR-1271 is
downregulated in MM
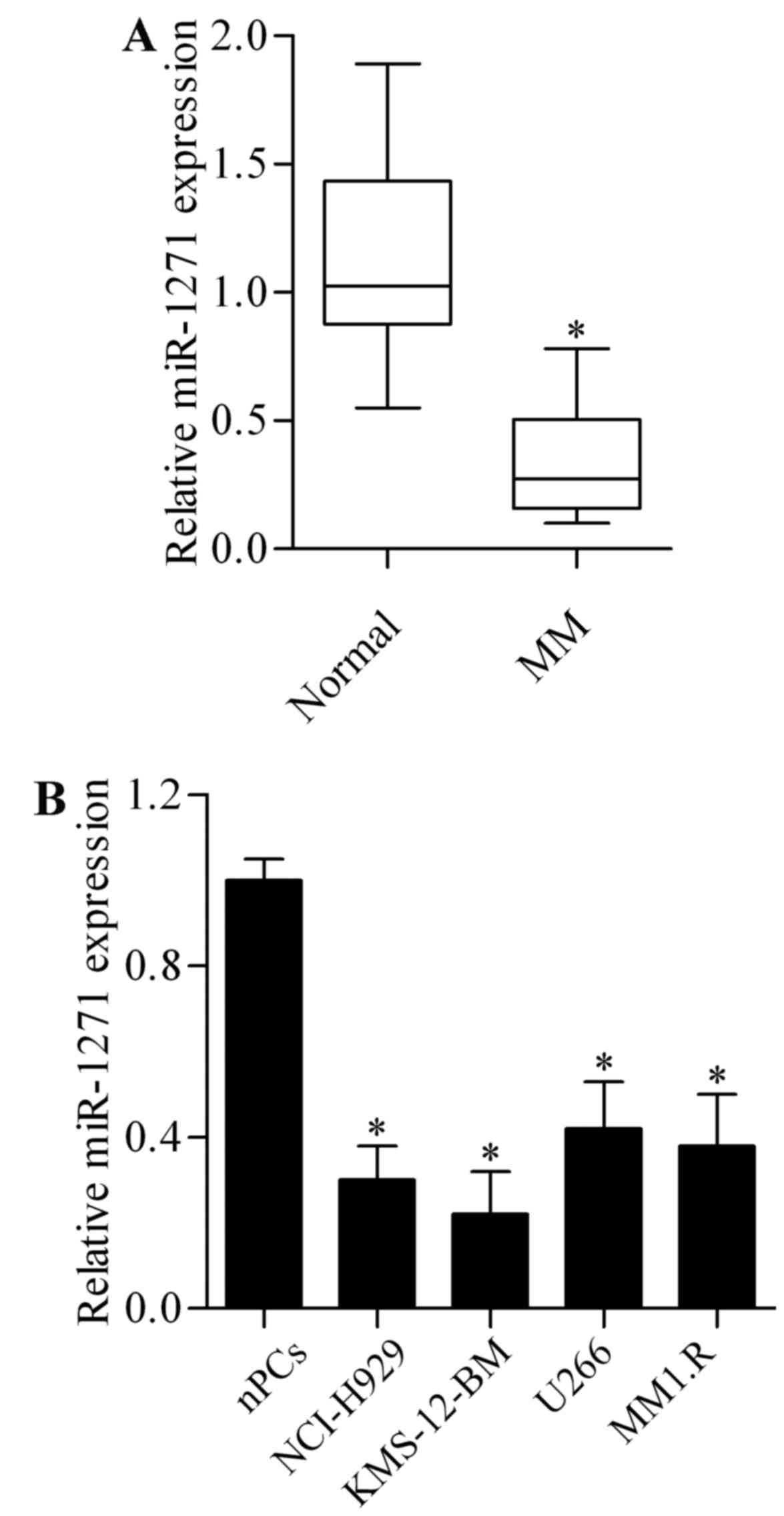
To investigate whether miR-1271 plays a role in MM,
we firstly examined the miR-1271 expression in primary MM cells
isolated from MM patient bone marrow samples. The results showed
that the expression of miR-1271 was significantly lower in MM
primary cells than in plasma cells from healthy donors (Fig. 1A). We then evaluated miR-1271
expression in MM cell lines including NCI-H929, KMS-12-BM, U266 and
MM1.R. As compared with normal plasma cells (nPCs), miR-1271 was
significantly decreased in MM cell lines (Fig. 1B). These results suggest a tumor
suppressor role of miR-1271 in MM.
miR-1271 inhibits MM cell
proliferation
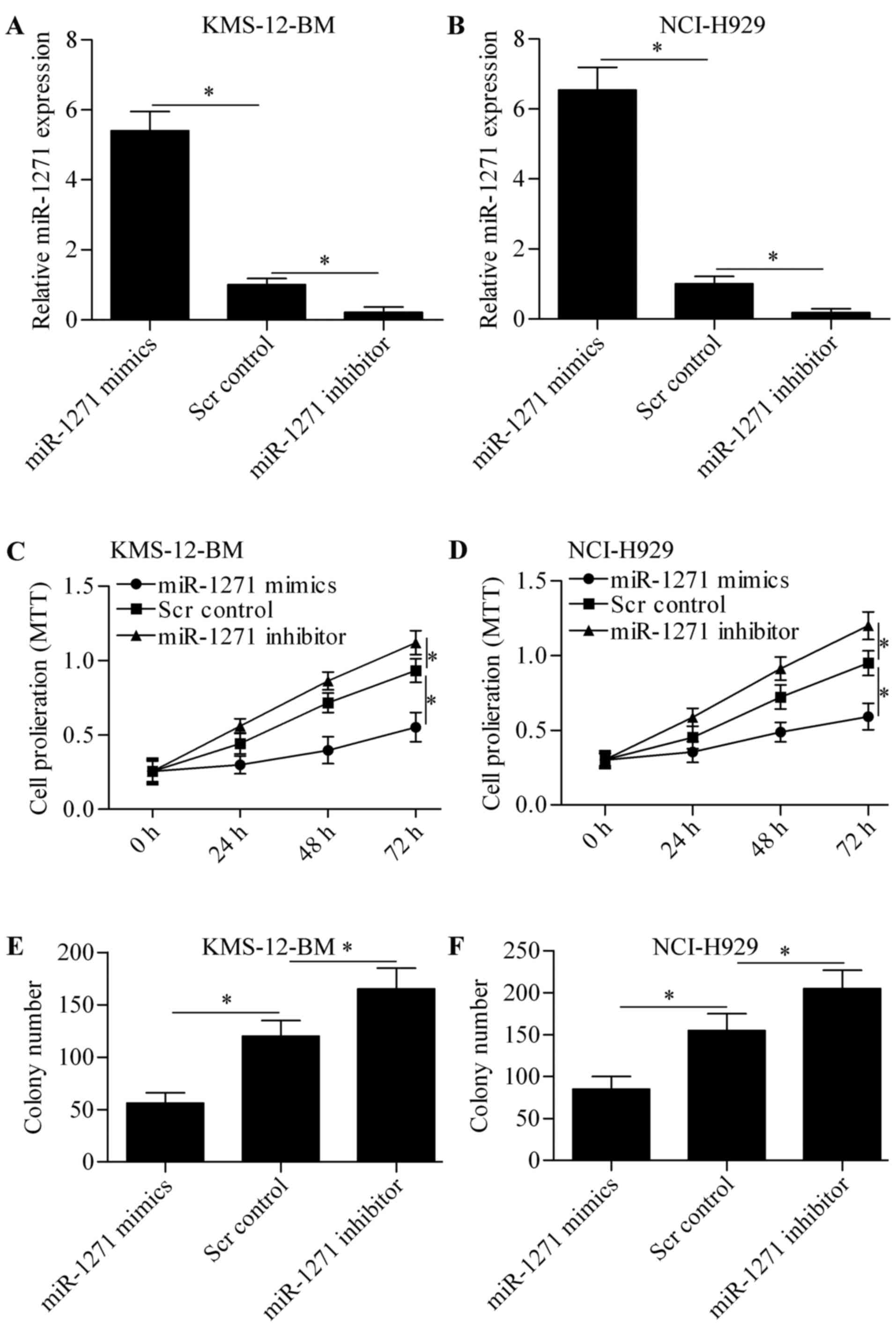
To explore the biological role of miR-1271 in MM, we
performed gain-of- and loss-of-function experiments in KMS-12-BM
and NCI-H929 cells by transfection of miR-1271 mimics or miR-1271
inhibitor. The results showed that transfection of miR-1271
significantly increased the expression of miR-1271 in KMS-12-BM
(Fig. 2A) and NCI-H929 (Fig. 2B) cells, whereas miR-1271 inhibitor
markedly decreased miR-1271 expression. We then detected the effect
of miR-1271 on cell proliferation by MTT assay. The results showed
that overexpression of miR-1271 significantly suppressed MM cell
proliferation and miR-1271 inhibition markedly promoted cell
proliferation (Fig. 2C and D).
Furthermore, the colony growth of KMS-12-BM (Fig. 2E) and NCI-H929 (Fig. 2F) cells was also significantly
decreased or increased by miR-1271 overexpression or miR-1271
inhibition, respectively. These data indicate that miR-1271 inhibit
MM cell proliferation.
miR-1271 induces MM cell
apoptosis
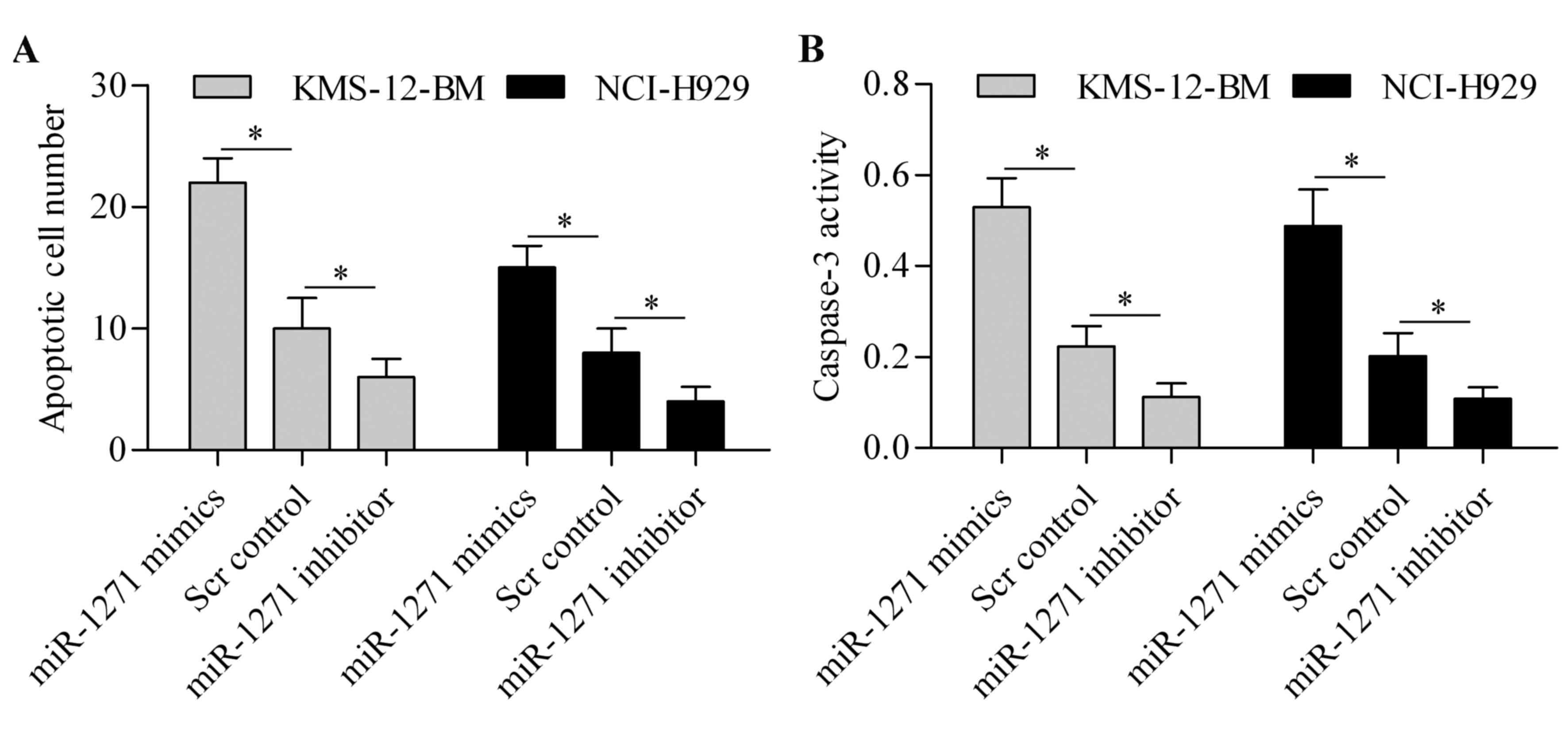
To further investigate the function of miR-1271 in
MM, we then evaluated its effect on cell apoptosis. TUNEL assay
showed that miR-1271 overexpression markedly induced cell apoptosis
of MM cells, whereas miR-1271 inhibition markedly suppressed MM
cell apoptosis (Fig. 3A). Moreover,
caspase-3 activity assay showed that the activity of caspase-3 was
significantly upregulated by miR-1271 overexpression but decreased
by miR-1271 inhibition (Fig. 3B).
These results suggest that miR-1271 induces apoptosis of MM
cells.
miR-1271 targets the 3-UTR of SMO
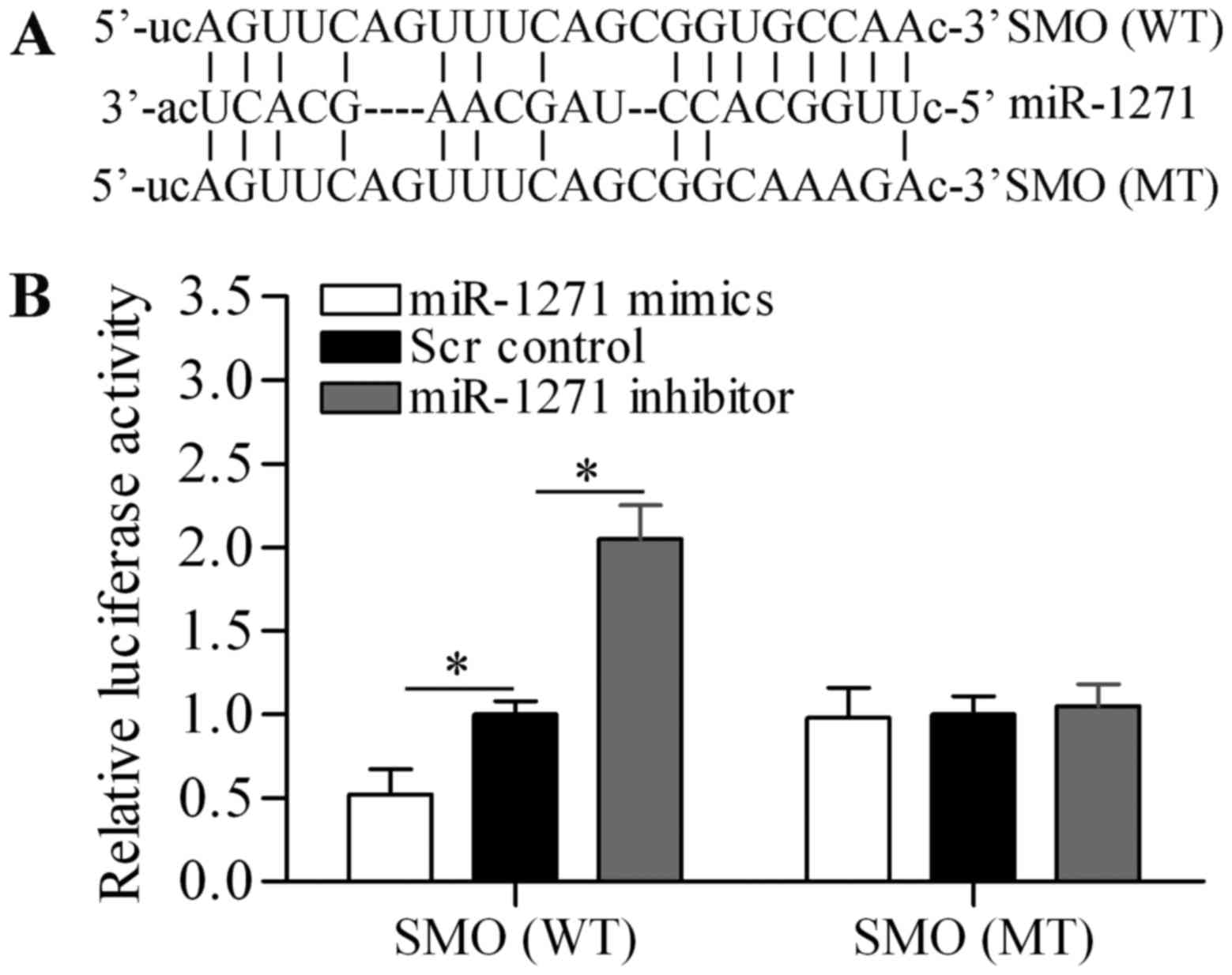
To understand the underlying mechanism by which
miR-1271 inhibits proliferation and induces apoptosis of MM cells,
we used bioinformatics analysis to seek the potential targets of
miR-1271. Notably, we found that SMO, an important oncogene
(42), was predicted as a potential
target gene of miR-1271 (Fig. 4A).
To verify the targeting relationship, luciferase reporter vectors
containing wild-type (WT) or mutant (MT) SMO 3-UTR were
constructed. The Dual-luciferase reporter assay showed that
miR-1271 mimics significantly decreased the luciferase activity of
SMO 3-UTR (WT), whereas miR-1271 inhibitor significantly increased
the luciferase activity of SMO 3-UTR (WT) (Fig. 4B). However, neither miR-1271 mimics
nor miR-1271 inhibitor showed significant effect on the luciferase
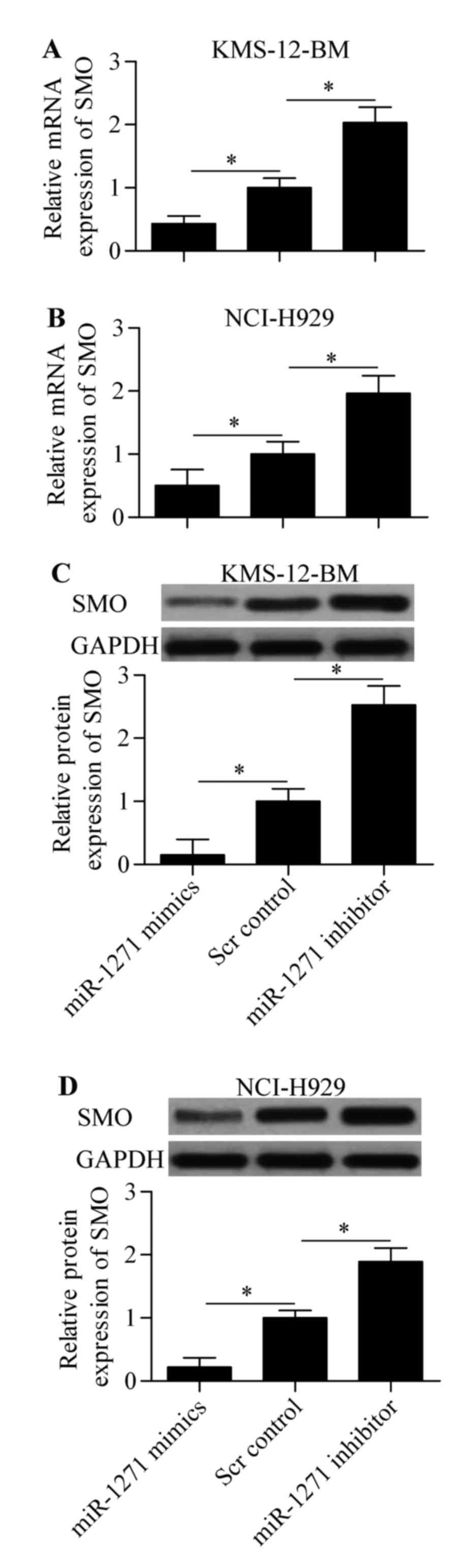
activity of SMO 3-UTR (WT). Subsequently, we performed RT-qPCR and
western blot analysis to detect the direct effect of miR-1271 on
SMO expression. The results showed that expression of both mRNA
(Fig. 5A and B) and protein
(Fig. 5C and D) was significantly
decreased by miR-1271 mimics or increased by miR-1271 inhibitor in
MM cells. Taken together, these results indicate that miR-1271
inhibits SMO expression by directly targeting the 3-UTR of SMO.
miR-1271 inhibits the HH signaling
pathway
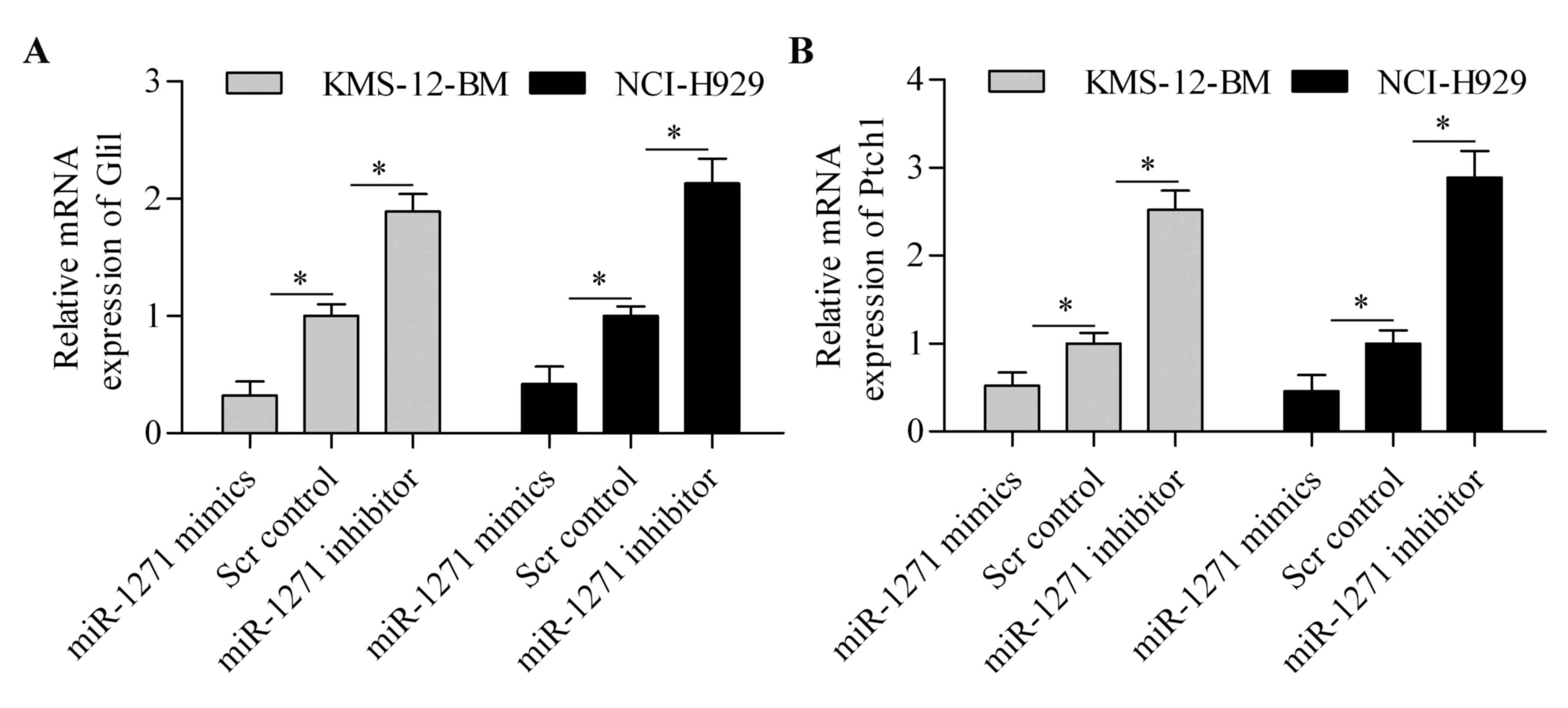
Because SMO is the critical regulator of HH
signaling pathway, we speculated that miR-1271 might affect the HH
signaling pathway. To test this hypothesis, we detected the effect
of miR-1271 on Gli1 and Ptch1 expression. The results showed that
the mRNA expression of Gli1 (Fig.
6A) and Ptch1 (Fig. 6B) was
significantly downregulated by miR-1271 overexpression. By
contrast, suppression of miR-1271 markedly increased the expression
of Gli1 (Fig. 6A) and Ptch1
(Fig. 6B) in MM cells. These
results suggest that miR-1271 regulates the HH signaling
pathway.
Overexpression of SMO reverses the
effect of miR-1271
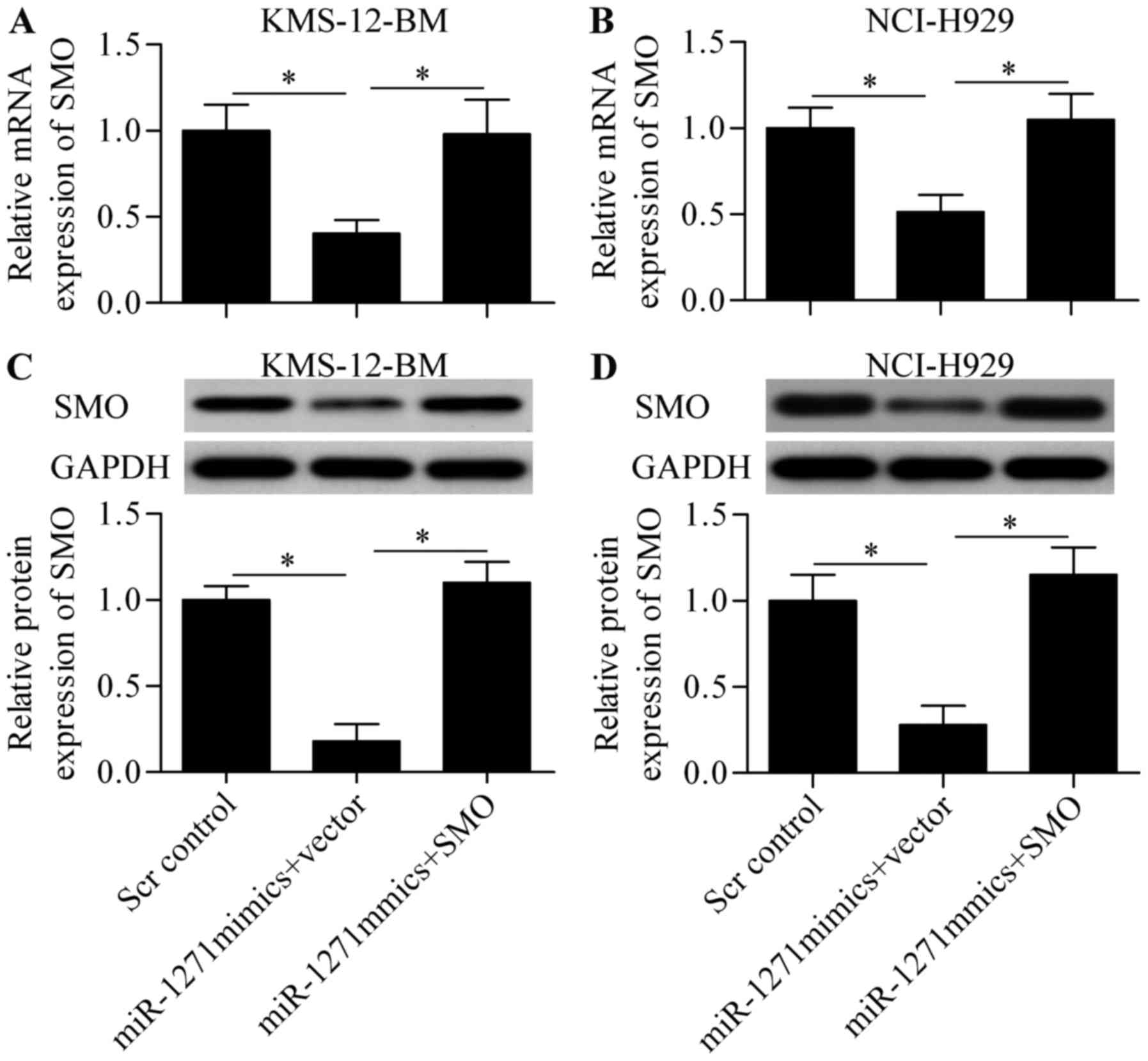
To validate whether miR-1271 functions through SMO,
we constructed a SMO-overexpressing vector harboring no 3-UTR of
SMO and performed a rescue experiment. The cells were
co-transfected with miR-1271 mimics and SMO-overexpressing vector.
The results showed that the decreased mRNA (Fig. 7A and B) and protein (Fig. 7C and D) expression induced by
miR-1271 was significantly restored by SMO-overexpressing vector
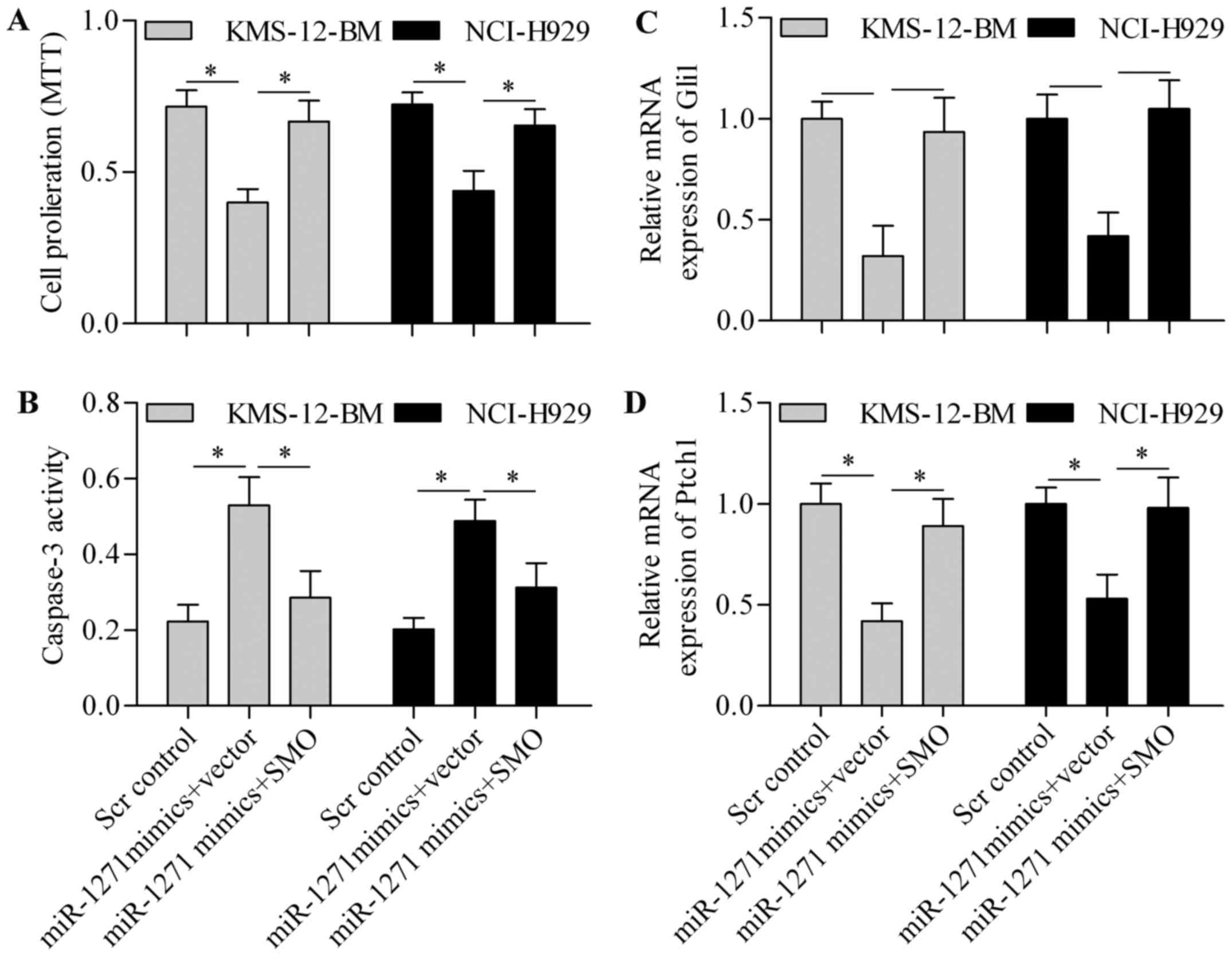
transfection. Notably, overexpression of SMO expression
significantly restored MM cell proliferation, which was suppressed
by miR-1271 (Fig. 8A). Furthermore,
the promotion effect of miR-1271 on cell apoptosis was markedly
reversed by SMO overexpression (Fig.
8B). In addition, the inhibitory effect of miR-1271 on Gli1
(Fig. 8C) and Ptch1 (Fig. 8D) expression was restored by SMO
overexpression, thereby implying that SMO overexpression reversed
the inhibitory effect of miR-1271 on HH signaling pathway.
Discussion
In the present study, we established a tumor
suppressor role of miR-1271 in MM. The expression of miR-1271 was
significantly downregulated in MM cells. Importantly,
overexpression of miR-1271 inhibited proliferation and promoted
apoptosis of MM cells. Further data demonstrated that SMO was a
direct target gene of miR-1271 that overexpression of miR-1271
inhibited SMO expression. Suppression of SMO by miR-1271
overexpression also significantly inhibited the HH signaling
pathway. However, these effects of miR-1271 overexpression were
markedly reversed by SMO overexpression. This study for the first
time report the expression and function of miR-1271 in MM.
miRNAs play an important role in tumorigenesis
acting as either oncogenes or tumor suppressors, representing an
attractive therapeutic target (43). In recent years, miR-1271 has been
reported as tumor-associated genes that are involved in various
cancer types and functions through different target genes. A
functional screening identifies that miR-1271 is a tumor suppressor
in hepatocellular carcinoma that is decreased in tumor samples and
inhibits tumor cell growth through targeting glypican-3 (44). In gastric cancer, miR-1271 is
downregulated and inversely correlated with tumor size, tumor stage
and lymph node metastasis (40).
Yang et al (45)reported
that miR-1271 inhibits gastric cancer cell proliferation and
promotes the sensitization to cisplatin-induced apoptosis through
targeting insulin-like growth factor 1 receptor, insulin receptor
substrate 1, serine/threonine-protein kinase mTOR, and
anti-apoptotic protein Bcl-2. miR-1271 is decreased in oral
squamous cell carcinoma tissues and cell lines and overexpression
of miR-1271 inhibits cell proliferation, colony formation,
migration, and invasion of oral squamous cell carcinoma cells
through targeting anaplastic lymphoma kinase (46). More recently, Liu et al
(37) reported that miR-1271
functioned as tumor suppressor in pancreatic cancer through
targeting ZEB1 and TWIST1. Tumor suppressor role of miR-1271 is
also found in ovarian (39) and
lung cancer (38) through targeting
cyclin G1 or mTOR, respectively. Interestingly, an oncogenic role
of miR-1271 is also reported that miR-1271 promotes cell
proliferation and invasion of non-small cell lung cancer cells
through inhibiting homeobox A5 (47). However, in this study, we have
demonstrated that miR-1271 is downregulated in MM cells and
overexpression of miR-1271 inhibits proliferation and induces
apoptosis of MM cells, supporting a tumor suppressor role of
miR-1271.
SMO is an activator of the HH signaling pathway that
functions as oncogene in various cancers (42). Overexpression of SMO is associated
with aberrant activation of HH signaling pathway (42). Peacock et al (33) reported that HH signaling pathway
maintains a tumor stem cell trait of MM cells. In MM cells,
inhibition of SMO induces a decrease in cell viability and inhibits
the HH signaling pathway (48).
Inhibiting HH signaling pathway inhibits proliferation of MM cells
(34,36). A new drug named vismodegib involves
the inhibition of HH pathway and shows promising results in the
treatment of medulloblastoma and basal-cell carcinoma (49). Therefore, strategies inhibiting HH
signaling represent promising and effective anticancer intervention
for MM.
miRNAs have emerged as promising tools for cancer
treatment because of the inhibitory effect of miRNAs on gene
expression. Indeed, increasing evidence has accumulated on the
potential miRNAs that can directly target and inhibit SMO
expression and the HH signaling pathway. miR-388-3p has been
reported to target SMO to inhibit liver cancer cell invasion
(50) and colorectal cancer cell
growth and invasion (51,52). In chronic myeloid leukemia cells,
inhibition of SMO by miR-326 inhibits cell proliferation as well as
oncogenic HH pathway (53).
Similarly, Du et al (54)
reported that miR-326 inhibits glioma biological behavior and
stemness through targeting and inhibiting SMO-mediated HH signaling
pathway. These findings support the notion that targeting
SMO-mediated HH signaling by specific miRNAs is a promising
strategy for MM. In this study, we identified miR-1271 as a novel
miRNA that can target SMO and inhibit the HH signaling pathway in
MM suggesting a novel and promising molecular target for MM
treatment.
In conclusion, the present study for the first time
demonstrated a tumor suppressive role of miR-1271 in MM. We have
elucidated that miR-1271 inhibits proliferation and induces
apoptosis of MM cells through targeting and inhibition of SMO,
leading to the inhibition of HH signaling pathway. Our findings not
only improve the understanding of MM pathogenesis, but also provide
a potential and promising molecular target for MM therapy
development.
Glossary
Abbreviations
Abbreviations:
|
miRNAs
|
microRNA
|
|
MM
|
multiple myeloma
|
|
SMO
|
smoothened
|
|
UTR
|
untranslated region
|
|
HH
|
Hedgehog
|
|
Gli1
|
glioma-associated oncogene homolog
1
|
References
|
1
|
Hatzimichael E, Dasoula A, Benetatos L,
Syed N, Dranitsaris G, Crook T and Bourantas K: Study of specific
genetic and epigenetic variables in multiple myeloma. Leuk
Lymphoma. 51:2270–2274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kyle RA and Rajkumar SV: Multiple myeloma.
Blood. 111:2962–2972. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Agarwal A and Mahadevan D: Novel targeted
therapies and combinations for the treatment of multiple myeloma.
Cardiovasc Hematol Disord Drug Targets. 13:2–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rossi M, Tagliaferri P and Tassone P:
MicroRNAs in multiple myeloma and related bone disease. Ann Transl
Med. 3:3342015.PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Z, Zhang G, Yu W, Gao N and Peng J:
miR-186 inhibits cell proliferation in multiple myeloma by
repressing Jagged1. Biochem Biophys Res Commun. 469:692–697. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang B, Yin JJ and Zhan XR: MiR-301a
promotes cell proliferation by directly targeting TIMP2 in multiple
myeloma. Int J Clin Exp Pathol. 8:9168–9174. 2015.PubMed/NCBI
|
|
11
|
Lu Y, Wu D, Wang J, Li Y, Chai X and Kang
Q: miR-320a regulates cell proliferation and apoptosis in multiple
myeloma by targeting pre-B-cell leukemia transcription factor 3.
Biochem Biophys Res Commun. 473:1315–1320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saha MN, Abdi J, Yang Y and Chang H:
MiRNA-29a as a tumor suppressor mediates PRIMA-1Met-induced
anti-myeloma activity by targeting c-Myc. Oncotarget. 7:7149–7160.
2016.PubMed/NCBI
|
|
13
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137 and miR-197 induce apoptosis and suppress
tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer
Res. 21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Q, Yan W, Bai Y, Xu H, Fu C, Zheng
W, Zhu Y and Ma J: Synthetic miR-145 mimic inhibits multiple
myeloma cell growth in vitro and in vivo. Oncol Rep. 33:448–456.
2015.PubMed/NCBI
|
|
16
|
Seckinger A, Meißner T, Moreaux J, Benes
V, Hillengass J, Castoldi M, Zimmermann J, Ho AD, Jauch A,
Goldschmidt H, et al: miRNAs in multiple myeloma - a survival
relevant complex regulator of gene expression. Oncotarget.
6:39165–39183. 2015.PubMed/NCBI
|
|
17
|
Sedlaříková L, Bešše L, Novosadová S,
Kubaczková V, Radová L, Staník M, Krejčí M, Hájek R and Ševčíková
S: MicroRNAs in urine are not biomarkers of multiple myeloma. J
Negat Results Biomed. 14:162015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Besse L, Sedlarikova L, Kryukov F,
Nekvindova J, Radova L, Slaby O, Kuglik P, Almasi M, Penka M,
Krejci M, et al: Circulating serum MicroRNA-130a as a novel
putative marker of extramedullary myeloma. PLoS One.
10:e01372942015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ingham PW and McMahon AP: Hedgehog
signaling in animal development: Paradigms and principles. Genes
Dev. 15:3059–3087. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hooper JE and Scott MP: Communicating with
Hedgehogs. Nat Rev Mol Cell Biol. 6:306–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mann RK and Beachy PA: Novel lipid
modifications of secreted protein signals. Annu Rev Biochem.
73:891–923. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taipale J, Cooper MK, Maiti T and Beachy
PA: Patched acts catalytically to suppress the activity of
Smoothened. Nature. 418:892–897. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lum L and Beachy PA: The Hedgehog response
network: Sensors, switches, and routers. Science. 304:1755–1759.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Beachy PA, Karhadkar SS and Berman DM:
Tissue repair and stem cell renewal in carcinogenesis. Nature.
432:324–331. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kinzler KW, Bigner SH, Bigner DD, Trent
JM, Law ML, OBrien SJ, Wong AJ and Vogelstein B: Identification of
an amplified, highly expressed gene in a human glioma. Science.
236:70–73. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee Y, Miller HL, Jensen P, Hernan R,
Connelly M, Wetmore C, Zindy F, Roussel MF, Curran T, Gilbertson
RJ, et al: A molecular fingerprint for medulloblastoma. Cancer Res.
63:5428–5437. 2003.PubMed/NCBI
|
|
27
|
Katoh Y and Katoh M: Hedgehog target
genes: Mechanisms of carcinogenesis induced by aberrant hedgehog
signaling activation. Curr Mol Med. 9:873–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Suzman DL and Antonarakis ES: Clinical
implications of Hedgehog pathway signaling in prostate cancer.
Cancers (Basel). 7:1983–1993. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang L, Walter V, Hayes DN and Onaitis M:
Hedgehog-GLI signaling inhibition suppresses tumor growth in
squamous lung cancer. Clin Cancer Res. 20:1566–1575. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Onishi H and Katano M: Hedgehog signaling
pathway as a new therapeutic target in pancreatic cancer. World J
Gastroenterol. 20:2335–2342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lindemann RK: Stroma-initiated hedgehog
signaling takes center stage in B-cell lymphoma. Cancer Res.
68:961–964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tam M, Lin P, Hu P and Lennon PA:
Examining Hedgehog pathway genes GLI3, SHH, and PTCH1 and the p53
target GLIPR1/GLIPR1L1/GLIPR1L2 gene cluster using fluorescence in
situ hybridization uncovers GLIPR1/GLIPR1L1/GLIPR1L2 deletion in 9%
of patients with multiple myeloma. J Assoc Genet Technol.
36:111–114. 2010.PubMed/NCBI
|
|
33
|
Peacock CD, Wang Q, Gesell GS,
Corcoran-Schwartz IM, Jones E, Kim J, Devereux WL, Rhodes JT, Huff
CA, Beachy PA, et al: Hedgehog signaling maintains a tumor stem
cell compartment in multiple myeloma. Proc Natl Acad Sci USA.
104:4048–4053. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Z, Xu J, He J, Zheng Y, Li H, Lu Y,
Qian J, Lin P, Weber DM, Yang J, et al: A critical role of
autocrine sonic hedgehog signaling in human CD138+
myeloma cell survival and drug resistance. Blood. 124:2061–2071.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
de la Puente P, Muz B, Azab F, Luderer M
and Azab AK: Molecularly targeted therapies in multiple myeloma.
Leukemia Res Treat. 2014:9765672014.
|
|
36
|
Agarwal JR, Wang Q, Tanno T, Rasheed Z,
Merchant A, Ghosh N, Borrello I, Huff CA, Parhami F and Matsui W:
Activation of liver X receptors inhibits hedgehog signaling,
clonogenic growth, and self-renewal in multiple myeloma. Mol Cancer
Ther. 13:1873–1881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu H, Wang H, Liu X and Yu T: miR-1271
inhibits migration, invasion and epithelial-mesenchymal transition
by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem
Biophys Res Commun. 472:346–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Z, Niu X, Li C, Sheng S and Lu S:
Inhibition of the growth of non-small cell lung cancer by
miRNA-1271. Am J Transl Res. 7:1917–1924. 2015.PubMed/NCBI
|
|
39
|
Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li
C and Wang X: MiR-1271 inhibits ovarian cancer growth by targeting
cyclin G1. Med Sci Monit. 21:3152–3158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: MiR-1271 inhibits cell proliferation, invasion
and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hönemann D, Chatterjee M, Savino R,
Bommert K, Burger R, Gramatzki M, Dörken B and Bargou RC: The IL-6
receptor antagonist SANT-7 overcomes bone marrow stromal
cell-mediated drug resistance of multiple myeloma cells. Int J
Cancer. 93:674–680. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rimkus TK, Carpenter RL, Qasem S, Chan M
and Lo HW: Targeting the sonic Hedgehog signaling pathway: Review
of smoothened and GLI inhibitors. Cancers (Basel). 8:82016.
View Article : Google Scholar
|
|
43
|
Chira P, Vareli K, Sainis I, Papandreou C
and Briasoulis E: Alterations of MicroRNAs in solid cancers and
their prognostic value. Cancers (Basel). 2:1328–1353. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maurel M, Jalvy S, Ladeiro Y, Combe C,
Vachet L, Sagliocco F, Bioulac-Sage P, Pitard V, Jacquemin-Sablon
H, Zucman-Rossi J, et al: A functional screening identifies five
microRNAs controlling glypican-3: Role of miR-1271 down-regulation
in hepatocellular carcinoma. Hepatology. 57:195–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang M, Shan X, Zhou X, Qiu T, Zhu W, Ding
Y, Shu Y and Liu P: miR-1271 regulates cisplatin resistance of
human gastric cancer cell lines by targeting IGF1R, IRS1, mTOR, and
BCL2. Anticancer Agents Med Chem. 14:884–891. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kong D, Zhang G, Ma H and Jiang G:
miR-1271 inhibits OSCC cell growth and metastasis by targeting ALK.
Neoplasma. 62:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y, Xu L and Jiang L: miR-1271
promotes non-small-cell lung cancer cell proliferation and invasion
via targeting HOXA5. Biochem Biophys Res Commun. 458:714–719. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Blotta S, Jakubikova J, Calimeri T,
Roccaro AM, Amodio N, Azab AK, Foresta U, Mitsiades CS, Rossi M,
Todoerti K, et al: Canonical and noncanonical Hedgehog pathway in
the pathogenesis of multiple myeloma. Blood. 120:5002–5013. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sandhiya S, Melvin G, Kumar SS and Dkhar
SA: The dawn of hedgehog inhibitors: Vismodegib. J Pharmacol
Pharmacother. 4:4–7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang XH, Chen JS, Wang Q, Chen XL, Wen L,
Chen LZ, Bi J, Zhang LJ, Su Q and Zeng WT: miR-338-3p suppresses
invasion of liver cancer cell by targeting smoothened. J Pathol.
225:463–472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun K, Deng HJ, Lei ST, Dong JQ and Li GX:
miRNA-338-3p suppresses cell growth of human colorectal carcinoma
by targeting smoothened. World J Gastroenterol. 19:2197–2207. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ and
Li GX: MicroRNA-338-3p inhibits colorectal carcinoma cell invasion
and migration by targeting smoothened. Jpn J Clin Oncol. 44:13–21.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Babashah S, Sadeghizadeh M, Hajifathali A,
Tavirani MR, Zomorod MS, Ghadiani M and Soleimani M: Targeting of
the signal transducer Smo links microRNA-326 to the oncogenic
Hedgehog pathway in CD34+ CML stem/progenitor cells. Int
J Cancer. 133:579–589. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Du W, Liu X, Chen L, Dou Z, Lei X, Chang
L, Cai J, Cui Y, Yang D, Sun Y, et al: Targeting the SMO oncogene
by miR-326 inhibits glioma biological behaviors and stemness. Neuro
Oncol. 17:243–253. 2015. View Article : Google Scholar : PubMed/NCBI
|