Introduction
Osthole
[7-methoxy-8-(3-methyl-2-butenyl)-2H-1-benzopyran-2-one] is one of
effective compounds found in Cnidium monnieri. It has been
shown to exert multiple functions including anti-inflammatory and
antiproliferative effects (1–4).
Recently, it has been reported that osthole induces apoptosis in
hepatocellular carcinoma cells through inhibition of nuclear
factor-κB (NF-κB) activity and modulation of apoptosis-related
genes (2). Osthole was found to
suppress proliferation of ovarian cancer cells by promoting G2/M
arrest and to induce apoptosis (3).
In addition, osthole inhibited epithelial to mesenchymal transition
(EMT)-mediated metastasis through reduction of Snail-DNA-binding
activity and induction of E-cadherin expression (5), and suppressed migration and invasion
of lung cancer cells via inhibition of metalloproteinase (MMP)-2
and −9 levels (6). However, the
anticancer effect of osthole requires further elucidation.
Tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL) was identified to be a member of
the TNF ligand family. TRAIL has been shown to be effective in
inducing apoptosis through death receptor (DR)4 and/or DR5 in a
variety of tumor cells, but not normal cells (7,8). Upon
the binding of TRAIL to DR, DR recruits FAS-associated protein
death domain (FADD) and caspase-8, resulting in the formation of
the death-inducing signal complex (DISC) (9). However, the downregulation of DR
expression and the upregulation of anti-apoptotic proteins (c-FLIP,
Bcl-2, Bcl-xL and IAPs) cause resistance to TRAIL-induced apoptosis
in many cancer cell types (10–14).
Therefore, identification of TRAIL sensitizers is required to
overcome TRAIL resistance.
In the present study, we showed that osthole
enhances TRAIL-induced apoptotic cell death through downregulation
of c-FLIP expression. Osthole may be an efficient apoptosis
sensitizer that can overcome chemoresistance against TRAIL.
Materials and methods
Cell cultures and materials
Human renal carcinoma (Caki), human glioma (U251MG)
and human breast cancer (MDA-MB-231) cells were obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA). The
normal human skin fibroblast (HSF) cells were purchased from the
Korea Cell Line Bank (Seoul, Korea). The culture medium used
throughout these experiments was Dulbecco's modified Eagle's medium
(DMEM) containing 10% fetal bovine serum (FBS), 20 mM HEPES buffer
(all from Welgene, Daegu, Korea) and 100 µg/ml gentamicin (Gibco,
Grand Island, NY, USA). Osthole was purchased from Abcam
(Cambridge, MA, USA). Recombinant human TRAIL and z-VAD-fmk was
purchased from R&D Systems (Minneapolis, MN, USA). Anti-Mcl-1,
anti-Bcl-2, anti-cIAP2, anti-CHOP and anti-ATF4 antibodies were
purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Anti-MnSOD was purchased from Millipore Corp. (Billerica, MA, USA).
Anti-caspase-3 and anti-Grp78 antibodies were purchased from ENZO
(Ann Arbor, MI, USA). Anti-cytochrome c and anti-XIAP
antibodies were purchased from BD Biosciences (San Jose, CA, USA).
Anti-c-FLIP antibody was obtained from Alexis Corporation (San
Diego, CA, USA). Anti-PARP, anti-cleaved caspase-3, and anti-DR5
antibodies were purchased from Cell Signaling Technology (Beverly,
MA, USA). Anti-actin antibody was obtained from Sigma-Aldrich (St.
Louis, MO, USA). Other reagents were purchased from Sigma Chemical
Co. (St. Louis, MO, USA).
Flow cytometric analysis
For flow cytometry, the cells were resuspended in
100 µl of phosphate-buffered saline (PBS), and 200 µl of 95%
ethanol was added while the cells were being vortexed. Then, the
cells were incubated at 4̊C for 1 h, washed with PBS, resuspended
in 250 µl of 1.12% sodium citrate buffer (pH 8.4) with 12.5 µg of
RNase and incubated for an additional 30 min at 37̊C. The cellular
DNA was then stained by adding 250 µl of a propidium iodide
solution (50 µg/ml) to the cells for 30 min at room temperature.
The stained cells were analyzed by fluorescent-activated cell
sorting on a FACScan flow cytometer to determine the relative DNA
content, which was based on the red fluorescence intensity.
Western blot analysis
Cells were washed with cold PBS and lysed on ice in
50 µl of lysis buffer (50 mM Tris-HCl, 1 mM EGTA, 1% Triton X-100,
1 mM phenylmethylsulfonyl fluoride, pH 7.5). Lysates were
centrifuged at 10,000 × g for 15 min at 4̊C, and the supernatant
fractions were collected. Proteins were separated by SDS-PAGE and
transferred to an Immobilon-P membrane (Millipore Corp., Bedford,
MA, USA). Specific proteins were detected using an enhanced
chemiluminescence (ECL) western blot kit (Millipore Corp.)
according to the manufacturer's instructions (15,16).
4′,6′-Diamidino-2-phenylindole (DAPI) staining for
nuclei condensation and fragmentation. To examine cellular nuclei,
the cells were fixed with 1% paraformaldehyde on glass slides for
30 min at room temperature. After fixation, the cells were washed
with PBS and a 300 nM 4′,6′-diamidino-2-phenylindole solution
(Roche, Mannheim, Germany) was added to the fixed cells for 5 min.
After the nuclei were stained, the cells were examined by
fluorescence microscopy.
Cell death assessment by DNA
fragmentation assay
The cell death detection ELISA Plus kit (Boehringer,
Mannheim, Germany) was used for assessing apoptotic activity by
detecting fragmented DNA within the nucleus in the cells treated
with osthole and TRAIL alone, or the combination of osthole and
TRAIL. Briefly, each culture plate was centrifuged for 10 min at
200 × g, the supernatant was removed, and the pellet was lysed for
30 min. After centrifuging the plate again at 200 × g for 10 min,
the supernatant that contained the cytoplasmic histone-associated
DNA fragments was collected and incubated with an immobilized
anti-histone antibody. The reaction products were incubated with a
peroxidase substrate for 5 min and measured by spectrophotometry at
405 and 490 nm (reference wavelength) with a microplate reader. The
signals in the wells containing the substrate alone were subtracted
as the background.
Asp-Glu-Val-Asp-ase (DEVDase) activity
assay
To evaluate DEVDase activity, cell lysates were
prepared after their respective treatments with TRAIL in the
presence or absence of osthole. Assays were performed in 96-well
microtiter plates by incubating 20 µg of cell lysates in 100 µl of
reaction buffer (1% NP-40, 20 mM Tris-HCl, pH 7.5, 137 mM NaCl, 10%
glycerol) containing a caspase substrate
[Asp-Glu-Val-Asp-chromophore-p-nitroanilide (DVAD-pNA)] at 5 µM.
Lysates were incubated at 37̊C for 2 h. Thereafter, the absorbance
at 405 nm was measured with a spectrophotometer.
Determination of the mitochondrial
membrane potential by Rhodamine 123
Rhodamine 123 (Molecular Probes Inc., Eugene, OR,
USA) uptake by mitochondria is directly proportional to its
membrane potential. After treatment, the cells were incubated with
Rhodamine 123 (5 µM) for 5 min in the dark at 37̊C. The cells were
harvested and suspended in PBS. The mitochondrial membrane
potential (MMP) was subsequently analyzed using a flow cytometer
(Becton-Dickinson, Franklin Lakes, NJ, USA).
Analysis of cytochrome c release
The cells were harvested, washed once with ice-cold
PBS and gently lysed for 2 min in 80 µl ice-cold lysis buffer [250
mM sucrose, 1 mM EDTA, 20 mM Tris-HCl (pH 7.2), 1 mM DTT, 10 mM
KCl, 1.5 mM MgCl2, 5 µg/ml pepstatin A, 10 µg/ml
leupeptin and 2 µg/ml aprotinin]. Lysates were centrifuged at
12,000 × g at 4̊C for 10 min to obtain the supernatants (cytosolic
extracts free of mitochondria) and the pellets (fraction that
contains mitochondria). The resulting cytosolic fractions were used
for western blot analysis with an anti-cytochrome c
antibody.
c-FLIP constructs and stable cell
The human c-FLIP expression vector was constructed
as previously described (17). The
Caki cells were transfected in a stable manner with the pcDNA
3.1-c-FLIP(L) plasmid using Lipofectamine as prescribed by the
manufacturer (Invitrogen, Carlsbad, CA, USA). After 48 h of
incubation, the transfected cells were selected in cell culture
medium containing 700 µg/ml G418 (Invitrogen). After two or three
weeks, single independent clones were randomly isolated, and each
individual clone was plated separately. After clonal expansion,
cells from each independent clone were tested for c-FLIP expression
by immunoblotting and were used in the present study.
Statistical analysis
The data were analyzed using a one-way ANOVA and
post hoc comparisons (Student-Newman-Keuls) using the Statistical
Package for Social Sciences 22.0 software (SPSS, Inc., Chicago, IL,
USA). Statistical significance was determined at P≤0.05.
Results
Effect of osthole on TRAIL-mediated
apoptosis in human renal carcinoma Caki cells
We investigated whether osthole could sensitize
TRAIL-mediated apoptosis in human renal carcinoma Caki cells. Cells
were treated with osthole alone (20 and 30 µM), TRAIL alone (50
ng/ml), and combined treatment with osthole and TRAIL. Apoptosis
was determined using flow cytometric and western blot analyses. As
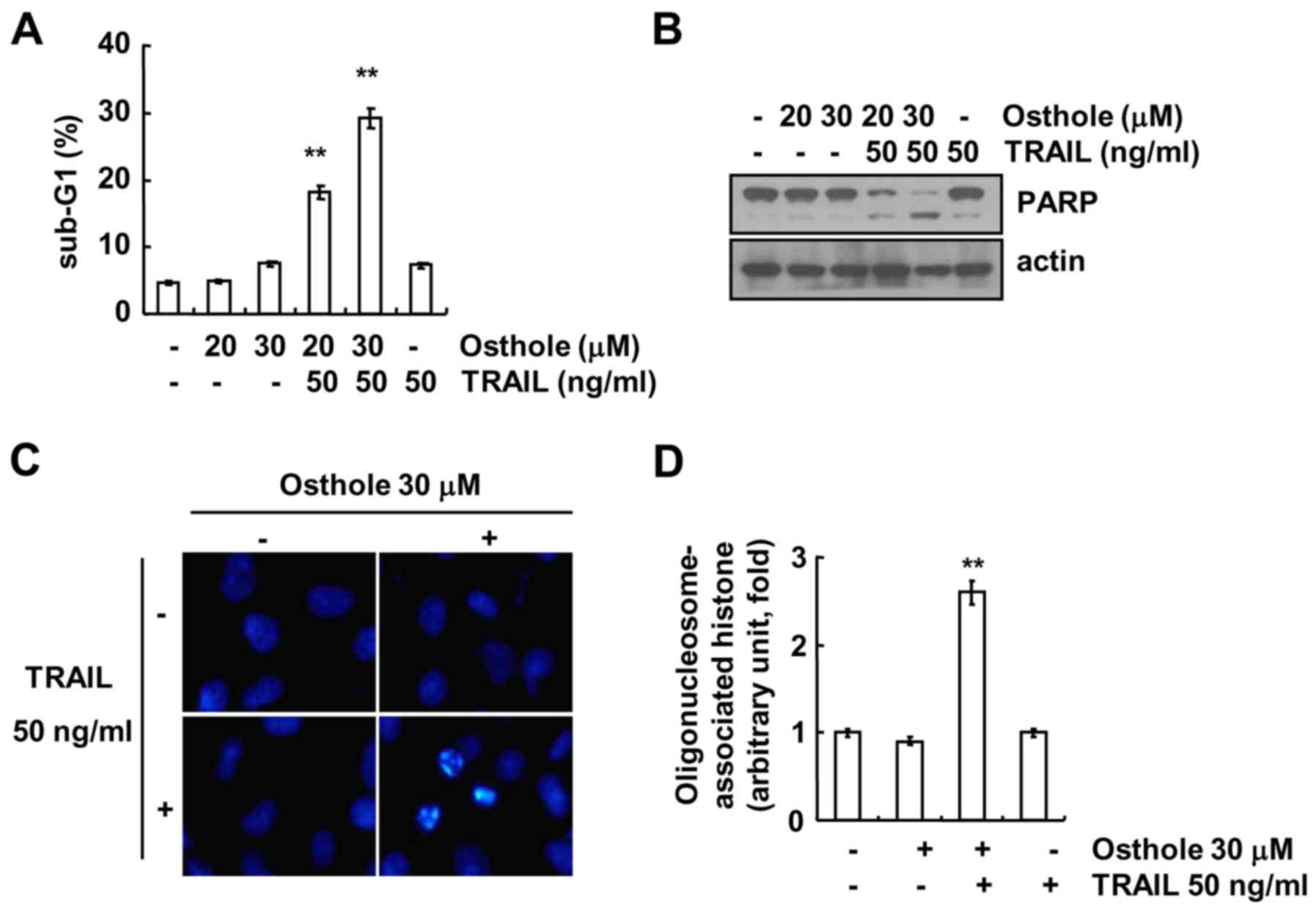
shown in Fig. 1A and B, combined
treatment with osthole and TRAIL markedly induced accumulation of
the sub-G1 population and cleavage of poly(ADP-ribose) polymerase
(PARP). However, treatment with osthole and TRAIL alone had no
effect on apoptosis. Next, we analyzed nuclear condensation and
deoxyribonucleic acid (DNA) fragmentation, which is a hallmark of
apoptosis. Osthole plus TRAIL induced nuclear condensation and DNA
fragmentation (Fig. 1C and D).
Effect of caspase activation on
osthole plus TRAIL-induced apoptosis
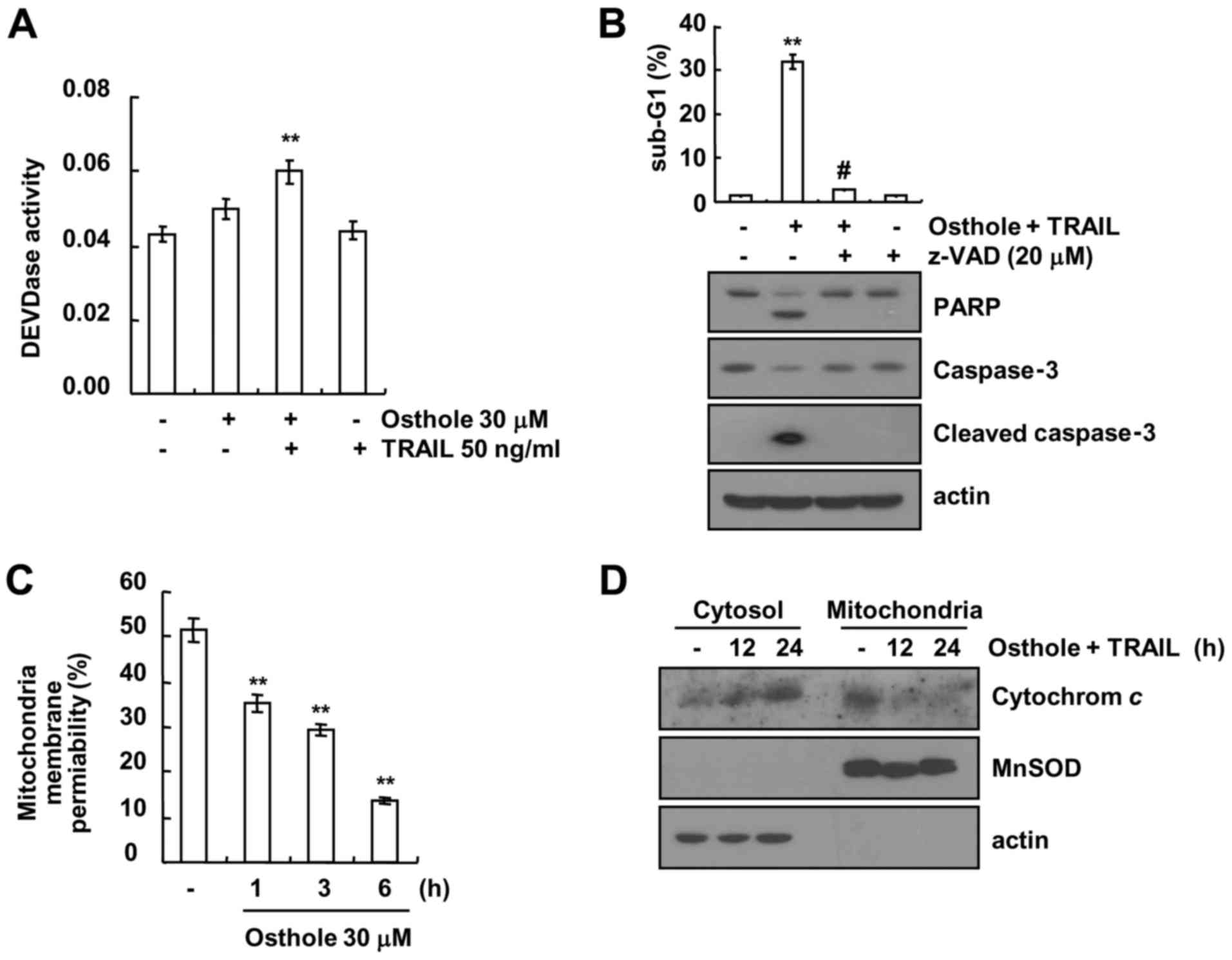
Next, we examined whether caspase activation plays a
critical role in osthole plus TRAIL-induced apoptosis. Combined
treatment with osthole and TRAIL increased caspase-3 activity
(Fig. 2A). To confirm the roles of
caspase activation in the osthole plus TRAIL-induced apoptosis, a
test was conducted to determine whether caspase inhibitors could
attenuate apoptosis. Th sub-G1 population and cleavage of PARP and
caspase-3 were completely prevented by pre-treatment with the
pan-caspase inhibitor, z-VAD-fmk (Fig.
2B). In addition, we examined whether the loss of MMP is
involved in osthole plus TRAIL-induced apoptosis, using Rhodamine
123 fluorescence dye. As shown in Fig.
2C, osthole markedly reduced MMP levels, and increased
cytosolic cytochrome c release following the combined
treatment with osthole and TRAIL (Fig.
2D). These results suggest that combined treatment with osthole
and TRAIL induced apoptosis in Caki cells through a
caspase-dependent pathway.
Effect of combined treatment with
osthole and TRAIL on expression of apoptosis-related proteins
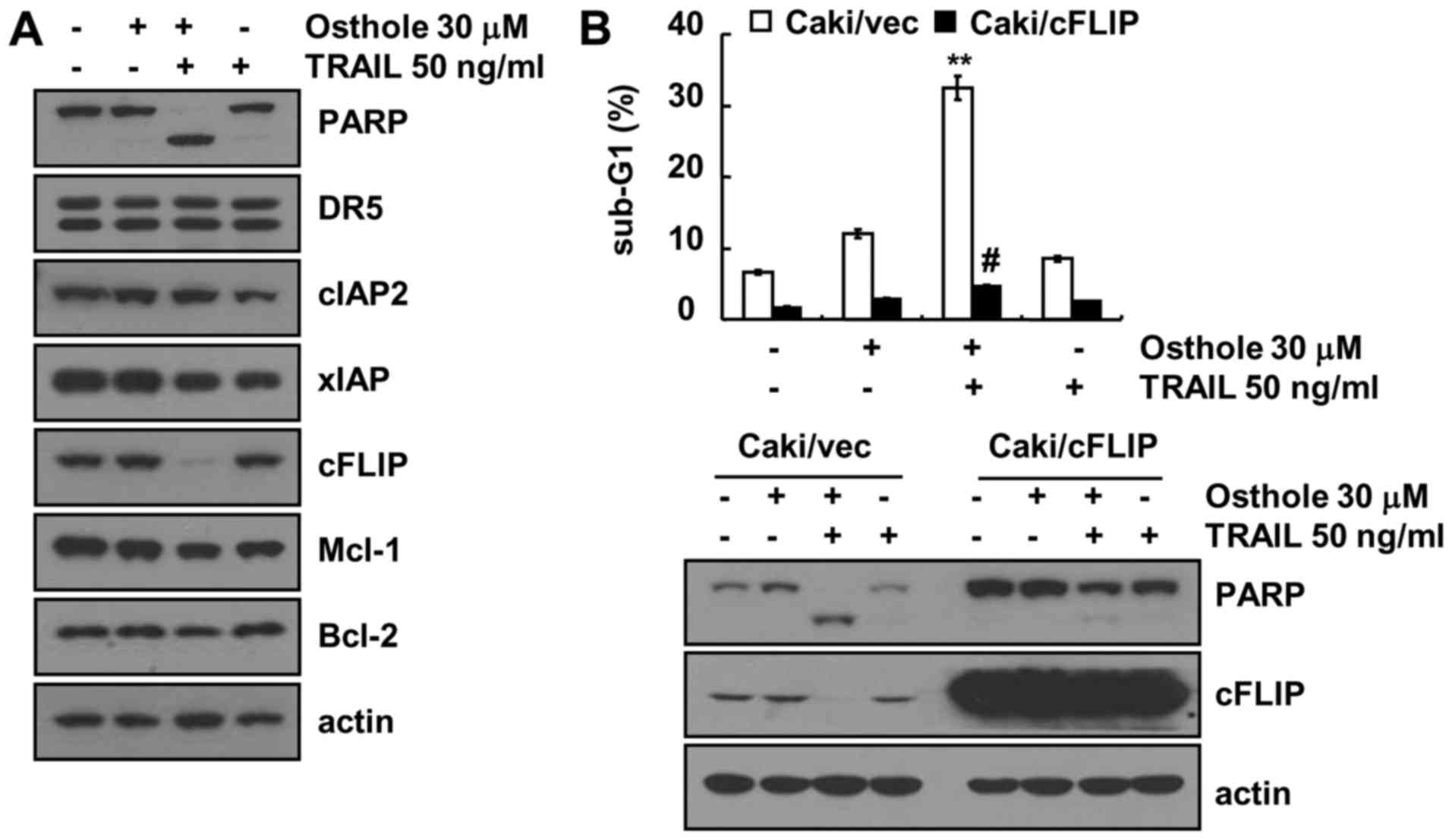
To identify the involvement of apoptosis-related
proteins in the combined treatment-induced apoptosis in Caki cells,
the expression patterns of anti-apoptotic and pro-apoptotic
proteins were investigated. Combined treatment markedly induced
downregulation of c-FLIP expression, whereas expression of
apoptosis-related proteins (DR5, cIAP1, XIAP, Mcl-1 and Bcl-2) did
not change (Fig. 3A). To
investigate the role of the downregulation of c-FLIP protein in
osthole plus TRAIL-induced apoptosis, we used c-FLIP-overexpressing
cells. Combined treatment with osthole and TRAIL induced apoptosis
in Caki/vector cells, while the sub-G1 population and PARP cleavage
were markedly blocked in the ectopic c-FLIP-expressing cells
(Fig. 3B). These data suggest that
the downregulation of c-FLIP expression plays a critical role in
the combined treatment with osthole and TRAIL-induced
apoptosis.
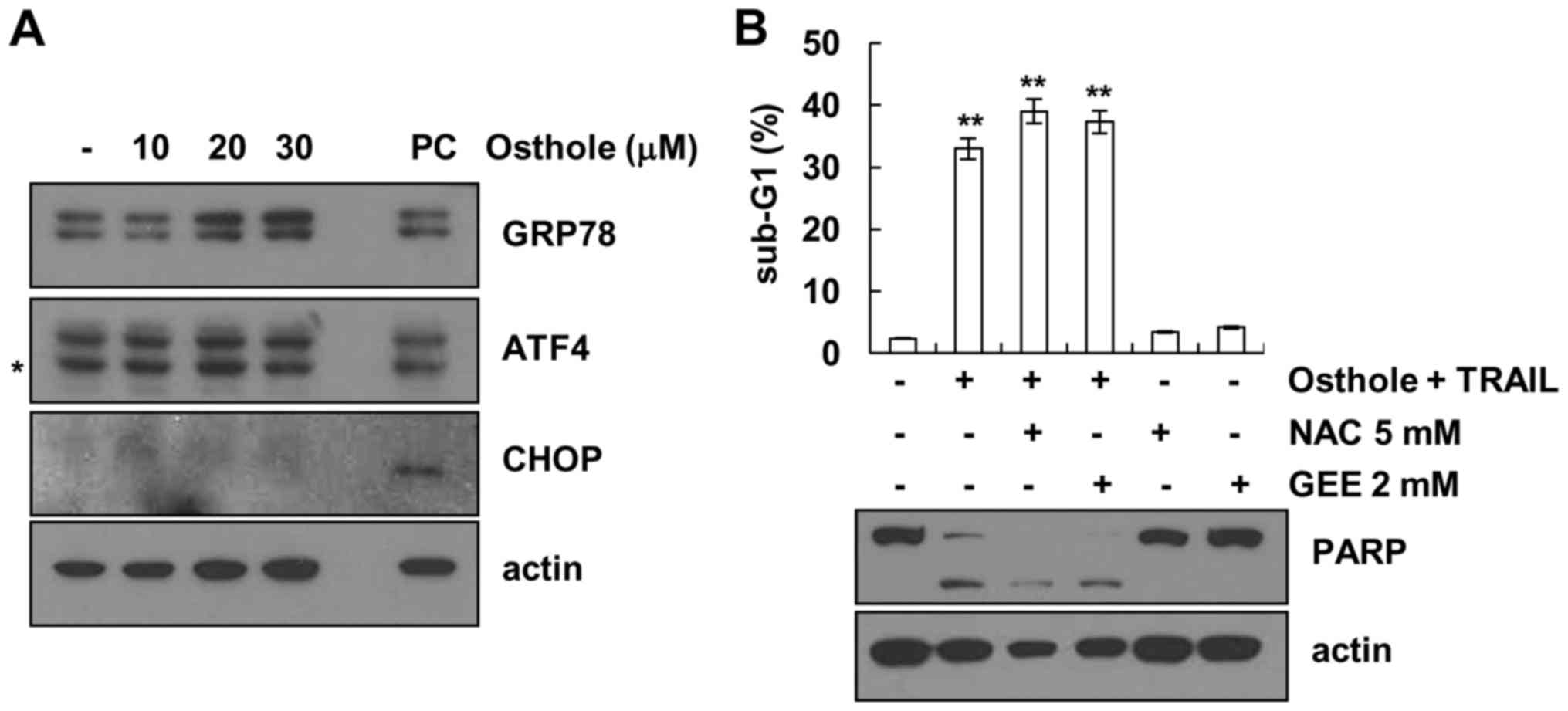
Effect of ER stress and ROS signaling
pathway on osthole-mediated TRAIL sensitization. Next, we
investigated whether osthole induces endoplasmic reticulum (ER)
stress
As shown in Fig. 4A,
protein levels of the 78 kDa glucose-regulated protein (Grp78)
increased by osthole in a dose-dependent manner. However, protein
levels of activating transcription factor 4 (ATF4) and
CCAAT-enhancer-binding protein homologous protein (CHOP) were not
altered in response to osthole. In addition, we investigated
whether reactive oxygen species (ROS) is involved in
osthole-mediated TRAIL sensitization. As shown in Fig. 4B, ROS scavengers (NAC and GEE) had
no effect on osthole plus TRAIL-induced apoptosis. Therefore,
osthole-mediated TRAIL sensitization is independent of ER stress
and ROS signaling.
Effect of combined treatment with
osthole and TRAIL on apoptosis in other cancer and normal
cells
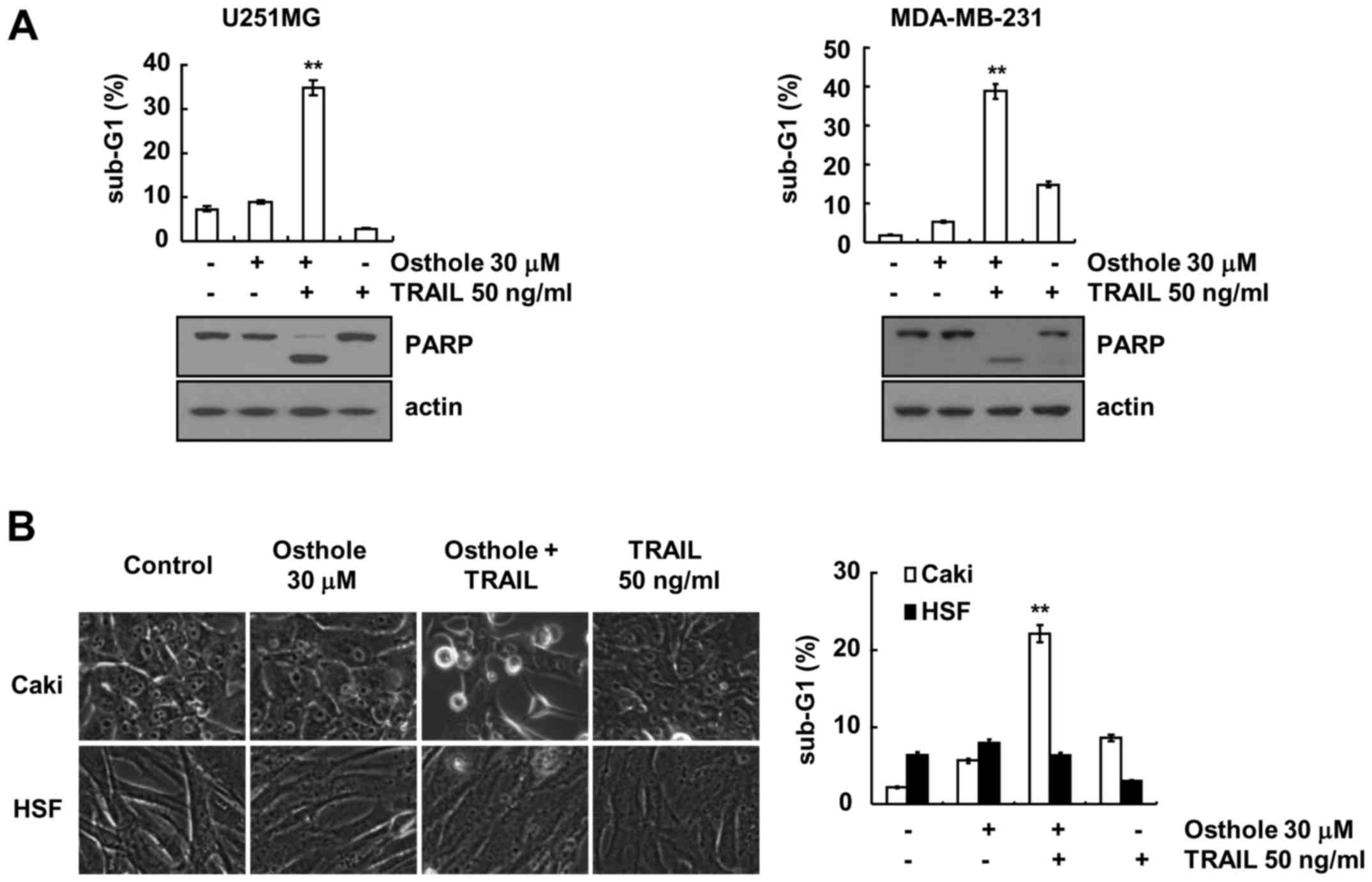
Next, we investigated whether osthole and TRAIL
enhanced apoptosis in other cancer and normal cells. As shown in
Fig. 5A, we found that combined
treatment with osthole and TRAIL enhanced the sub-G1 population and
PARP cleavage in U251MG (glioma) and MDA-MB-231 (breast cancer)
cells. In contrast, osthole plus TRAIL had no effect on
morphological changes and apoptotic cell death in HSF cells
(Fig. 5B). These data indicate that
osthole induces TRAIL-mediated apoptosis in cancer cells, but not
in normal cells.
Discussion
In the present study, we showed that osthole
promotes TRAIL-mediated apoptotic cell death through downregulation
of c-FLIP in human renal carcinoma Caki cells. Furthermore, osthole
markedly reduced MMP levels, and increased cytosolic cytochrome
c release following combined treatment with osthole and
TRAIL. Therefore, these data suggest that osthole could be an
effective TRAIL sensitizer.
Recently, Zhang et al reported that osthole
significantly inhibited hepatocellular carcinoma growth in
vitro and in vivo through cell cycle arrest and induced
apoptosis by suppressing NF-κB activity and promoting the
expression of apoptosis-related genes (18). They used high concentrations
(IC50, >100 µM) of osthole (18). In the present study, a low
concentration of osthole (30 µM) did not induce apoptotic cell
death. However, the combined treatment with osthole (30 µM) and
TRAIL (50 ng/ml) caused apoptotic cell death in Caki, U251 MG and
MDA-MB-231 cells, but not normal cells. Previously several studies
have shown that osthole induces apoptotic cell death in many types
of cancer cells by various signaling pathways. Osthole induces cell
cycle arrest and antitumorigenesis via regulating the PTEN/Akt
pathway (19). In addition, osthole
significantly induces apoptosis by mitochondrial dysfunction via
upregulation of Bax and downregulation of Bcl-2 (20). In the present study, one of the
mechanisms of osthole-mediated TRAIL sensitization was found to be
downregulation of c-FLIP expression. As shown in Fig. 3B, ectopic expression of c-FLIP
markedly blocked apoptosis induced by the combined treatment of
osthole and TRAIL. Overexpression of c-FLIP has been observed in
multiple types of human cancer, and can protect against cell death
receptor-mediated apoptosis through inhibition of caspase-8
recruitment and death-inducing signaling complex (DISC) formation
(21–24).
In the present study, osthole induced Grp78
expression, while CHOP and ATF4 did not change. Upregulation of DR5
and Bim expression by CHOP and ATF4 plays important roles in ER
stress-mediated apoptosis (25).
However, osthole treatment did not induce upregulation of DR5, CHOP
and ATF4. Lv et al reported that osthole prevents tricalcium
phosphate particles-induced osteoclastogenesis and osteolysis in
vivo through inhibition of the ER stress signaling pathway
(26). Therefore, osthole-mediated
TRAIL sensitization is independent of ER stress signaling.
Collectively, these findings revealed that osthole
sensitized TRAIL-mediated apoptosis through the downregulation of
c-FLIP expression in human renal Caki cells. Therefore, osthole may
be an attractive sensitizer for TRAIL-resistant cancer cells.
Acknowledgements
The present study was supported by an NRF grant
funded by the Korea Government (MSIP) (2014R1A5A2010008), and a
2016 Scholar Research Grant from Keimyung University.
Glossary
Abbreviations
Abbreviations:
|
TRAIL
|
tumor necrosis factor-related
apoptosis-inducing ligand
|
|
ROS
|
reactive oxygen species
|
|
DISC
|
death-inducing signaling complex
|
|
HSF
|
human skin fibroblast
|
|
DR
|
death receptor
|
|
FADD
|
FAS-associated protein death
domain
|
|
MMP
|
mitochondrial membrane potential
|
References
|
1
|
Maas M, Deters AM and Hensel A:
Anti-inflammatory activity of Eupatorium perfoliatum L.
extracts, eupafolin, and dimeric guaianolide via iNOS inhibitory
activity and modulation of inflammation-related cytokines and
chemokines. J Ethnopharmacol. 137:371–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chung KS, Choi JH, Back NI, Choi MS, Kang
EK, Chung HG, Jeong TS and Lee KT: Eupafolin, a flavonoid isolated
from Artemisia princeps, induced apoptosis in human cervical
adenocarcinoma HeLa cells. Mol Nutr Food Res. 54:1318–1328. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu K, Park C, Chen H, Hwang J,
Thimmegowda NR, Bae EY, Lee KW, Kim HG, Liu H, Soung NK, et al:
Eupafolin suppresses prostate cancer by targeting
phosphatidylinositol 3-kinase-mediated Akt signaling. Mol Carcinog.
54:751–760. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SR, Park MJ, Lee MK, Sung SH, Park EJ,
Kim J, Kim SY, Oh TH, Markelonis GJ and Kim YC: Flavonoids of Inula
britannica protect cultured cortical cells from necrotic cell death
induced by glutamate. Free Radic Biol Med. 32:596–604. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen YC, Lee WJ, Tan P, Yang SF, Hsiao M,
Lee LM and Chien MH: By inhibiting snail signaling and miR-23a-3p,
osthole suppresses the EMT-mediated metastatic ability in prostate
cancer. Oncotarget. 6:21120–21136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu XM, Zhang Y, Qu D, Feng XW, Chen Y and
Zhao L: Osthole suppresses migration and invasion of A549 human
lung cancer cells through inhibition of matrix metalloproteinase-2
and matrix metallopeptidase-9 in vitro. Mol Med Rep.
6:1018–1022. 2012.PubMed/NCBI
|
|
7
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang S and El-Deiry WS: TRAIL and
apoptosis induction by TNF-family death receptors. Oncogene.
22:8628–8633. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kischkel FC, Lawrence DA, Chuntharapai A,
Schow P, Kim KJ and Ashkenazi A: Apo2L/TRAIL-dependent recruitment
of endogenous FADD and caspase-8 to death receptors 4 and 5.
Immunity. 12:611–620. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Walczak H, Bouchon A, Stahl H and Krammer
PH: Tumor necrosis factor-related apoptosis-inducing ligand retains
its apoptosis-inducing capacity on Bcl-2- or
Bcl-xL-overexpressing chemotherapy-resistant tumor
cells. Cancer Res. 60:3051–3057. 2000.PubMed/NCBI
|
|
11
|
Kelly MM, Hoel BD and Voelkel-Johnson C:
Doxorubicin pretreatment sensitizes prostate cancer cell lines to
TRAIL induced apoptosis which correlates with the loss of c-FLIP
expression. Cancer Biol Ther. 1:520–527. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ng CP, Zisman A and Bonavida B: Synergy is
achieved by complementation with Apo2L/TRAIL and actinomycin D in
Apo2L/TRAIL-mediated apoptosis of prostate cancer cells: Role of
XIAP in resistance. Prostate. 53:286–299. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin Z, McDonald ER III, Dicker DT and
El-Deiry WS: Deficient tumor necrosis factor-related
apoptosis-inducing ligand (TRAIL) death receptor transport to the
cell surface in human colon cancer cells selected for resistance to
TRAIL-induced apoptosis. J Biol Chem. 279:35829–35839. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y and Zhang B: TRAIL resistance of
breast cancer cells is associated with constitutive endocytosis of
death receptors 4 and 5. Mol Cancer Res. 6:1861–1871. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jo HS, Yeo HJ, Cha HJ, Kim SJ, Cho SB,
Park JH, Lee CH, Yeo EJ, Choi YJ, Eum WS, et al: Transduced
Tat-DJ-1 protein inhibits cytokines-induced pancreatic RINm5F cell
death. BMB Rep. 49:297–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han EH, Kim HG, Lee EJ and Jeong HG:
Endosulfan induces CYP1A1 expression mediated through aryl
hydrocarbon receptor signal transduction by protein kinase C.
Toxicol Res. 31:339–345. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim S, Lee TJ, Leem J, Choi KS, Park JW
and Kwon TK: Sanguinarine-induced apoptosis: Generation of ROS,
down-regulation of Bcl-2, c-FLIP, and synergy with TRAIL. J Cell
Biochem. 104:895–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang L, Jiang G, Yao F, He Y, Liang G,
Zhang Y, Hu B, Wu Y, Li Y and Liu H: Growth inhibition and
apoptosis induced by osthole, a natural coumarin, in hepatocellular
carcinoma. PLoS One. 7:e378652012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Yang L, Lu Y, Chen Y, Liu T, Peng
Y, Zhou Y, Cao Y, Bi Z, Liu T, et al: Osthole induces cell cycle
arrest and inhibits migration and invasion via PTEN/Akt pathways in
osteosarcoma. Cell Physiol Biochem. 38:2173–2182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding Y, Lu X, Hu X, Ma J and Ding H:
Osthole inhibits proliferation and induces apoptosis in human
osteosarcoma cells. Int J Clin Pharmacol Ther. 52:112–117. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oyarzo MP, Medeiros LJ, Atwell C,
Feretzaki M, Leventaki V, Drakos E, Amin HM and Rassidakis GZ:
c-FLIP confers resistance to FAS-mediated apoptosis in anaplastic
large-cell lymphoma. Blood. 107:2544–2547. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou XD, Yu JP, Liu J, Luo HS, Chen HX and
Yu HG: Overexpression of cellular FLICE-inhibitory protein (FLIP)
in gastric adenocarcinoma. Clin Sci. 106:397–405. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ryu BK, Lee MG, Chi SG, Kim YW and Park
JH: Increased expression of cFLIPL in colonic
adenocarcinoma. J Pathol. 194:15–19. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Pan X, Zhang H, Lei D, Liu D, Xu F
and Luan X: Overexpression of cFLIP in head and neck squamous cell
carcinoma and its clinicopathologic correlations. J Cancer Res Clin
Oncol. 134:609–615. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jung KJ, Min KJ, Bae JH and Kwon TK:
Carnosic acid sensitized TRAIL-mediated apoptosis through
down-regulation of c-FLIP and Bcl-2 expression at the post
translational levels and CHOP-dependent up-regulation of DR5, Bim,
and PUMA expression in human carcinoma caki cells. Oncotarget.
6:1556–1568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv S, Zhang Y, Yan M, Mao H, Pan C, Gan M,
Fan J and Wang G: Inhibition of osteolysis after local
administration of osthole in a TCP particles-induced osteolysis
model. Int Orthop. 40:1545–1552. 2016. View Article : Google Scholar : PubMed/NCBI
|