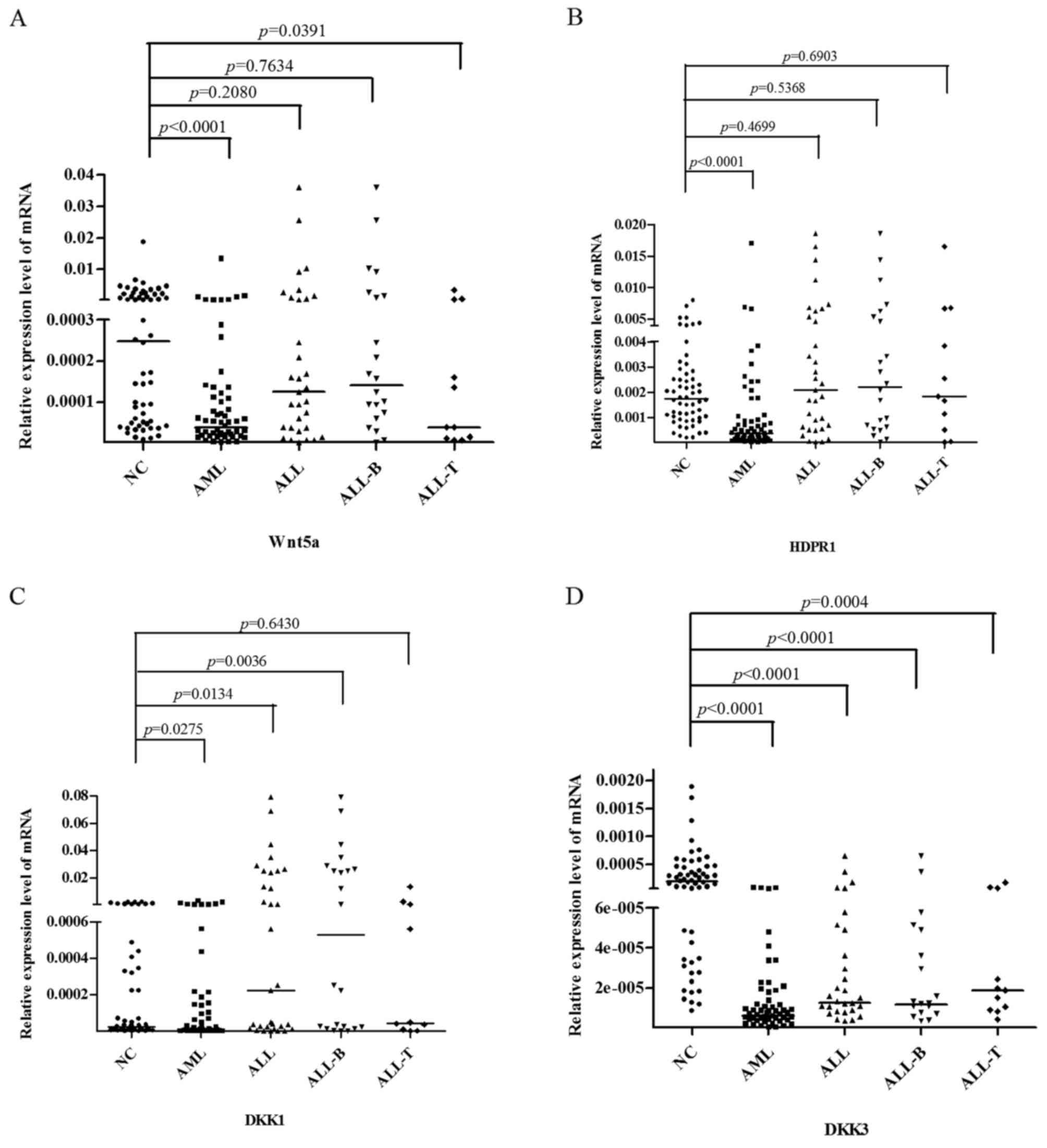
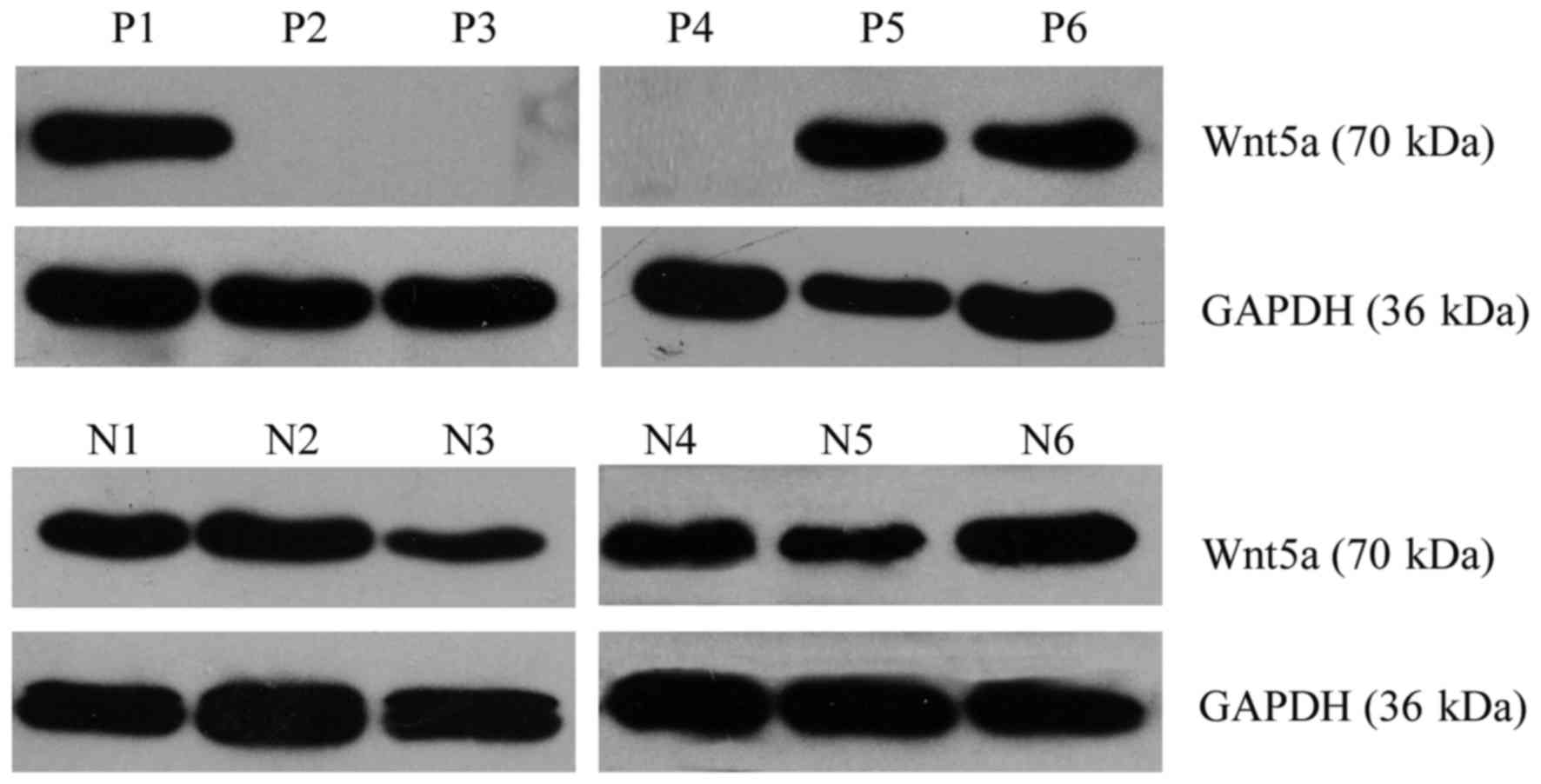
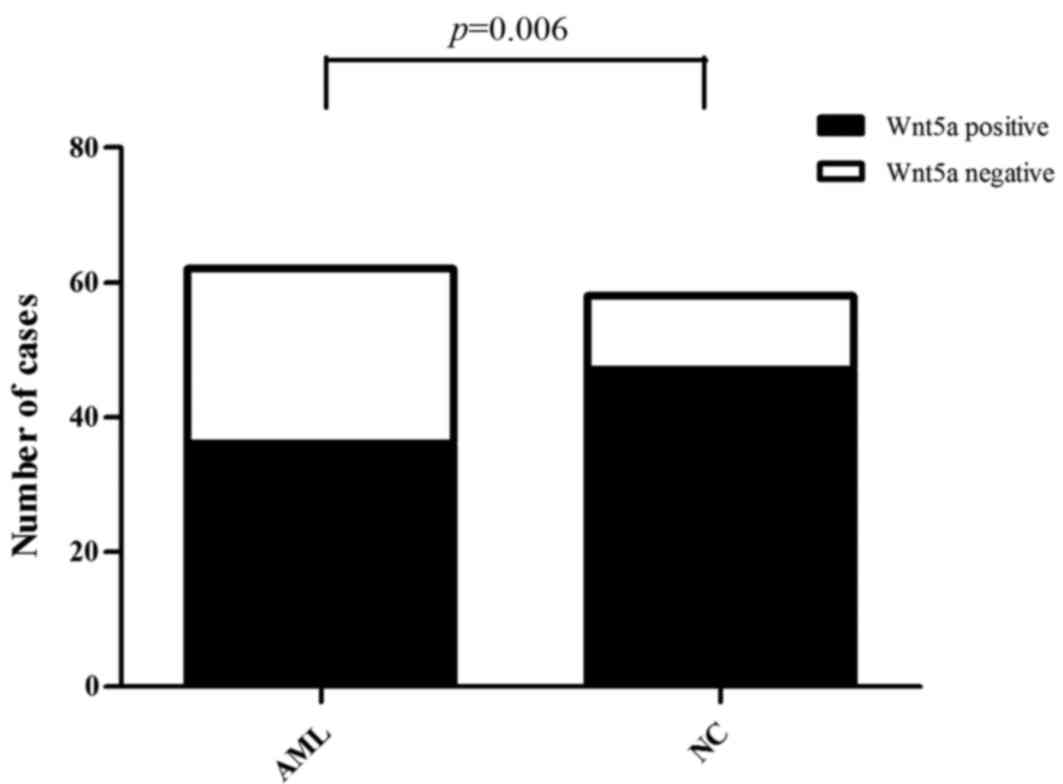
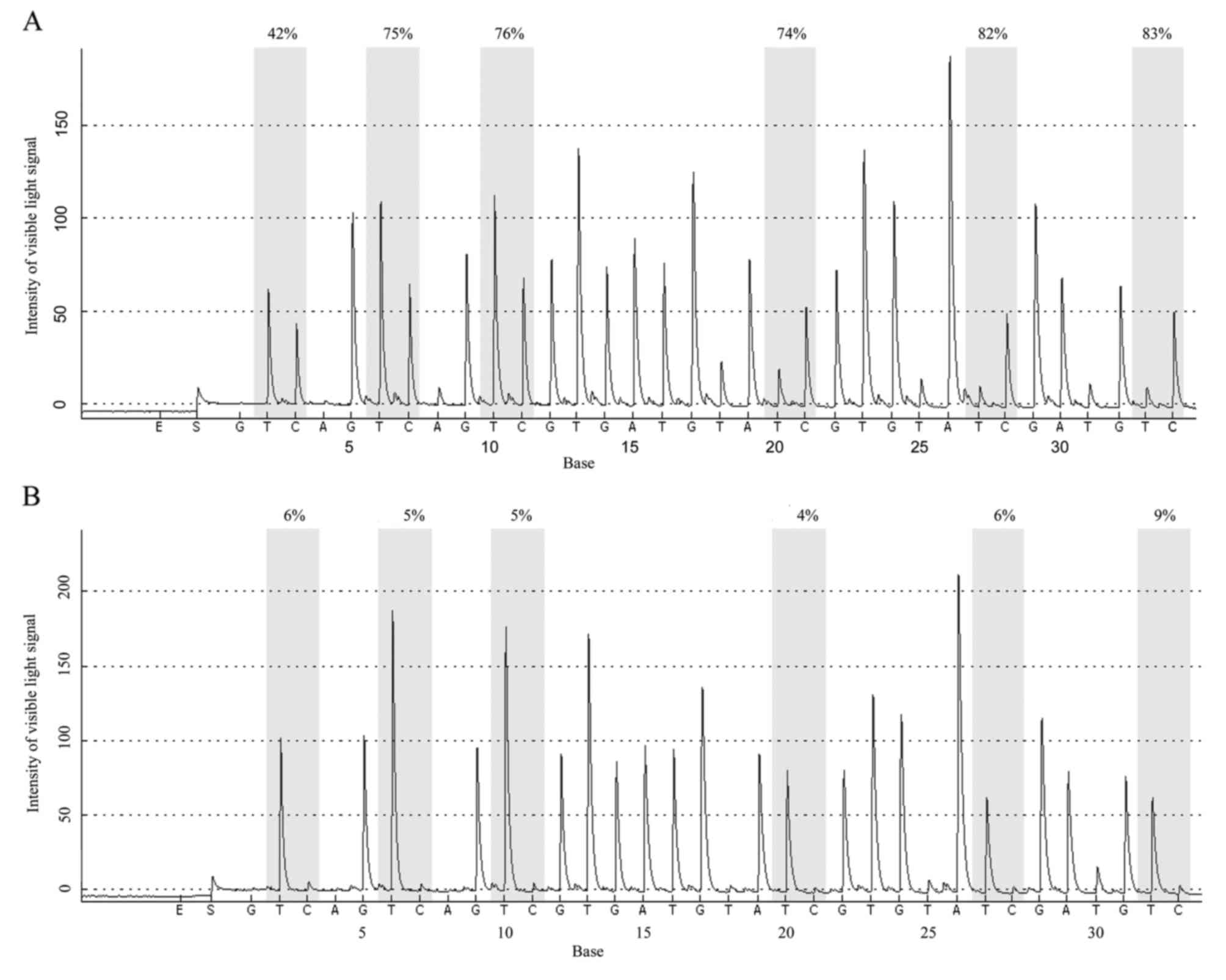
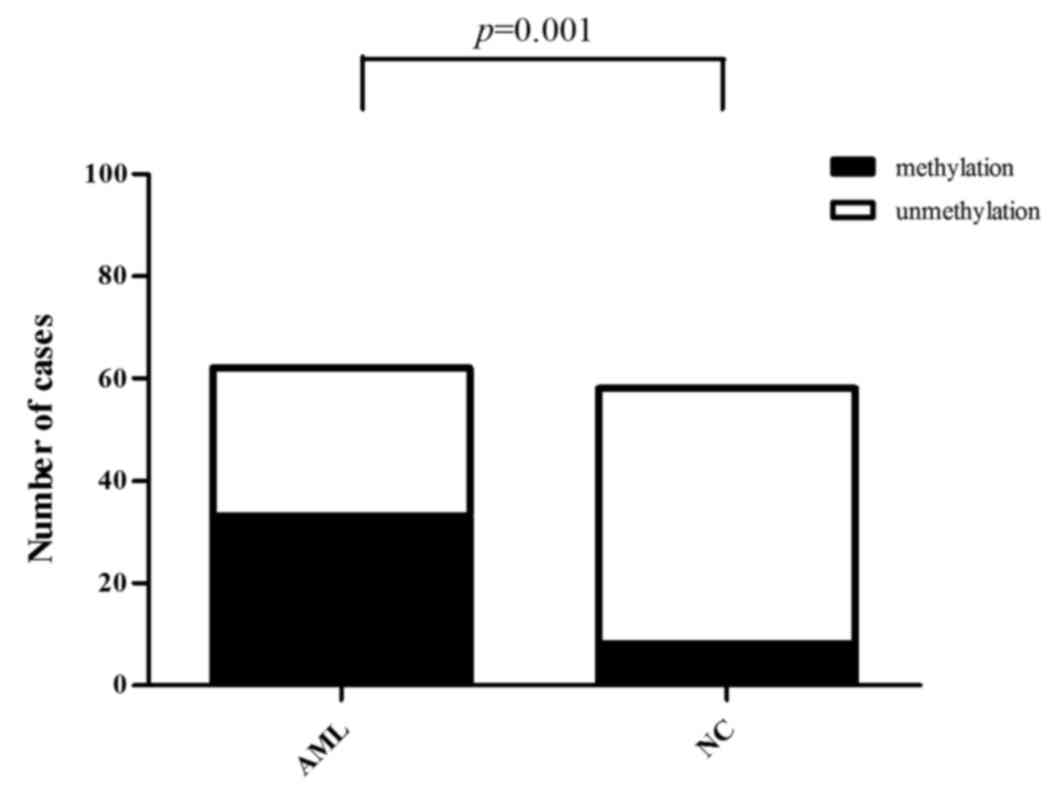
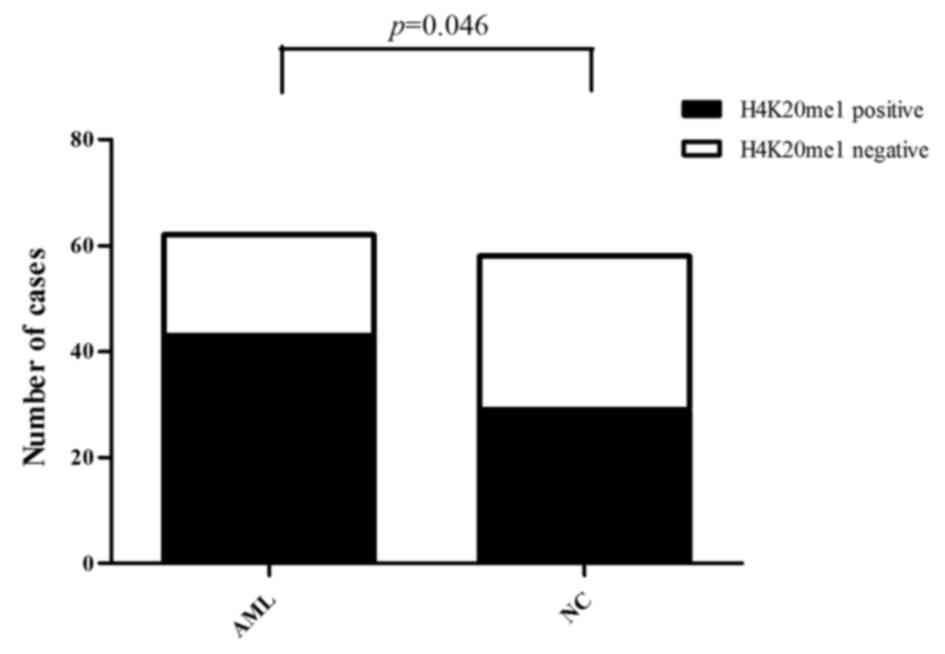
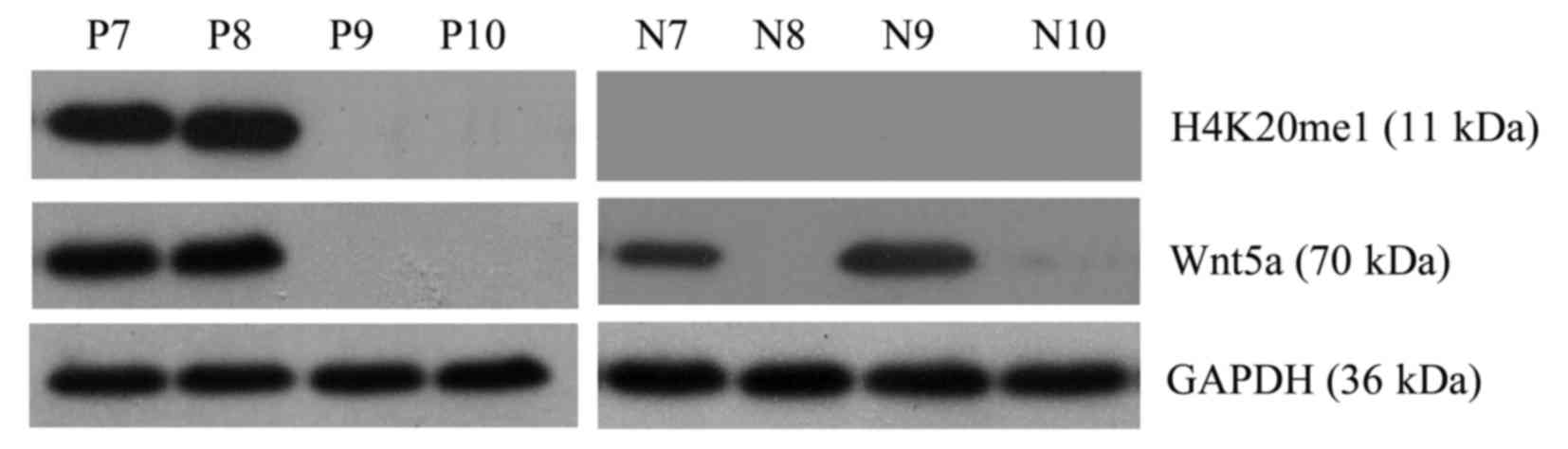
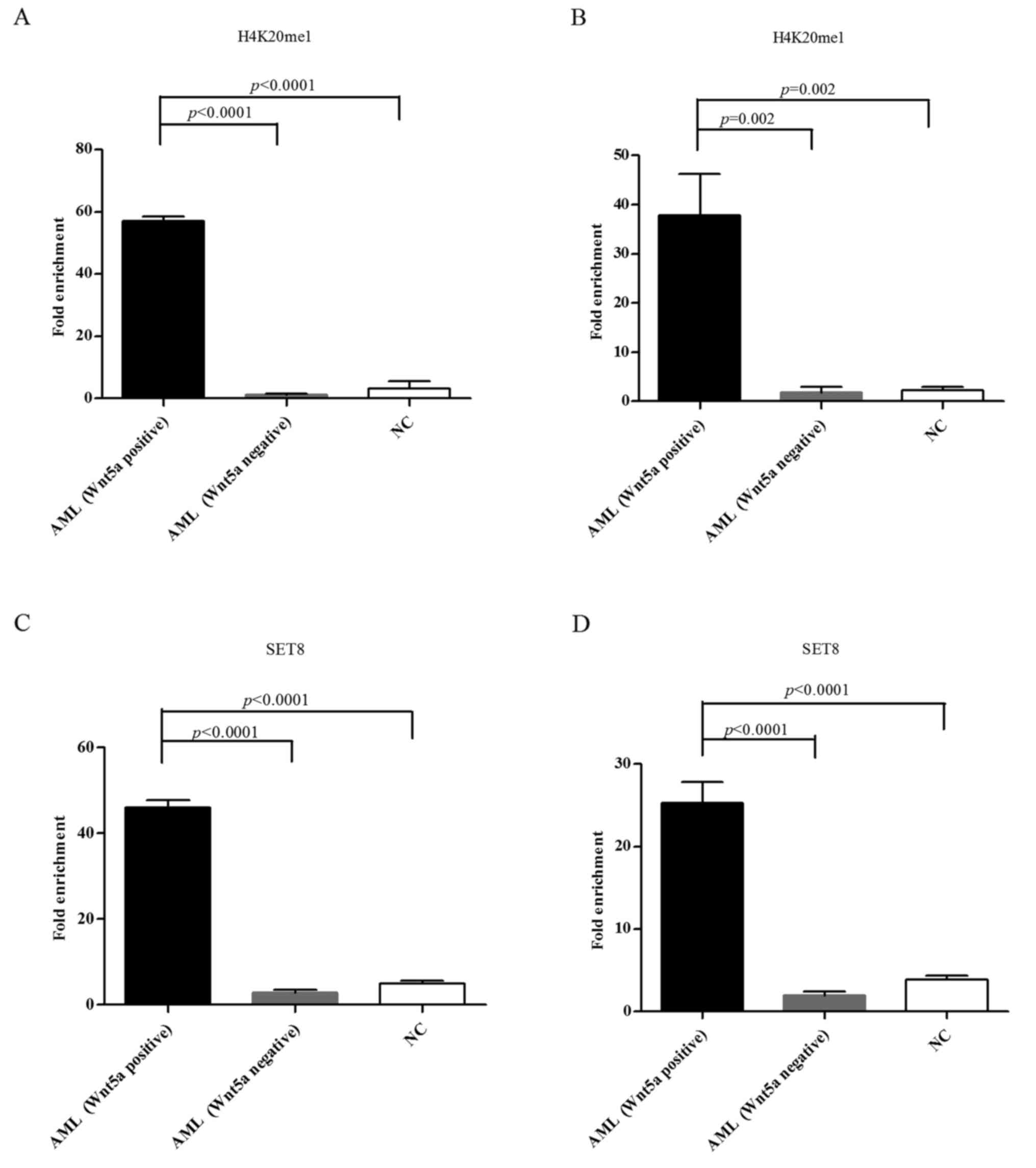
|
1
|
DeLange RJ, Fambrough DM, Smith EL and
Bonner J: Calf and pea histone IV. II. The complete amino acid
sequence of calf thymus histone IV; presence of
epsilon-N-acetyllysine. J Biol Chem. 244:319–334. 1969.PubMed/NCBI
|
|
2
|
Zhang Y and Reinberg D: Transcription
regulation by histone methylation: Interplay between different
covalent modifications of the core histone tails. Genes Dev.
15:2343–2360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oda H, Okamoto I, Murphy N, Chu J, Price
SM, Shen MM, Torres-Padilla ME, Heard E and Reinberg D:
Monomethylation of histone H4-lysine 20 is involved in chromosome
structure and stability and is essential for mouse development. Mol
Cell Biol. 29:2278–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perner J, Lasserre J, Kinkley S, Vingron M
and Chung HR: Inference of interactions between chromatin modifiers
and histone modifications: From ChIP-Seq data to
chromatin-signaling. Nucleic Acids Res. 42:13689–13695. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hori T, Shang WH, Toyoda A, Misu S, Monma
N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al: Histone
H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for
kinetochore assembly. Dev Cell. 29:740–749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yao L, Li Y, Du F, Han X, Li X, Niu Y, Ren
S and Sun Y: Histone H4 Lys 20 methyltransferase SET8 promotes
androgen receptor-mediated transcription activation in prostate
cancer. Biochem Biophys Res Commun. 450:692–696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, et al: PR-Set7 is a nucleosome-specific
methyltransferase that modifies lysine 20 of histone H4 and is
associated with silent chromatin. Mol Cell. 9:1201–1213. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Veloso A, Kirkconnell KS, Magnuson B,
Biewen B, Paulsen MT, Wilson TE and Ljungman M: Rate of elongation
by RNA polymerase II is associated with specific gene features and
epigenetic modifications. Genome Res. 24:896–905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Figueroa ME, Skrabanek L, Li Y, Jiemjit A,
Fandy TE, Paietta E, Fernandez H, Tallman MS, Greally JM, Carraway
H, et al: MDS and secondary AML display unique patterns and
abundance of aberrant DNA methylation. Blood. 114:3448–3458. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tolwinski NS and Wieschaus E: A nuclear
function for armadillo/β-catenin. PLoS Biol. 2:E952004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Claus R, Almstedt M and Lübbert M:
Epigenetic treatment of hematopoietic malignancies: In vivo targets
of demethylating agents. Semi Oncol. 32:511–520. 2005. View Article : Google Scholar
|
|
12
|
Picard F, Cadoret JC, Audit B, Arneodo A,
Alberti A, Battail C, Duret L and Prioleau MN: The spatiotemporal
program of DNA replication is associated with specific combinations
of chromatin marks in human cells. PLoS Genet. 10:e10042822014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bannister AJ, Schneider R and Kouzarides
T: Histone methylation: Dynamic or static? Cell. 109:801–806. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schübeler D, MacAlpine DM, Scalzo D,
Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill
LP, Turner BM, Delrow J, et al: The histone modification pattern of
active genes revealed through genome-wide chromatin analysis of a
higher eukaryote. Genes Dev. 18:1263–1271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang G, Lin JCY, Wei V, Yoo C, Cheng JC,
Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al:
Distinct localization of histone H3 acetylation and H3-K4
methylation to the transcription start sites in the human genome.
Proc Natl Acad Sci USA. 101:7357–7362. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ysebaert L, Chicanne G, Demur C, De Toni
F, Prade-Houdellier N, Ruidavets JB, Mansat-De Mas V, Rigal-Huguet
F, Laurent G, Payrastre B, et al: Expression of β-catenin by acute
myeloid leukemia cells predicts enhanced clonogenic capacities and
poor prognosis. Leukemia. 20:1211–1216. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|