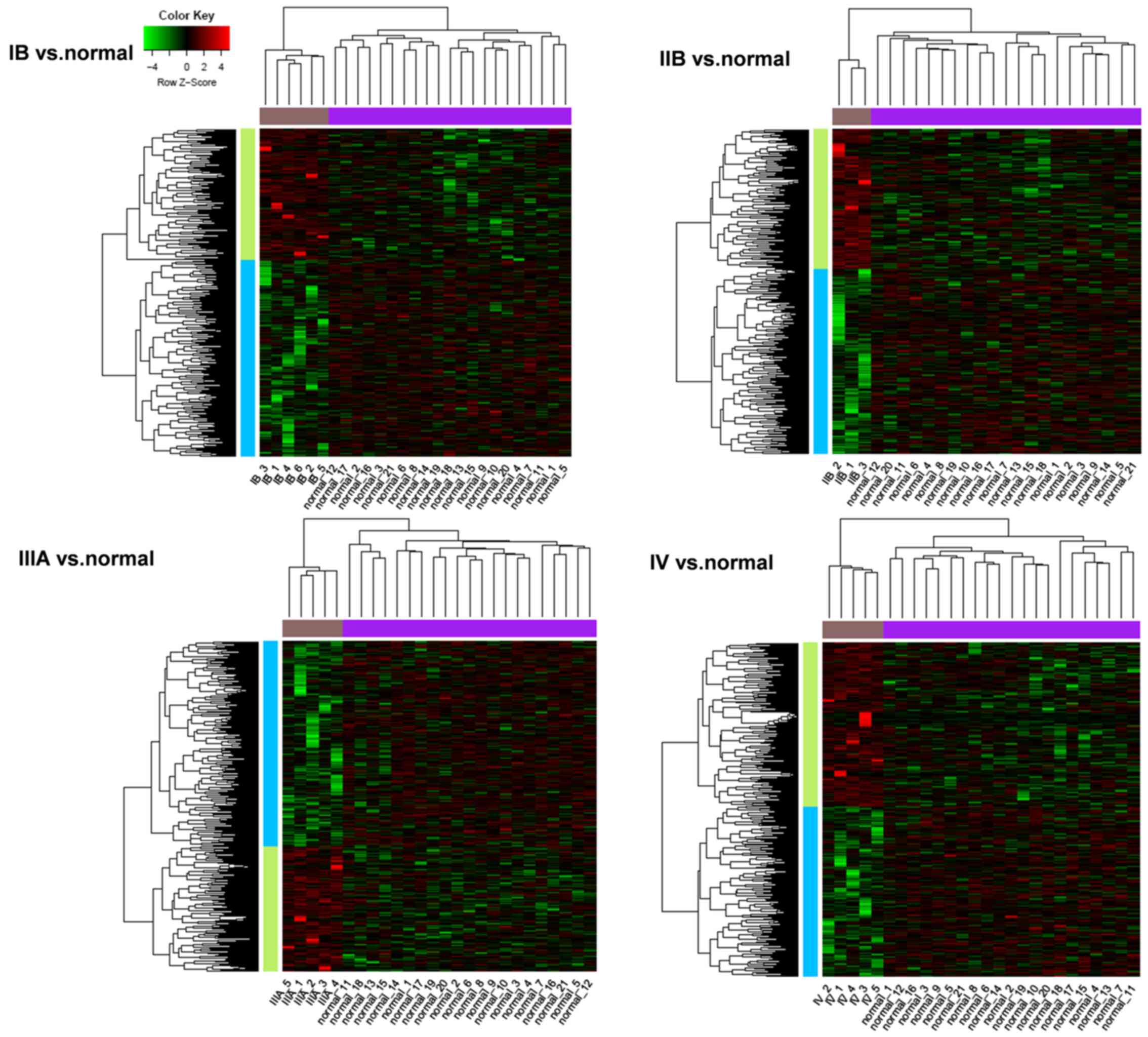
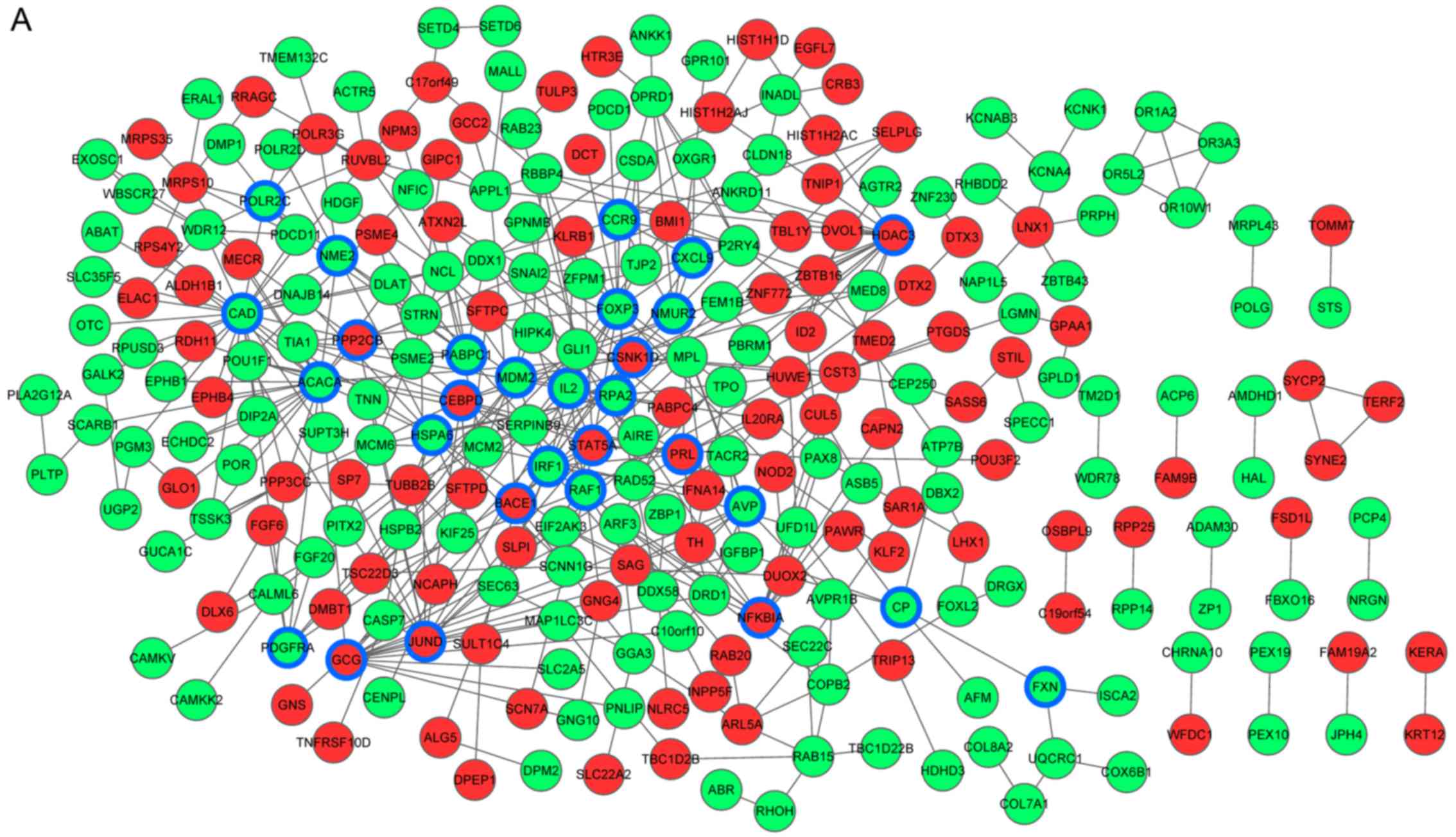
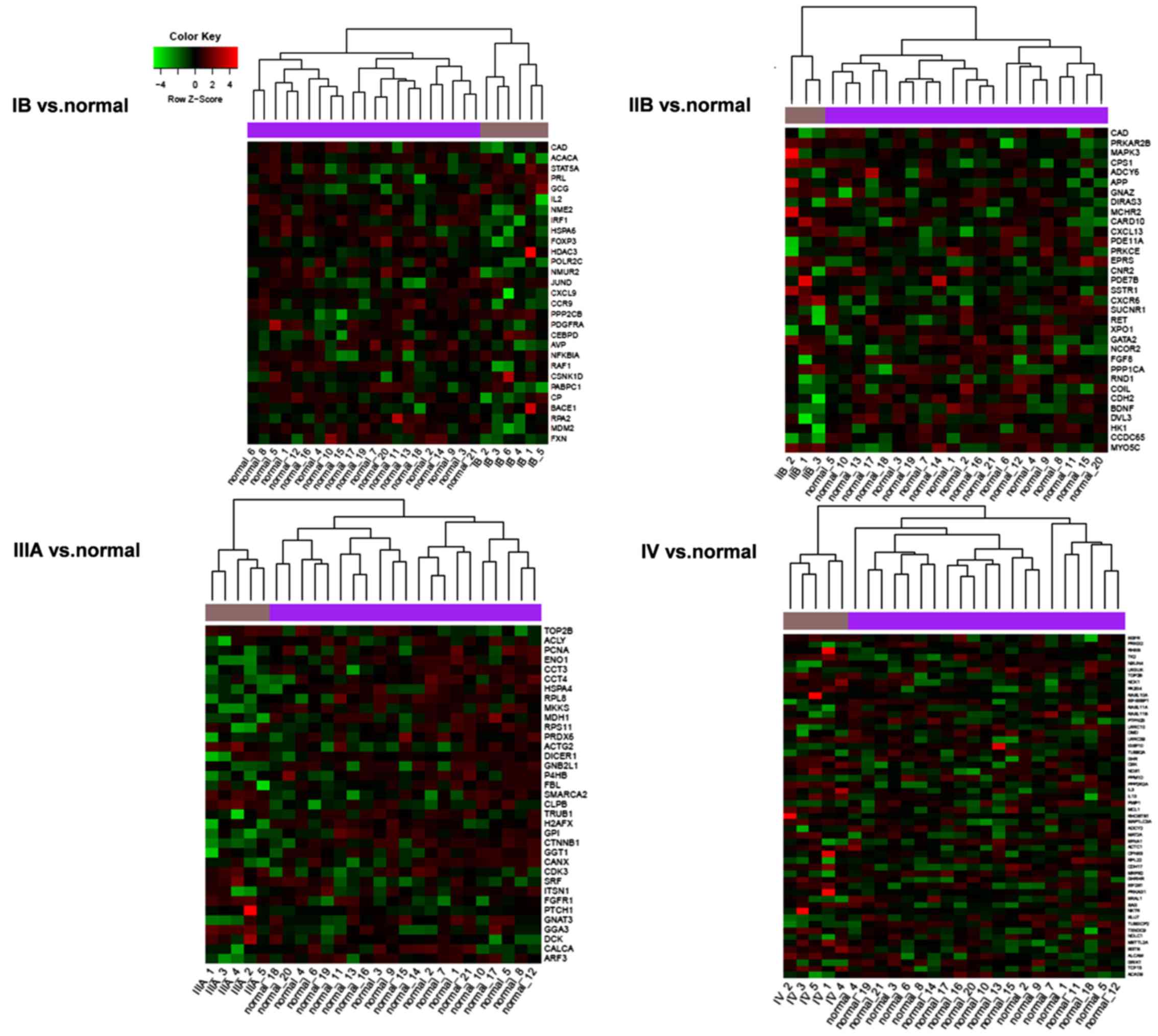
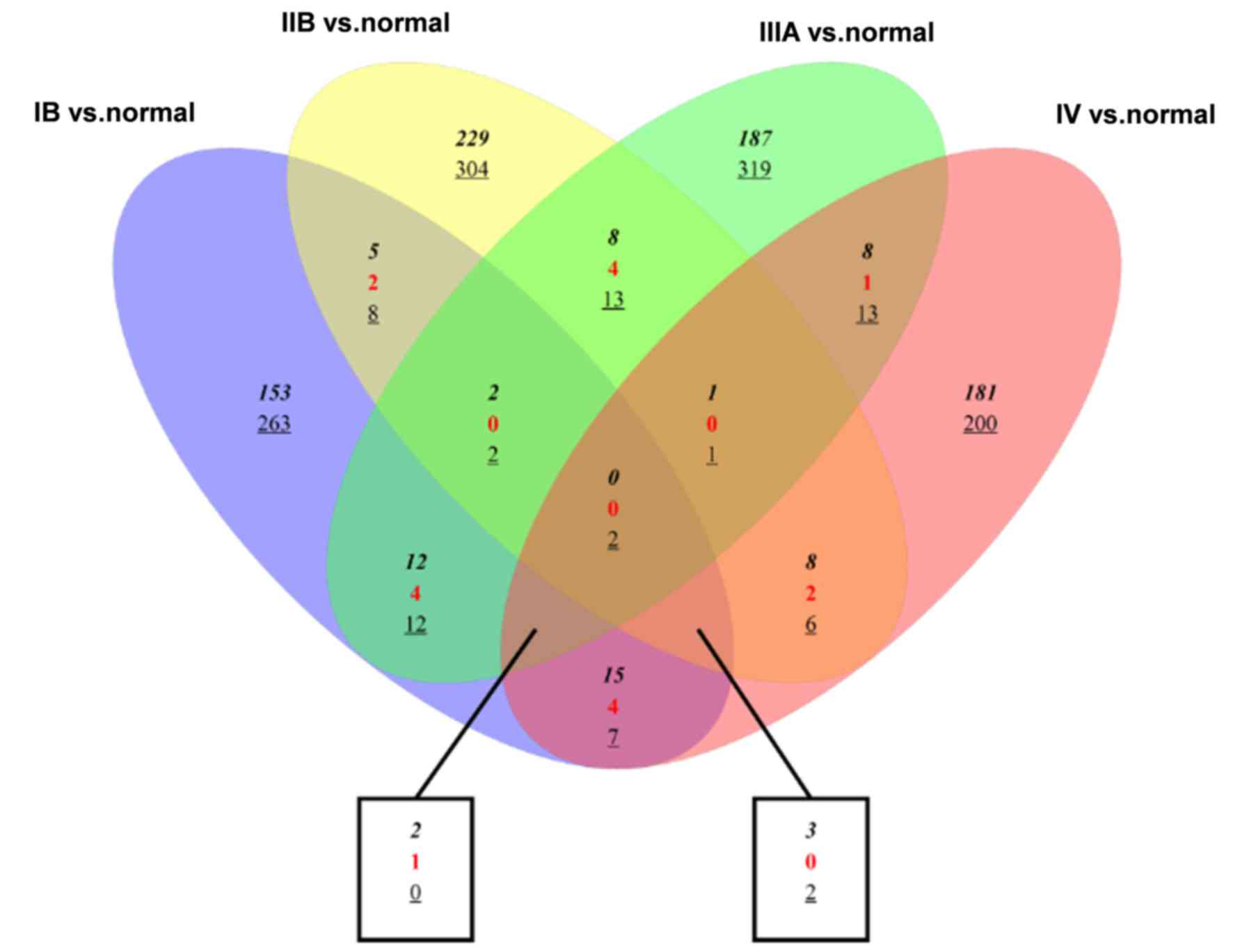
|
1
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Edwards BK, Noone AM, Mariotto AB, Simard
EP, Boscoe FP, Henley SJ, Jemal A, Cho H, Anderson RN, Kohler BA,
et al: Annual Report to the Nation on the status of cancer,
1975–2010, featuring prevalence of comorbidity and impact on
survival among persons with lung, colorectal, breast, or prostate
cancer. Cancer. 120:1290–1314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Strauss GMHJ, Maddaus MA, Johnstone DW,
Johnson EA, Watson DM, Sugarbaker DJ, Schilsky RA, Vokes EE and
Green MR: Adjuvant chemotherapy in stage IB non-small cell lung
cancer (NSCLC): Update of Cancer and Leukemia Group B (CALGB)
protocol 9633. In: ASCO Annual Meeting Abstract. 24:Suppl
20072006.
|
|
4
|
Winton T, Livingston R, Johnson D, Rigas
J, Johnston M, Butts C, Cormier Y, Goss G, Inculet R, Vallieres E,
et al: National Cancer Institute of Canada Clinical Trials Group;
National Cancer Institute of the United States Intergroup JBR.10
Trial Investigators: Vinorelbine plus cisplatin vs. observation in
resected non-small-cell lung cancer. N Engl J Med. 352:2589–2597.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang Q, RT II Dunn, Jayadev S, DiSorbo O,
Pack FD, Farr SB, Stoll RE and Blanchard KT: Assessment of
cisplatin-induced nephrotoxicity by microarray technology. Toxicol
Sci. 63:196–207. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu Y, Lemon W, Liu P-Y, Yi Y, Morrison C,
Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al: A gene
expression signature predicts survival of patients with stage I
non-small cell lung cancer. PLoS Med. 3:e4672006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lo FY, Chang JW, Chang IS, Chen YJ, Hsu
HS, Huang SF, Tsai FY, Jiang SS, Kanteti R, Nandi S, et al: The
database of chromosome imbalance regions and genes resided in lung
cancer from Asian and Caucasian identified by array-comparative
genomic hybridization. BMC Cancer. 12:2352012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smyth GK: limma: linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Springer; NY, New York: pp. 397–420. 2005,
View Article : Google Scholar
|
|
11
|
Warnes GR, Bolker B, Bonebakker L,
Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller
S, Schwartz M, et al: gplots: Various R programming tools for
plotting data. R package version. 2:2009.https://cran.r-project.org/web/packages/gplots
|
|
12
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Opsahl T, Agneessens F and Skvoretz J:
Node centrality in weighted networks: Generalizing degree and
shortest paths. Soc Networks. 32:245–251. 2010. View Article : Google Scholar
|
|
16
|
Goh KI, Oh E, Kahng B and Kim D:
Betweenness centrality correlation in social networks. Phys Rev E
Stat Nonlin Soft Matter Phys. 67:0171012003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Estrada E and Rodríguez-Velázquez JA:
Subgraph centrality in complex networks. Phys Rev E Stat Nonlin
Soft Matter Phys. 71:0561032005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cai H, Chen H, Yi T, Daimon CM, Boyle JP,
Peers C, Maudsley S and Martin B: VennPlex - a novel Venn diagram
program for comparing and visualizing datasets with differentially
regulated datapoints. PLoS One. 8:e533882013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang M, Zhang F, Jin G and Zhu J:
FastGCN: A GPU accelerated tool for fast gene co-expression
networks. PLoS One. 10:e01167762015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Davis AP, Grondin CJ, Lennon-Hopkins K,
Saraceni-Richards C, Sciaky D, King BL, Wiegers TC and Mattingly
CJ: The Comparative Toxicogenomics Database's 10th year
anniversary: Update 2015. Nucleic Acids Res. 43:D914–D920. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gibbons DL, Byers LA and Kurie JM:
Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 12:3–13.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Molina-Vila MA, Bertran-Alamillo J, Gascó
A, Mayo-de-las-Casas C, Sánchez-Ronco M, Pujantell-Pastor L,
Bonanno L, Favaretto AG, Cardona AF, Vergnenègre A, et al:
Nondisruptive p53 mutations are associated with shorter survival in
patients with advanced non-small cell lung cancer. Clin Cancer Res.
20:4647–4659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ilić D, Almeida EA, Schlaepfer DD, Dazin
P, Aizawa S and Damsky CH: Extracellular matrix survival signals
transduced by focal adhesion kinase suppress p53-mediated
apoptosis. J Cell Biol. 143:547–560. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun W, Zhang K, Zhang X, Lei W, Xiao T, Ma
J, Guo S, Shao S, Zhang H, Liu Y, et al: Identification of
differentially expressed genes in human lung squamous cell
carcinoma using suppression subtractive hybridization. Cancer Lett.
212:83–93. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
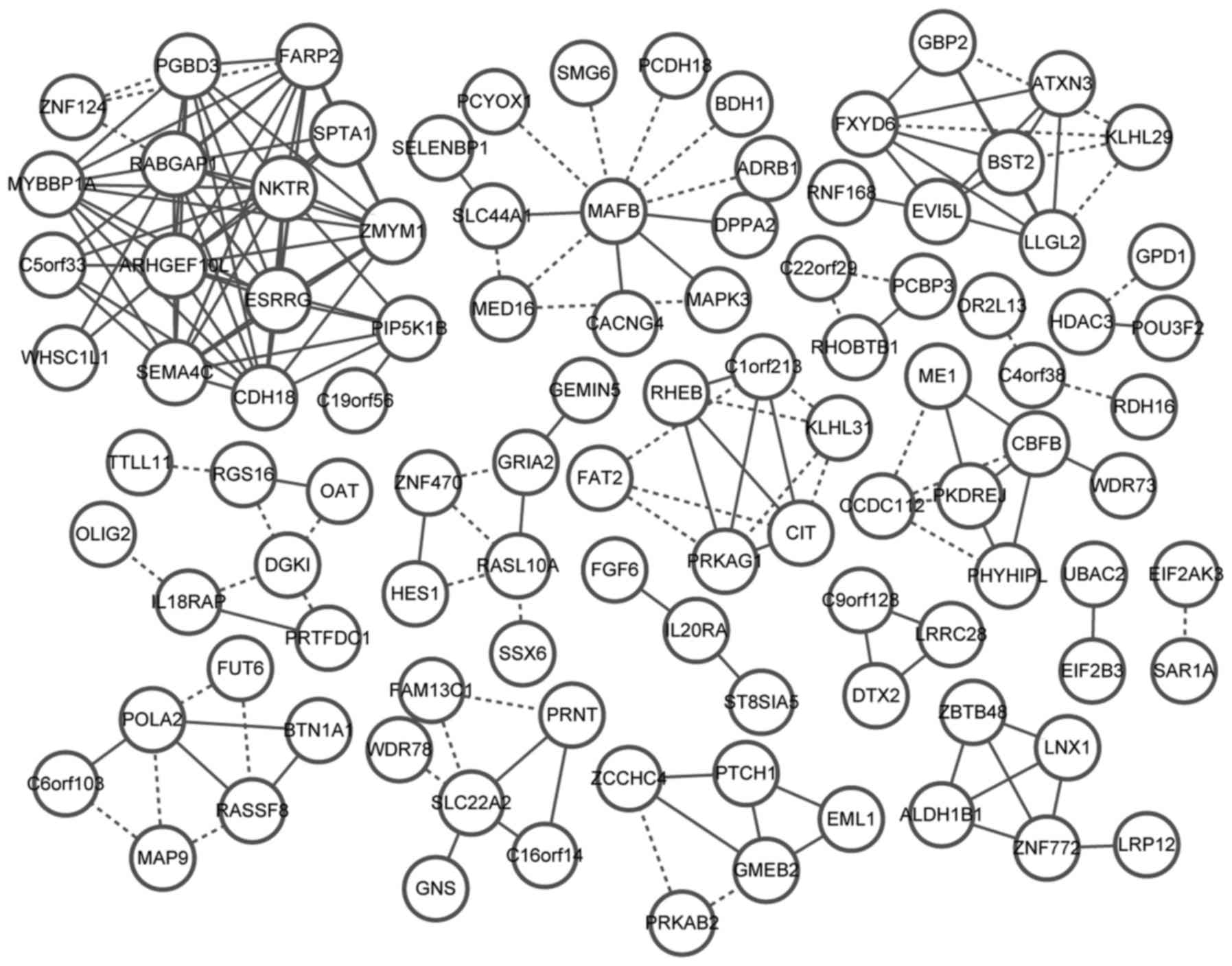
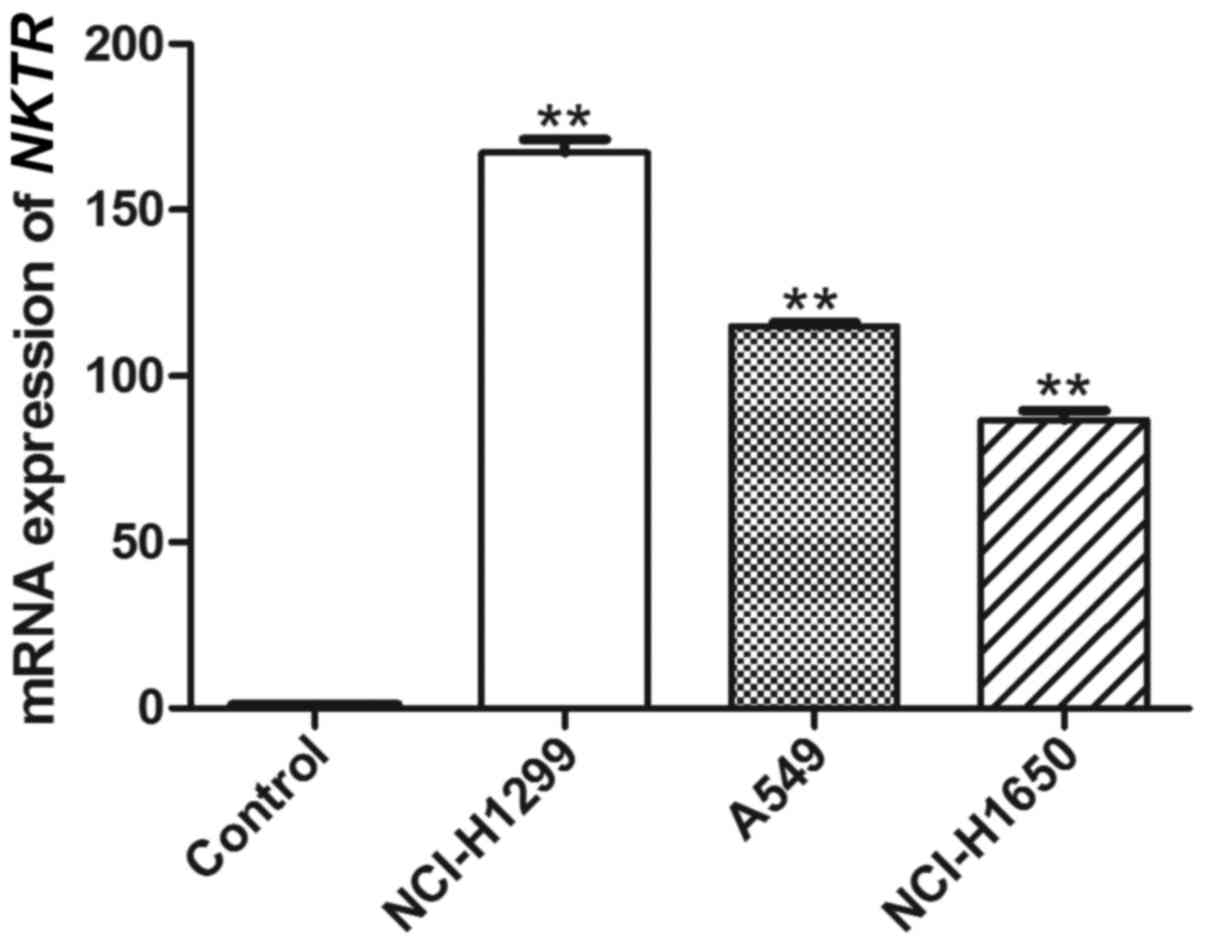
Eldon MA, Staschen CM, Viegas T and
Bentley M: NKTR-102, a novel PEGylated-irinotecan conjugate,
results in sustained tumor growth inhibition in mouse models of
human colorectal and lung tumors that is associated with increased
and sustained tumor SN38 exposureAacr-Nci-Eortc International
Conference on Molecular Targets and Cancer Therapeutics. San
Franciso: 2007
|
|
27
|
Ramadori G, Konstantinidou G,
Venkateswaran N, Biscotti T, Morlock L, Galié M, Williams NS,
Luchetti M, Santinelli A, Scaglioni PP and Coppari R: Diet-induced
unresolved ER stress hinders KRAS-driven lung tumorigenesis. Cell
Metab. 21:117–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stueckle TA, Lu Y, Davis ME, Wang L, Jiang
BH, Holaskova I, Schafer R, Barnett JB and Rojanasakul Y: Chronic
occupational exposure to arsenic induces carcinogenic gene
signaling networks and neoplastic transformation in human lung
epithelial cells. Toxicol Appl Pharmacol. 261:204–216. 2012.
View Article : Google Scholar : PubMed/NCBI
|