Introduction
Colorectal carcinoma (CRC) is the second most
frequent cause of cancer-related deaths (1–4).
Although the survival of patients with CRC has improved along with
marked advances in diagnostic and therapeutic modalities, prognosis
for CRC remains poor (5).
Accordingly, there is an urgent need to develop new strategies for
treating CRC.
Ribosome biogenesis by eukaryotes is a complex,
subtly-regulated, and energy-consuming process (6). RNA polymerases I–III (RNAPs) and over
200 assembly factors (including accessory proteins and small
nucleolar RNAs (snoRNAs) are required to manufacture the ribosome
in the nucleolus (7,8). Underlying the intricateness of
ribosome biogenesis are many pre-rRNA processing events that make
the terminal mature rRNAs, the large catalytic subunits (LSU), and
the small recognition subunits (SSU) of the ribosome (9). In eukaryotic cells, a large
ribonucleoprotein (RNP), called the SSU processome, is involved in
forming the SSU of the ribosome (10). The SSU processome is assembled
co-transcriptionally with the 47S precursor pre-rRNA in the
nucleolus (11), and is a complex
of the U3 snoRNA and over 70 associated proteins, including the U3
proteins (UTPs). The SSU processome is required for the assembly of
the ribosomal SSU and for the maturation of the 18S rRNA SSU
component (12). Studies have
demonstrated that some defects in the assembly of ribosomes,
notably arising from mutations in various factors involved in
ribosome biogenesis and components of the SSU processome, results
in human diseases, the so-called ribosomopathies (7).
A mutation in the SSU processome component,
hUTP4/Cirhin, results in North American Indian childhood cirrhosis
(NAIC/CIRH1A; OMIM: 604901); a severe autosomal recessive
intrahepatic cholestasis. Cirhin (NP_116219) is encoded by the
CIRH1A gene (13). All NAIC
patients have a homozygous mutation in CIRH1A that changes the
conserved Arg565 to Trp (R565W) in Crihin (14). Presently, the only effective
treatment for the disease is liver transplantation (15). A study showed that knockdown of
CIRH1A caused biliary defects in zebrafish (16). However, no functional information is
available so far for CIRH1A in human CRC.
In the present study, we analyzed the level of
CIRH1A expression in pairs of colon and para-rectum adenocarcinomas
in silico and in CRC cell lines in vitro.
Subsequently, we transduced the RKO CRC cell line with a
lentivirus-delivered small interfering RNA (siRNA) to study the
impact CIRH1A knockdown has on the growth of human CRC cells in
vitro.
Materials and methods
In silico expression analyses
RNAseq and RNAseqV2 data derived from 23 paired
samples of colon and rectum adenocarcinomas were obtained from The
Cancer Genome Atlas (TCGA) database (17). Data were normalized using trimmed
mean of M-values (18) and quality
controlled according to the observed biological coefficient of
variation (19). Negative binomial
dispersion was performed for assessing differences in gene
expression (20–22). A P-value of <0.05 and fold
changes >2.0 between carcinoma and para-carcinomas were
considered statistically significant.
Cell lines
HCT116, RKO, LoVo, and HT-29 human colorectal cell
lines were obtained from the Shanghai Cell Bank (Shanghai, China).
Cell lines were maintained in RPMI-1640 medium (Gibco®,
Shanghai, China) supplemented with 10% fetal bovine serum (FBS;
Zhejiang Tianhang Biotech Co. Ltd., Huzhou, China), 100 U/ml
penicillin and 0.1 mg/ml streptomycin (Sangon Biotech Co. Ltd.,
Shanghai, China) at 37°C in a 5% CO2 incubator.
RT-qPCR
Total RNA was extracted using the TRIzol®
reagent (Invitrogen, Shanghai, China) and reverse transcribed into
cDNA with a PrimeScript® 1st Strand cDNA Synthesis kit
(Takara, Dalian, China) completed according to the manufacturer's
instructions. Next, 1 µl of cDNA was used as a template for
real-time quantitative PCR (qPCR). The sequences for CIRH1A primers
were 5′-TGAGTCTCGGGCTACAGAAG-3′ (forward) and
5′-GCATACTTGATGTTTAACGCCTG-3′ (reverse). The sequences for GAPDH
internal control primers were 5′-TGACTTCAACAGCGACACCCA-3′ (forward)
and 5′-CACCCTGTTGCTGTAGCCAAA-3′ (reverse). Each qPCR occurred over
an initial denaturation at 95°C for 20 sec, followed with 45 cycles
of denaturation at 95°C for 5 sec and extension 60°C for 30 sec.
The PCR products of CIRH1A and GAPDH were 114 and 121 bp,
respectively. All samples were examined in triplicates. Relative
quantitation of gene expression was calculated as described
previously (23).
Construction of recombinant lentiviral
vector and cell transduction
A siRNA that targets the human UTP4/CIRH1A
gene (Genbank no. NM_032830) with view of specifically knocking
down RIHR1A expression was designed from the full-length
UTP4/CIRH1A sequence by GeneChem Co. Ltd. (Shanghai, China).
The siRNA sequence was TTG TGA AGA GCC ATC TCA T. For testing
knockdown efficiencies, the stem-loop oligonucleotides were
synthesized and inserted into a lentivirus-based pGV115-GFP
(GeneChem Co. Ltd.) with AgeI/EcoRI sites. Lentivirus
particles were prepared as described previously (24).
For cell transduction, RKO cells (2×105
cells/well) were cultured in 6-well plates and infected with either
a CIRH1A-siRNA (shCIRH1A) lentivirus or negative control (shCtrl)
lentivirus at a multiplicity of infection (MOI) of 20. Cells were
incubated in a 5% CO2 incubator at 37°C for 5 days.
After 72 h of transduction, cells were observed under a
fluorescence microscope (MicroPublisher 3.3RTV; Olympus, Tokyo,
Japan). After 5 days of transduction, the knockdown efficiency was
determined with qPCR and western blotting.
Western blotting
The expression of CIRH1A was determined at the
protein level by immunostaining with a specific anti-CIRH1A
antibody. After 48 h of lentiviral infection, cells were lysed
using lysis buffer (50 mM Tris, pH 7.4, 150 mMNaCl, 1% SDS, 1 mM
EDTA, 1% NP-40) containing 1 mM PMSF (Sangon Biotech Co. Ltd.) for
30 min on ice. The lysates were centrifuged at 10,000 × g for 10
min at 4°C, and the supernatants were collected. Protein
concentration was determined using a BCA Protein assay kit (Sangon
Biotech Co. Ltd.). Next, 10 mg protein sample of each treatment was
separated using 12.5% SDS-PAGE as per the Laemmli method (25), and transferred to polyvinylidene
difluoride (PVDF) membrane (Sangon Biotech Co. Ltd.).
Membranes were incubated with mouse anti-FLAG
(Sigma-Aldrich®, Shanghai, China) or anti-GAPDH
antibodies (1:1,000 dilution, Santa Cruz Biotechnology, Santa Cruz,
CA, USA) at 4°C overnight. Membranes were then subsequently
developed with a horseradish peroxidase (HRP)-conjugated goat
anti-mouse IgG (1:1,500 dilution, Santa Cruz Biotechnology) at 37°C
for 1 h and was detected with EasyBlot ECL kit (Sangon
Biotech).
Cell growth assay
Cell growth was measured by multiparametric
high-content screening (HCS) performed with slight modifications to
the protocol described previously (26). Briefly, infected RKO cells within
the logarithmic growth phase were seeded in 96-well plates (2,000
cells/well) and incubated for 5 days at 37°C in a 5% CO2
incubator. At least 800 cells/well in the plates were counted using
the Cellomics ArrayScan™ VT1 HCS automated reader (Cellomics Inc.
Pittsburgh, PA, USA) for measuring cell growth each day for all 5
days of growth. Each experiment was performed in triplicate.
Methyl-thiazol-tetrazolium (MTT)
assay
Infected RKO cells (2×103 cells) were
reseeded into 96-well plate suspended in 100 µl medium per well,
and cultured at 37°C. The proliferation of cells was detected at
days 1, 2, 3, 4 and 5. Briefly, 20 µl MTT (5 mg/ml, Sigma-Aldrich,
USA) per well was added and incubated for 4 h at 37°C. After
removing the cell media, 150 µl dimethyl sulfoxide (DMSO,
Sigma-Aldrich) was added to each well for dissolution of the
crystals. The absorbance was measured at 570 nm.
Cell cycle distribution and
apoptosis
Cell cycle distribution or apoptosis was analyzed
using flow cytometry as described previously (27). Briefly, RKO cells were infected with
shCIRH1A or shCtrl plasmids and incubated at 37°C for 1, 2, 3, 4 or
5 days. At each time point, cells were collected, washed twice with
ice-cold PBS, fixed with 0.5 ml ice-cold 70% ethanol for 1 h at
4°C, and stained with propidium iodide (50 µg/ml; Sigma-Aldrich in
the presence of RNase A (100 µg/ml; Sangon Biotech). The cell cycle
distribution was alluded from the DNA content analyzed with a BD
FACSCalibur Flow Cytometer (BD Biosciences, San Diego, CA, USA).
Each experiment was performed in triplicate.
Cell apoptosis with the Annexin V-APC
stain detection by flow cytometry
Briefly, RKO cells (1,000 cells/well) were cultured
in 6-well plates. After 48 h of infection with either an shCIRH1A
or shCtrl plasmid, cells were collected and washed twice with
ice-cold PBS. The cell concentrations were adjusted to
1×106/ml with 1X staining buffer (Sangon Biotech), of
which 100 µl of cell suspension was stained with 5 µl Annexin V-APC
(BD Biosciences) for 15 min at room temperature in the dark. Cells
were analyzed using flow cytometry within 1 h of staining. Each
experiment was performed in triplicate.
Colony formation assay
CIRH1A-siRNA and control cells were resuspended in
RPMI-1640 medium at logarithmic growth phase. Cells were seeded
onto 6-well plates at a density of 800 cells/well. The cells were
incubated over a period of 14 days. Cell colonies were photographed
by fluorescence microscopy (MicroPublisher 3.3RTV; Olympus, Tokyo,
Japan). The cells were fixed with paraformaldehyde (1 ml/well;
Sangon Biotech) for 30 min. The cells were washed with PBS and then
stained with 500 µl Giemsa (Sangon Biotech) for 20 min. Then, the
cells were washed with ddH2O several times and left to
dry at room temperature. A digital camera was used for imaging and
to obtain colony counts.
Statistical analyses
Statistical analyses were performed with SPSS
version 16.0 for Windows (SPSS, Chicago, IL, USA). Data are
expressed as the mean ± SD. Raw data were submitted to Student's
t-test to analyze for differences between two groups. A P-value of
<0.05 was considered statistically significant.
Results
CIRH1A gene expression is markedly
higher in colorectal cancer over para-carcinoma tissue
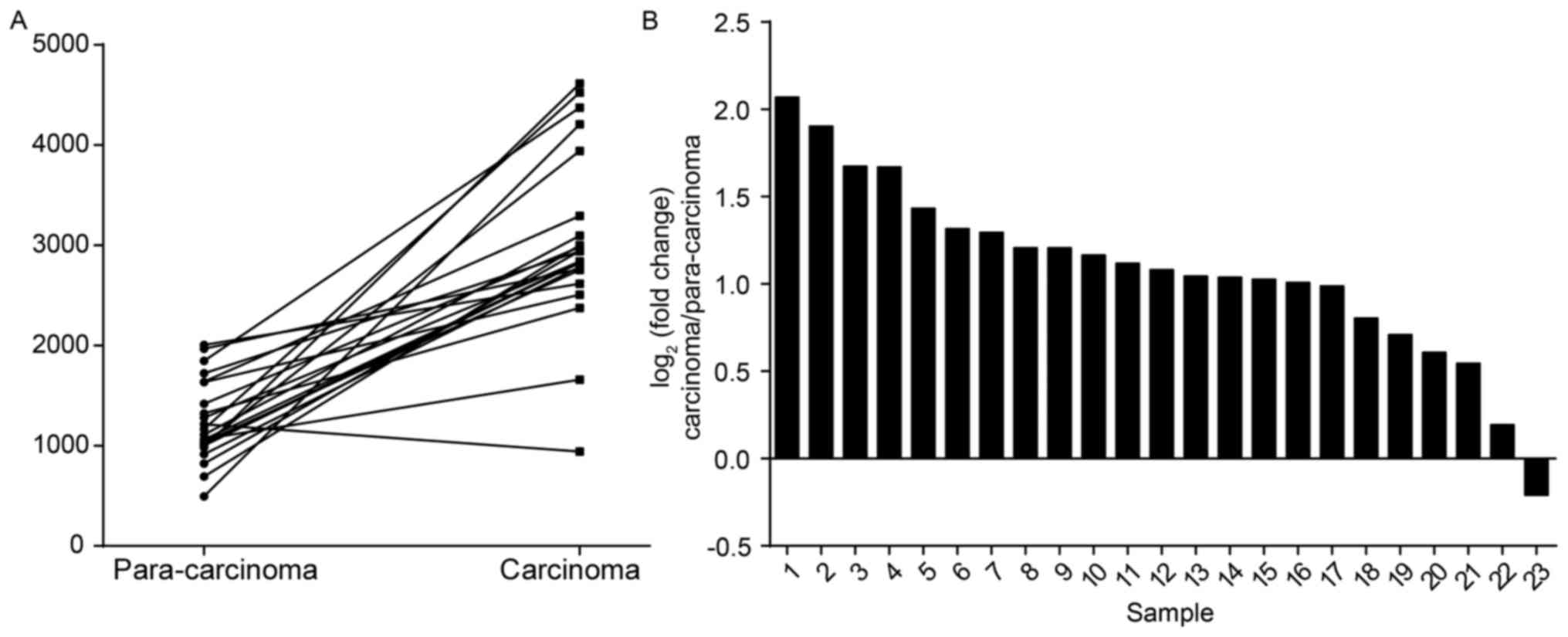
To examine a possible link of CIRH1A and colorectal
cancer, we analyzed the gene expression data of 23 colorectal
cancer cases with paired para-carcinoma tissues from The Cancer
Genome Atlas (TCGA). The data revealed a highly significant
correlation between CIRH1A mRNA expression and colorectal cancer
(Fig. 1A; P<0.01). The average
expression of CIRH1A was more than 2-fold greater in CRC tissues
over para-carcinomas (Fig. 1B).
CIRH1A mRNA detection in four
colorectal cancer cells
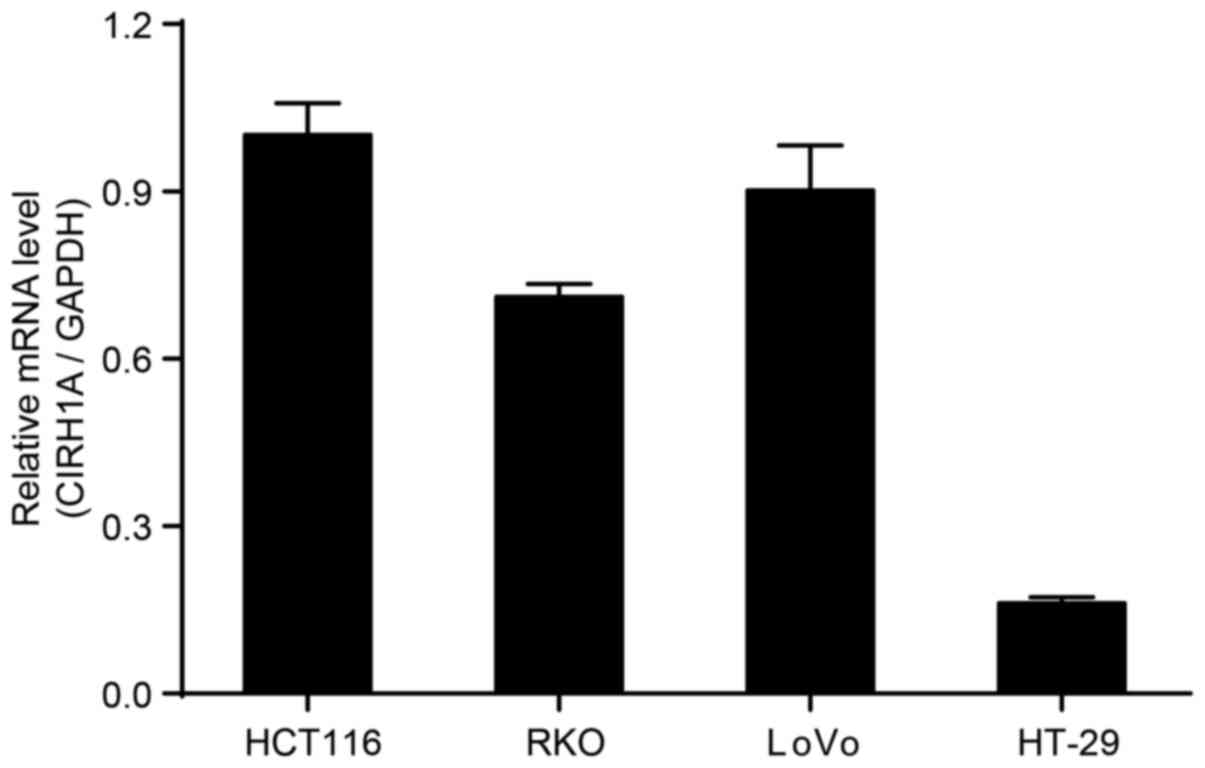
We determined the expression of CIRH1A mRNA in the
HT119, RKO, LoVo, and HT-29 CRC cell lines by RT-PCR. The data
showed that CIRH1A mRNA was highly expressed in HT119, RKO,
and LoVo cell lines (Fig. 2).
Lentivirus-mediated knockdown of
CIRH1A in RKO cells
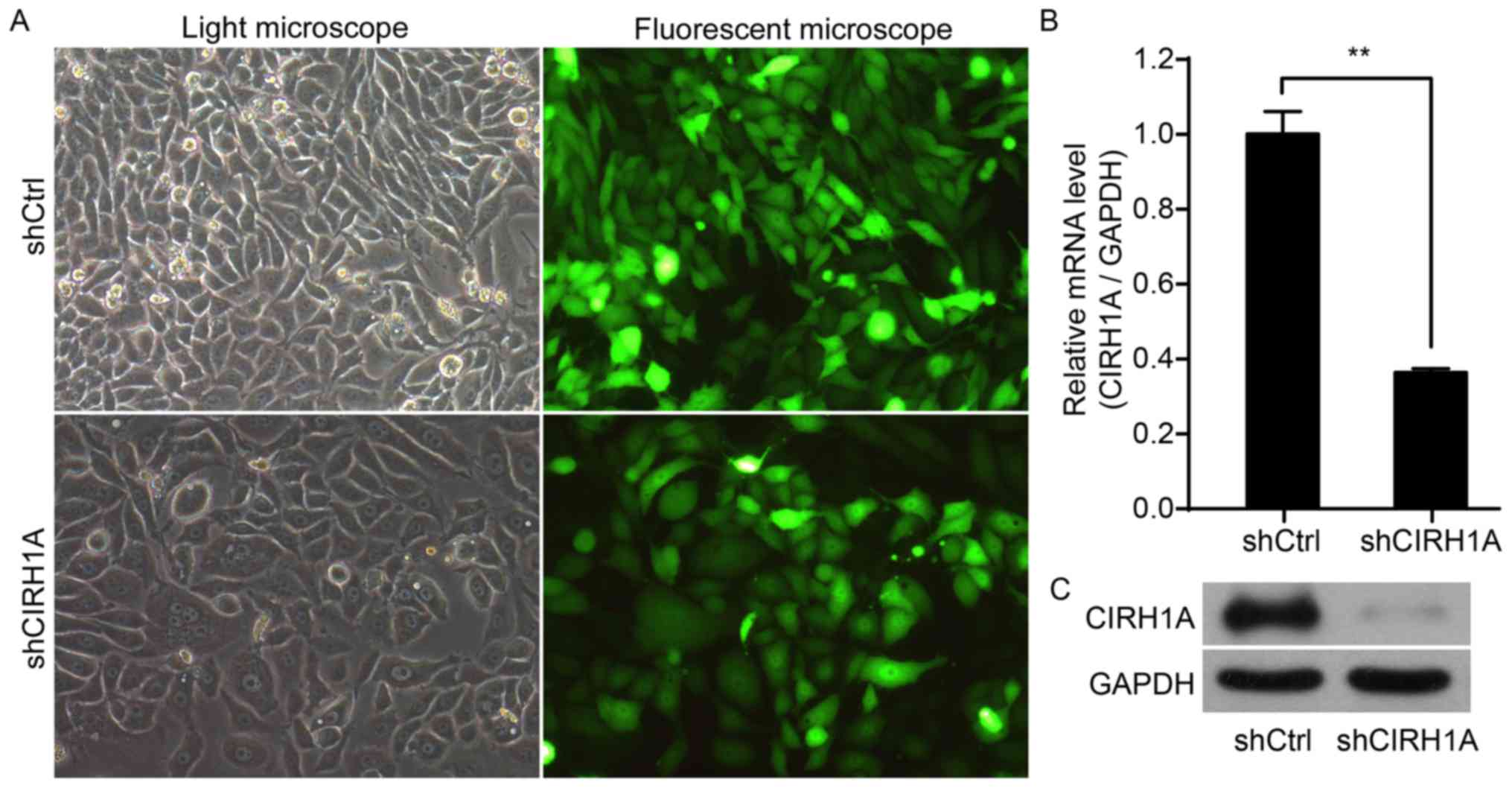
To explore the role of CIRH1A, we knocked down the
expression of CIRH1A in the RKO cell line. At 3 days
post-infection, >80% of the cells were successfully infected
with either a shCIRH1A lentivirus or shCtrl lentivirus (Fig. 3A). As determined by qPCR at 5 days
post-infection, shCIRH1A-infected cultures had significantly lower
levels of CIRH1A mRNA compared to levels in control cultures
infected with a shCtrl lentivirus (Fig.
3B). Western blotting for the CIRH1A protein confirmed that
CIRH1A levels were greatly reduced in cells infected with a
shCIRH1A payload, thereby indicating an effective knockdown of the
target gene (Fig. 3C).
Knocking down CIRH1A in RKO cells
inhibits cell proliferation
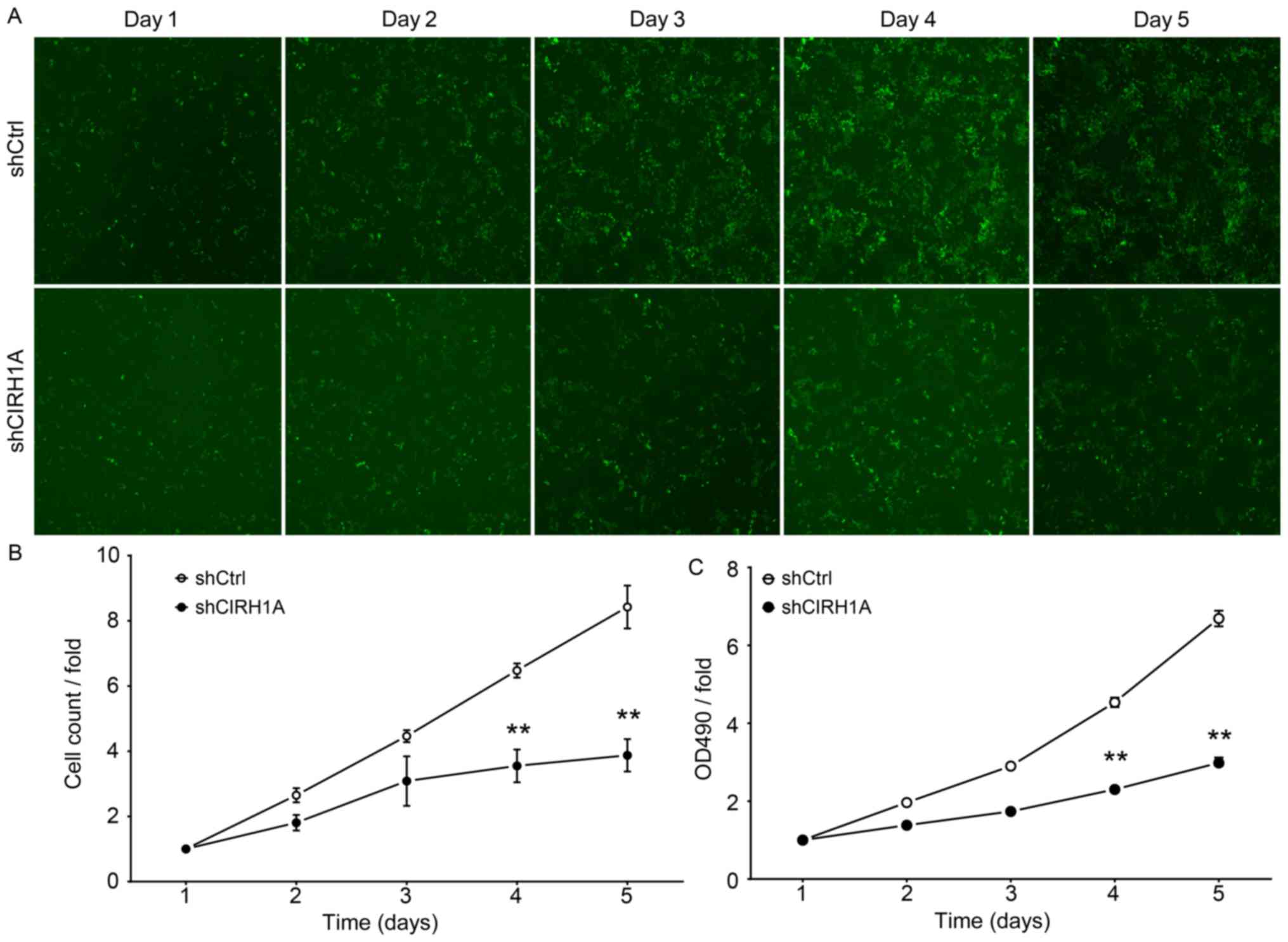
To examine the influence CIRH1A has on cell growth,
RKO cells expressing either a shCIRH1A lentivirus or shCtrl
lentivirus were seeded in 96-well plates and analyzed by Cellomics
every day for 5 days. shCtrl-transduced cells expanded markedly
over the 5 days of the experiment, while the number of
shCIRH1A-transduced cells increased only slightly by comparison
(Fig. 4). The data from this
experiment suggest that CIRH1A knockdown significantly
inhibited the proliferation of RKO cells.
The effect of CIRH1A protein reduction on RKO cell
proliferation was also determined with MTT assay. Although shCtrl
and shCIRH1A cells had similar growth on days 1–3, cells transduced
with shCIRH1A had significantly suppressed growth on days 4 and 5
by comparison (Fig. 4C). Seemingly
the growth of RKO cells in vitro is dependent on CIRH1A
expression.
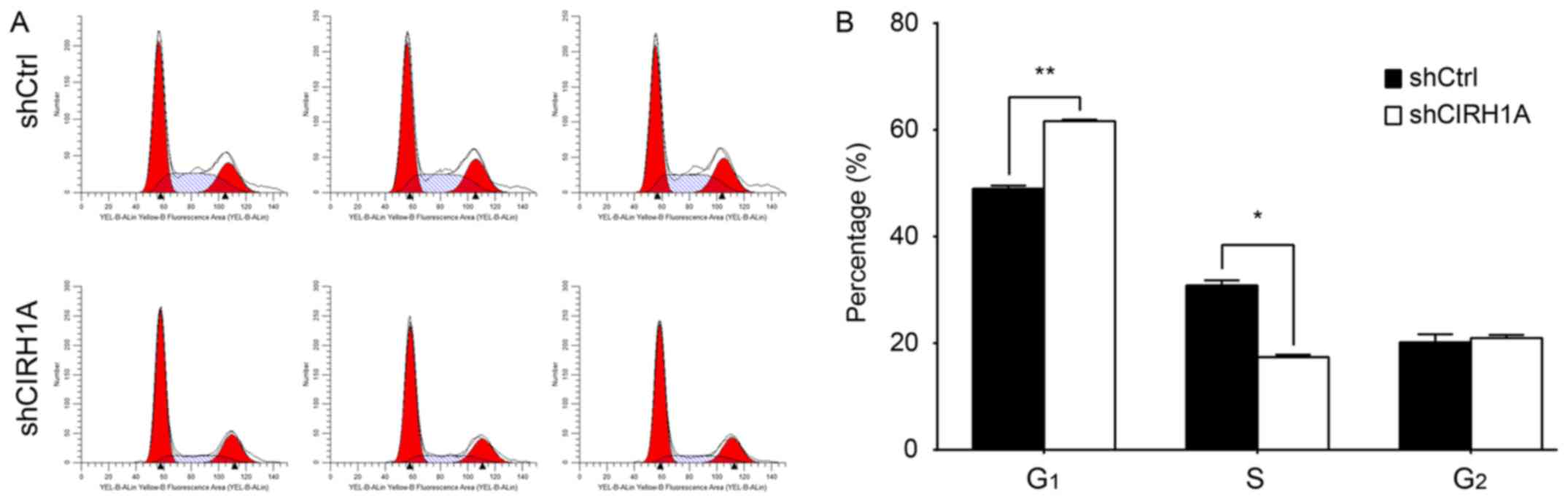
Knockdown of CIRH1A leads to cell
cycle arrest
To determine the necessity of CIRH1A for cell cycle
progression, we determined the cell cycle distribution of RKO cells
with intact or knockdown expression of CIRH1A (Fig. 5A). The shCtrl group displayed the
following distribution: G1 phase, 48.98±0.55%; S phase,
30.84±0.96%; G2 phase, 20.17±1.49%; whilst the shCIRH1A
group displayed the following: G1 phase, 61.65±0.25%; S
phase, 17.38±0.45%; G2 phase, 20.97±0.57%.
shCIRH1A-lentivirus cultures had a significant decrease in the
percentage of cells in the S phase (P<0.01) and an increase in
the percentage of cells in G1 phase (P<0.01) relative
to control cultures (Fig. 5B).
Taken together, the data suggest that CIRH1A regulates cell growth
and blocks cell cycle progression in the G1 phase.
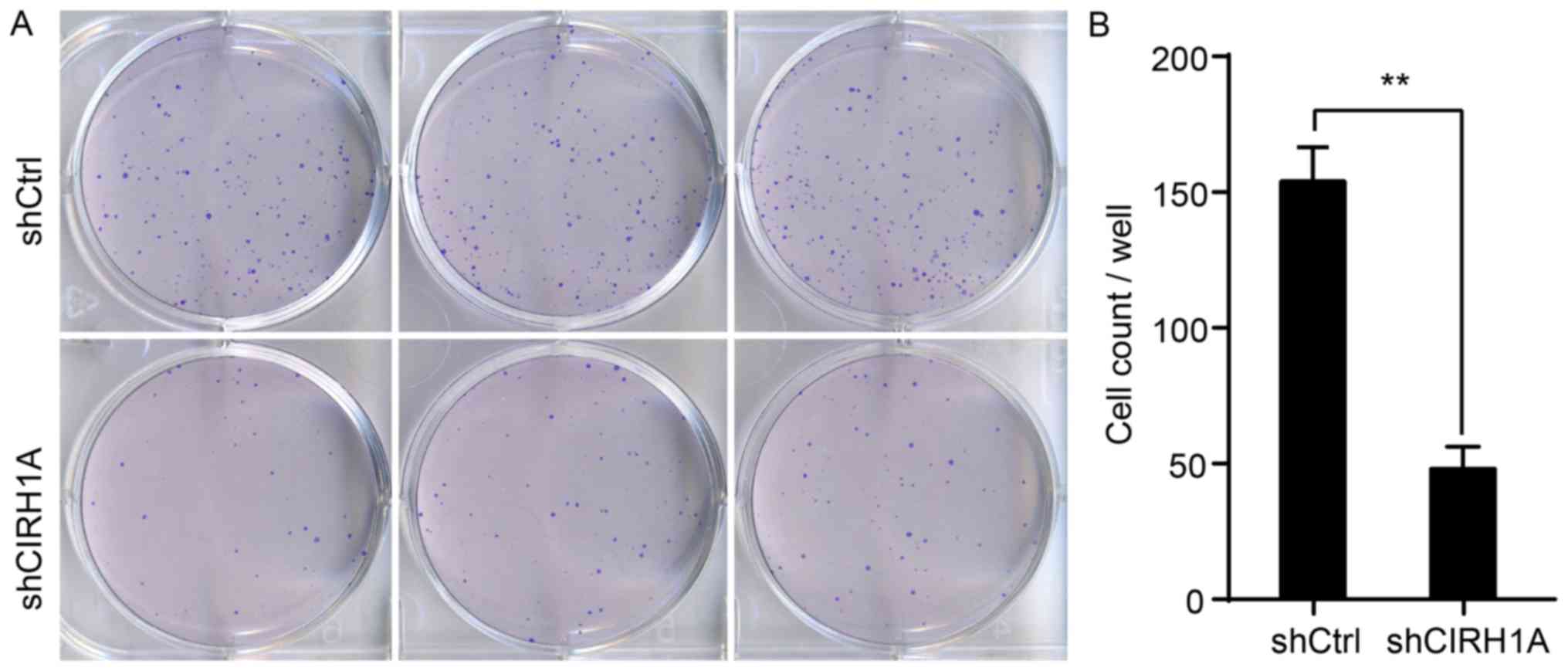
CIRH1A knockdown inhibits colony
formation in RKO cells
Finally, we labelled RKO cells with the Giemsa stain
to measure the effects of CIRH1A knockdown on formation of
RKO cell colonies (Fig. 6A). As
presented in Fig. 6B, the cell
number in a single colony was significantly fewer in the shCIRH1A
group compared to the shCtrl group (shCtrl: 154±12 vs. shCIRH1A:
48±8; P<0.01). This result indicates that reducing CIRH1A
expression endogenously can significantly inhibit the growth of
colorectal carcinomas.
Discussion
North American Indian childhood cirrhosis
(NAIC/CIRH1A) (OMIM: 604901) is an infrequent, autosomal recessive
familial cholestasis found exclusively in Canadian Ojibway-Cree
children. NAIC patients suffer from neonatal jaundice, progressing
to biliary cirrhosis and portal hypertension (7). Liver transplantation is the only known
treatment (15). CIRH1A, the
human homolog of yeast Utp4, was located to chromosome 16q22
(28). Analyses for single
nucleotide polymorphisms have revealed that NAIC patients present
with a R565W mutation in human Cirhin encoded with CIRH1A
(14).
Studies show that Cirhin might play various roles in
different organisms. As a member of the t-Utp/UtpA subcomplex of
the SSU processome, for example, the yeast ‘equivalent’ of human
Cirhin, Utp4 was required for pre-rRNA processing and
transcription, and for the assembly of the SSU processome (10,11).
Moreover, an Utp4 mutation did not affect ribosome biogenesis in
yeast (6). However, the orthologue
in human, Cirhin is only required for pre-18S rRNA processing, but
not for pre-rRNA transcription (29). Cirhin in mouse (mCirhin) is
expressed not only in fetal liver, but also in other developing
tissues (14). Knockout (−/−) of
mCirhin (also known as TEX292) is lethal to embryos (30), while heterozygotes (+/−) are
phenotypically normal (31). Yeast
two-hybrid (Y2H) analysis of a human liver cDNA library revealed
Cirhin interacts with the nucleolar protein NOL11 (32). Further functional analysis revealed
that NOL11 is required for pre-rRNA processing and transcription,
as well as for maintaining a normal nucleolar morphology. In
another study, human Cirhin interacted with Cirip, which is
required for transcription of the HIV-1 LTR enhancer element
(31). However, CIRH1A
expression and its function in human cancers, and CRC in
particular, have not been studied hitherto.
In the present study, we first determined the
expression levels of CIRH1A mRNA in silico using clinical
and molecular data extracted from the online TCGA and in
vitro by profiling four CRC cell lines. The data showed first
that CIRH1A mRNA was highly expressed in carcinoma compared with
paired para-carcinomas, and second that it was overexpressed in the
HCT116, RKO, and LoVo cell lines. Thereafter, in order to assess
the contribution of CIRH1A to CRC cell lines, we constructed
a shCIRH1A lentiviral vector, which efficiently silenced
CIRH1A in infected RKO cells. Compared to shCtrl-infected
cells, shCIRH1A-treated cells showed decreased proliferation and a
significant increase in the proportion of cells in G1
phase. Furthermore, we found that knockdown of CIRH1A
increased apoptosis in RKO cells. Taken together, the data suggest
that CIRH1A plays a novel role in promoting the growth of
CRCs in addition to its known function in ribosomal biogenesis. A
further study to validate the anti-apoptotic role of CIRH1A
in tumorigenesis of CRC is ongoing.
In conclusion, we have demonstrated here that the
downregulation of CIRH1A expression within RKO CRC cells by
RNAi inhibited their proliferation and induced apoptosis.
Accordingly, knockdown of CIRH1A by lentivirus-siRNA may be
a putative therapeutic approach for treating colorectal cancers
that overexpress CIRH1A.
References
|
1
|
Khan K, Cunningham D and Chau I: Targeting
angiogenic pathways in colorectal cancer: Complexities, challenges
and future directions. Curr Drug Targets. 18:56–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qian WF, Guan WX, Gao Y, Tan JF, Qiao ZM,
Huang H and Xia CL: Inhibition of STAT3 by RNA interference
suppresses angiogenesis in colorectal carcinoma. Braz J Med Biol
Res. 44:1222–1230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Freed EF and Baserga SJ: The C-terminus of
Utp4, mutated in childhood cirrhosis, is essential for ribosome
biogenesis. Nucleic Acids Res. 38:4798–4806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sondalle SB and Baserga SJ: Human diseases
of the SSU processome. Biochim Biophys Acta. 1842:758–764. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Doudna JA and Rath VL: Structure and
function of the eukaryotic ribosome: The next frontier. Cell.
109:153–156. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Henras AK, Soudet J, Gérus M, Lebaron S,
Caizergues-Ferrer M, Mougin A and Henry Y: The post-transcriptional
steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci.
65:2334–2359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dragon F, Gallagher JE, Compagnone-Post
PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE,
Shabanowitz J, Osheim Y, et al: A large nucleolar U3
ribonucleoprotein required for 18S ribosomal RNA biogenesis.
Nature. 417:967–970. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gallagher JE, Dunbar DA, Granneman S,
Mitchell BM, Osheim Y, Beyer AL and Baserga SJ: RNA polymerase I
transcription and pre-rRNA processing are linked by specific SSU
processome components. Genes Dev. 18:2506–2517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Phipps KR, Charette J and Baserga SJ: The
small subunit processome in ribosome biogenesis - progress and
prospects. Wiley Interdiscip Rev RNA. 2:1–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu B, Mitchell GA and Richter A: Nucleolar
localization of cirhin, the protein mutated in North American
Indian childhood cirrhosis. Exp Cell Res. 311:218–228. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chagnon P, Michaud J, Mitchell G, Mercier
J, Marion JF, Drouin E, Rasquin-Weber A, Hudson TJ and Richter A: A
missense mutation (R565W) in cirhin (FLJ14728) in North American
Indian childhood cirrhosis. Am J Hum Genet. 71:1443–1449. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Drouin E, Russo P, Tuchweber B, Mitchell G
and Rasquin-Weber A: North American Indian cirrhosis in children: A
review of 30 cases. J Pediatr Gastroenterol Nutr. 31:395–404. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilkins BJ, Lorent K, Matthews RP and Pack
M: p53-mediated biliary defects caused by knockdown of cirh1a, the
zebrafish homolog of the gene responsible for North American Indian
Childhood Cirrhosis. PLoS One. 8:e776702013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Muzny DM, Bainbridge MN, Chang K, Dinh HH,
Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF,
et al: Cancer Genome Atlas Network: Comprehensive molecular
characterization of human colon and rectal cancer. Nature.
487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Robinson MD and Oshlack A: A scaling
normalization method for differential expression analysis of
RNA-seq data. Genome Biol. 11:R252010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu D, Huber W and Vitek O: Shrinkage
estimation of dispersion in Negative Binomial models for RNA-seq
experiments with small sample size. Bioinformatics. 29:1275–1282.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robinson MD and Smyth GK: Small-sample
estimation of negative binomial dispersion, with applications to
SAGE data. Biostatistics. 9:321–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lund SP, Nettleton D, McCarthy DJ and
Smyth GK: Detecting differential expression in RNA-sequence data
using quasi-likelihood with shrunken dispersion estimates. Stat
Appl Genet Mol Biol. 11:307–314. 2012.
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lois C, Hong EJ, Pease S, Brown EJ and
Baltimore D: Germline transmission and tissue-specific expression
of transgenes delivered by lentiviral vectors. Science.
295:868–872. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Su Z, Huang Y, Sun T, Chen S, Wu
T, Chen G, Xie X, Li B and Du Z: The Zfx gene is expressed in human
gliomas and is important in the proliferation and apoptosis of the
human malignant glioma cell line U251. J Exp Clin Cancer Res.
30:1142011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Milner AE, Levens JM and Gregory CD: Flow
cytometric methods of analyzing apoptotic cells. Methods Mol Biol.
80:347–354. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bétard C, Rasquin-Weber A, Brewer C,
Drouin E, Clark S, Verner A, Darmond-Zwaig C, Fortin J, Mercier J,
Chagnon P, et al: Localization of a recessive gene for North
American Indian childhood cirrhosis to chromosome region 16q22-and
identification of a shared haplotype. Am J Hum Genet. 67:222–228.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prieto JL and McStay B: Recruitment of
factors linking transcription and processing of pre-rRNA to NOR
chromatin is UBF-dependent and occurs independent of transcription
in human cells. Genes Dev. 21:2041–2054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Richter A, Mitchell GA and Rasquin A:
North American Indian childhood cirrhosis (NAIC). Med Sci (Paris).
23:1002–1007. 2007.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu B, Mitchell GA and Richter A: Cirhin
up-regulates a canonical NF-kappaB element through strong
interaction with Cirip/HIVEP1. Exp Cell Res. 315:3086–3098. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Freed EF, Prieto JL, McCann KL, McStay B
and Baserga SJ: NOL11, implicated in the pathogenesis of North
American Indian childhood cirrhosis, is required for pre-rRNA
transcription and processing. PLoS Genet. 8:e10028922012.
View Article : Google Scholar : PubMed/NCBI
|