Introduction
Colorectal cancer (CRC) is one of the most prevalent
gastrointestinal tumors and has a high mortality rate (1). It is the third leading cause of
cancer-related deaths in the US (2), and the number of CRC-related deaths in
China was ~159,300 in 2012 (3).
Although, the mortality rate has declined in recent decades due to
the improvement of diagnosis and treatment methods, it still
remains high since present therapies are limited in treating
advanced CRC.
It is known that a majority of human malignancies
are associated with mutations of the cancer genome. Either oncogene
activation or tumor-suppressor gene inactivation leads to the
initiation or acceleration, respectively, of the malignancy
(4). With the improvement of array
techniques, more and more genes have been reported to be involved
with various types of carcinomas. Among these genes, atonal homolog
8 (ATOH8) has been identified as a new cancer-related gene. ATOH8
is a transcription factor from the basic-helix-loop-helix (bHLH)
family. It is comprised of 321 characteristic amino acids with a
bHLH domain and has been reported to participate in embryogenesis
(5) and the development of various
tissues such as the nervous system (6), pancreas (7), kidney (8), endothelial cells (9), and retina and muscle tissues (10,11).
Recently, it was reported that the ATOH8 gene copy number is
altered in multiple malignant disorders. For instance, in
hepatocellular carcinoma (HCC) (12), nasopharyngeal carcinoma (NPC)
(13) and bladder cancer (14) ATOH8 mRNA expression was
downregulated. However, in glioblastoma multiforme (15), prostate carcinoma (16) and breast cancer (17), the expression of ATOH8 was
upregulated and considered to be a candidate oncogene.
The significance of ATOH8 alterations in CRC remains
unknown. In the present study, we used immunohistochemistry to
investigate the expression of ATOH8 in CRC and its correlation with
clinicopathological features. We also conducted functional assays
in vitro to explore the function of this gene in CRC.
Materials and methods
Patients and tissues
The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Wenzhou Medical
University, and informed consent was provided by each patient.
Formalin-fixed, paraffin-embedded samples from 106 paired colon
cancer tissues and peritumoral tissues were obtained from the
Department of Pathology of the Second Affiliated Hospital of
Wenzhou Medical University between 2010-2013. No patients received
neoadjuvant chemotherapy before surgery. The histological grade was
determined based on World Health Organization classification
criteria by two independent pathologists. In addition, the clinical
stage was determined according to Dukes classification system. The
study consisted of 68 males and 38 females ranging in age from 24
to 91 years old (61.86±14.6). The clinicopathological features of
the patients are summarized in Table
I. The serum carcinoembryonic antigen (CEA) level was assessed
by means of radioimmunoassay (RIA) (CEA-RIA kit; CIS
Biointernational, China).
 | Table I.Relationship between ATOH8 expression
and clinicopathological features. |
Table I.
Relationship between ATOH8 expression
and clinicopathological features.
|
|
| ATOH8 |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Parameter | Low | High | P-value |
|---|
| Age (years) | ≥60 | 22 | 26 | 0.918 |
|
| <60 | 26 | 32 |
| Sex | Male | 27 | 41 | 0.123 |
|
| Female | 21 | 17 |
| Tumor site | Right colon | 18 | 29 | 0.197 |
|
| Left colon | 30 | 29 |
| Size | >5 cm | 17 | 26 | 0.433 |
|
| ≤5 cm | 30 | 33 |
| Histological
grade | High to
moderate | 24 | 33 | 0.478 |
|
| Low | 24 | 25 |
|
| Dukes stage | A-B | 28 | 28 | 0.333 |
|
| C-D | 20 | 30 |
|
| Serum CEA
(ng/ml) | <5 | 38 | 34 | 0.036 |
|
| ≥5 | 10 | 24 |
|
| Mean survival
time |
| 61.8 | 51.5 | 0.022 |
| (95%
CI) (months) |
|
(56.7–67.0) |
(45.2–57.7) |
|
Follow-up
Patients were followed up with physical examination
and assessment of their serum CEA level every 3–6 months in the
first two years regularly. Abdominal computed tomography and
colonoscopy were conducted every 6–12 months for surveillance of
recurrence. In addition, we called upon the patients every three
months to assess their level of discomfort. The overall survival
(OS) time was defined as the time from surgery to death (the final
event defined as death caused by the tumor or from complications)
or the time from surgery to the last follow-up of patients. The
median follow-up was 49.4 months (ranging from 1–71 months).
Immunohistochemistry and staining
evaluation
Tissue sections (4-µm thick) from paraffin-embedded
colon cancer and corresponding peritumoral samples were placed onto
slides coated with polylysine. ATOH8 expression in colon cancer
samples was examined using immunohistochemistry. In brief, sections
were immersed in xylene for 10 min 3 times, and subsequently
immersed in 100% ethanol for 5 min twice for deparaffination, and
then submerged in 95, 85 and 75% ethanol for 5 min/concentration at
room temperature for rehydration. For antigen retrieval, the slides
were immersed in 10 mM citrate buffer (pH 6.0), and heated to 120̊C
and at a pressure of 103 kPa for 2 min. Then, the slides were
immersed in 3% hydrogen peroxide (20 min, room temperature) to
block endogenous peroxidase activity and 10% normal goat serum (30
min, room temperature) to block non-specific binding. The slides
were then washed with phosphate-buffered saline (PBS) and treated
with primary polyclonal rabbit ATOH8 antibody (ab106377; diluted
1:100; Abcam, Cambridge, MA, USA) at 4̊C in a humidified incubator
overnight. Then, the slides were washed with PBS and incubated with
a secondary antibody (goat anti-rabbit IgG; Zhongshan Bio Co.,
Ltd., Beijing, China) for 18 min followed by
streptavidin-peroxidase conjugate for 22 min at room temperature.
Subsequently, the samples were stained with a 3,5-diaminobenzidine
(DAB) substrate kit (Zhongshan Bio Co., Ltd.) and haematoxylin for
counterstaining. All of the samples were evaluated by two
independent pathologists who were blinded to the clinical data of
the patients. The inconsistent cases were reassessed on a
double-headed microscope. Immunohistochemical evaluation of ATOH8
protein expression was performed as previously described, including
staining intensity and extent (18). The intensity score was as follows:
0, negative; 1, weak; 2, moderate; and 3, strong. The extent of
positivity score was quantified from the percentage of positive
tumor cells: 0, <5%; 1, ≥5–25%; 2, ≥26–50%; 3, ≥51–75%; and 4,
≥76%. The final score was determined by multiplying the intensity
and extent scores, which ranged between 0 and 12; scores ≤4
indicated low expression.
Quantitative RT-PCR
Total RNA was extracted from cultured cells with
TRIzol reagent (Superfec Bio Co., Ltd., Shanghai, China) according
to the manufacturer's instructions. Then, an M-MLV reverse
transcriptase kit (Promega Bio Co., Ltd., Beijing, China) was used
for reverse transcription. To determine the transcripts of the
target genes, quantitative real-time polymerase chain reaction
(qRT-PCR) was conducted with a SYBR Premix Ex Taq (RibBio Co.,
Ltd., Guangzhou, China) on an Agilent MX3000p Bioanalyzer (Applied
Biosystems Co., Ltd., CA, USA). qRT-PCR conditions were as follows:
95̊C for 30 sec; 45 cycles of 95̊C for 5 sec, 60̊C for 30 sec, and
95̊C for 15 sec; 55̊C for 30 sec; and 95̊C for 15 sec. Three
independent experiments were run in triplicate. The relative amount
of ATOH8 mRNA was normalized to the control GAPDH. The primer
sequences were as follows: ATOH8, 5′-TGGGCAGAAGCTGTCCAAACT-3′ and
GTGGTCGGCACTGTAGTCAAG, and GAPDH, 5′-TGACTTCAACAGCGACACCCA-3′ and
5′-CACCCTGTTGCTGTAGCCAAA-3′.
Western blotting
CRC cells were harvested and washed with cold PBS
and lysed with radio-immunoprecipitation assay (RIPA) lysis buffer
(Beyotime Bio Co., Ltd., Haimen, China). Cell debris was
centrifuged and total protein in the supernatants was assessed with
a BCA protein assay kit (Beyotime). Amounts equal to 50 µg of
protein were separated on a 10% SDS-PAGE gel, then transferred to a
polyvinylidene fluoride (PVDF) membrane (Beyotime) and incubated
with the ATOH8 (ab106377; diluted 1:800) and GAPDH (ab8245; diluted
1:5,000) (both from Abcam) primary antibodies at 4̊C overnight.
Then, the membrane was incubated with the HRP-labelled goat
anti-rabbit IgG secondary antibody (diluted 1:1,000; Beyotime) at
room temperature for 2 h. Proteins were visualized with enhanced
chemiluminescence detection reagents (Applygen Technologies,
Beijing, China). The relative amount of ATOH8 protein was
normalized to the control GAPDH.
Cell culture and lentiviral
infection
Four types of human colon carcinoma cell lines
(HCT116, SW620, RKO and LoVo) were purchased from the Shanghai Cell
Bank of the Chinese Academy of Sciences. All of the cancer cell
lines were cultured in high-glucose Dulbecco's modified Eagles
medium (DMEM) with 10% foetal bovine serum (FBS) (Atlanta
Biologicals, Lawrenceville, GA, USA) at 37̊C in 5% CO2
moist incubator. All experiments were performed on exponentially
growing cells. Based on the sequence of ATOH8 (NM_032827.6), we
designed three different small interfering RNAs (siRNAs) (siRNA#1,
5′-CGTCAATTTCACACGTAAT-3′; siRNA#2, 5′-ACGGCCTTAAGAAGCTCAA-3′; and
siRNA#3, 5′-TGAGGATCGCCTGTAACTA-3′), and a negative control (NC)
siRNA (5′-TTCTCCGAACGTGTCACGT-3′). The ATOH8-specific and negative
control lentiviral packaging plasmids were purchased from GeneChem
Biotechnologies, Co., Ltd. (Shanghai, China). The lentiviral
vectors which expressed green fluorescent protein (GFP) were
transfected into the CRC cell lines using the Lipofectamine 2000
reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the
manufacturer's instructions. Α fluorescence microscope
(Olympus-BX53; Olympus, Tokyo, Japan) confirmed whether the target
cells were transfected with the lentiviral vectors.
Cell proliferation and cytotoxicity
assay
Cells were seeded into a 96-well plate at a density
of 2.0×104 cells/ml and incubated in a humidified
environment of 5% CO2 at 37°C. On day 1–5, the medium
was removed and 20 µl of 5 mg/ml MTT (Genview, Tallahassee, FL,
USA) was added to each well, respectively. Four hours later, the
MTT reagent was removed and replaced with 100 µl of dimethyl
sulfoxide (DMSO) to stop the reaction, and then the plates were
agitated for 5 min. Subsequently, the absorbance of the samples was
assessed at 490 nm with a microplate reader.
Cell chemosensitivity to 5-fluorouracil (5-FU) was
determined by MTT assay. Approximately 2×104 cells/well
were seeded in 96-well plates, allowed to attach overnight, and
exposed to different concentrations of 5-FU from 1 to 150 µg/ml for
24 h. Then, an MTT assay was performed as aforementioned.
Cell apoptosis analysis
Following five days of transfection, apoptosis was
assessed by the Annexin V-APC apoptosis detection kit (eBioscience,
San Diego, CA, USA) according to the manufacturer's instructions.
At first, cells seeded in 6-well dishes were trypsinized, collected
and incubated in 80% ethanol at 4̊C overnight. Then, the cells were
stained with Annexin V-APC at room temperature in the dark. The
apoptosis rate was analysed by flow cytometry (BD Biosciences, San
Diego, CA, USA) within 1 h. Cells positive for Annexin V-APC were
considered to be apoptotic cells.
Cell cycle analysis
SW620 cells (2×106) were seeded in 6-cm
dishes and trypsinized, collected and washed with PBS twice. Prior
to staining with propidium iodide (PI), the cells were incubated in
80% ethanol at 4̊C overnight. Subsequently, the stained cells were
assessed by flow cytometry. The fraction of cells in the G0/G1, S
and G2/M phases was analysed using CellQuest software programs.
Cell wound-healing assay
In the cell motility assay, stably-transfected
ATOH8-siRNA cells were cultured until cell confluence reached 90%.
Then, a 200 µl pipette tip was used to draw a wound at the bottom
of each well. The cells were washed with PBS, and then further
cultured in 0.5% FBS at 37̊C with 5% CO2. Wound closure
was observed after 8 and 24 h. We used the relative migration rate
to evaluate the motility of cells. Relative migration rate =
distance of migration/the width at 0 h (%). Distance of migration =
the width at 0 h - the width at time (mm).
Statistical analysis
All statistical analyses were performed with SPSS
22.0 (SPSS, Inc., Chicago, IL, USA). For numerical variables, the
data was expressed as the means ± SEM. The significance of the
differences between the values was determined by Student's t-test.
The correlation between ATOH8 and the clinicopathological features
was assessed by Chi-squared and Fisher's exact tests. The OS time
was calculated by the Kaplan-Meier method, and the differences were
analysed by the log-rank test. A P-value <0.05 was considered to
indicate a statistically significant result.
Results
Expression of ATOH8 in colon cancer
patients and its correlation with clinicopathological
characteristics
To explore the role of ATOH8 in colon cancer, we
surveyed the expression of ATOH8 in 106 paired colon cancer samples
using immunohistochemistry. Our results indicated that ATOH8
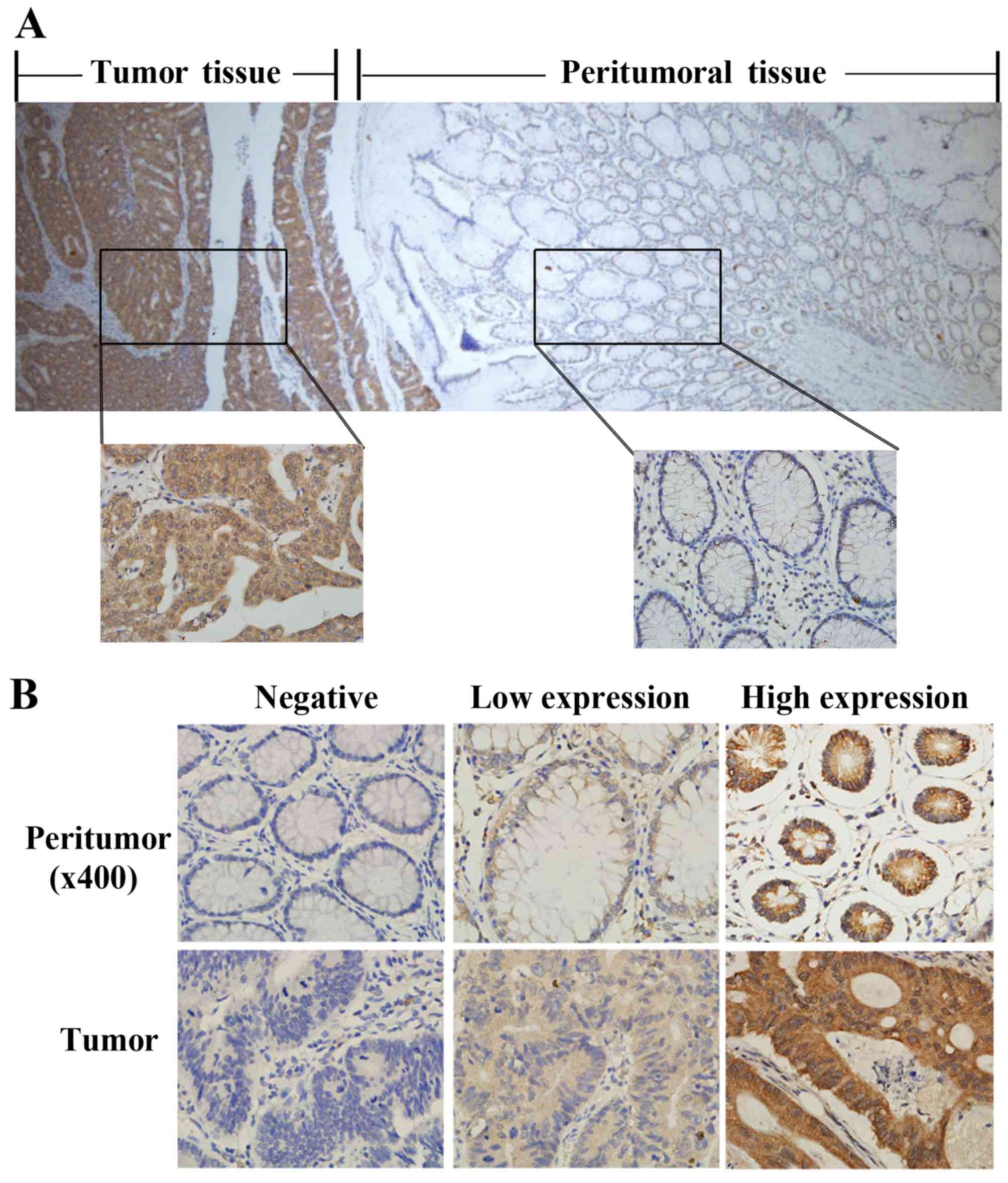
displayed either diffuse or localized patterns in the cytoplasm of
both cancer and peritumoral tissues (Fig. 1A and B). According to our evaluation
criteria, the ATOH8 protein was highly expressed in 58/106 (54.7%)
of colon cancer samples but only 7/106 (6.6%) of peritumoral
samples (P=0.000). Based on the level of ATOH8 expression in the
tumor-cell cytoplasm, patients were divided into an ATOH8 low
expression group (including negative and low expression) and a high
expression group (high expression; Fig.
1B). We explored the relationship between the expression level
of ATOH8 and clinicopathological features. High expression of ATOH8
was significantly associated with a high serum CEA level, but there
was no connection between the expression level of ATOH8 with age,
gender, tumor location, tumor size, histological grade or Dukes'
stage (Table I). We also compared
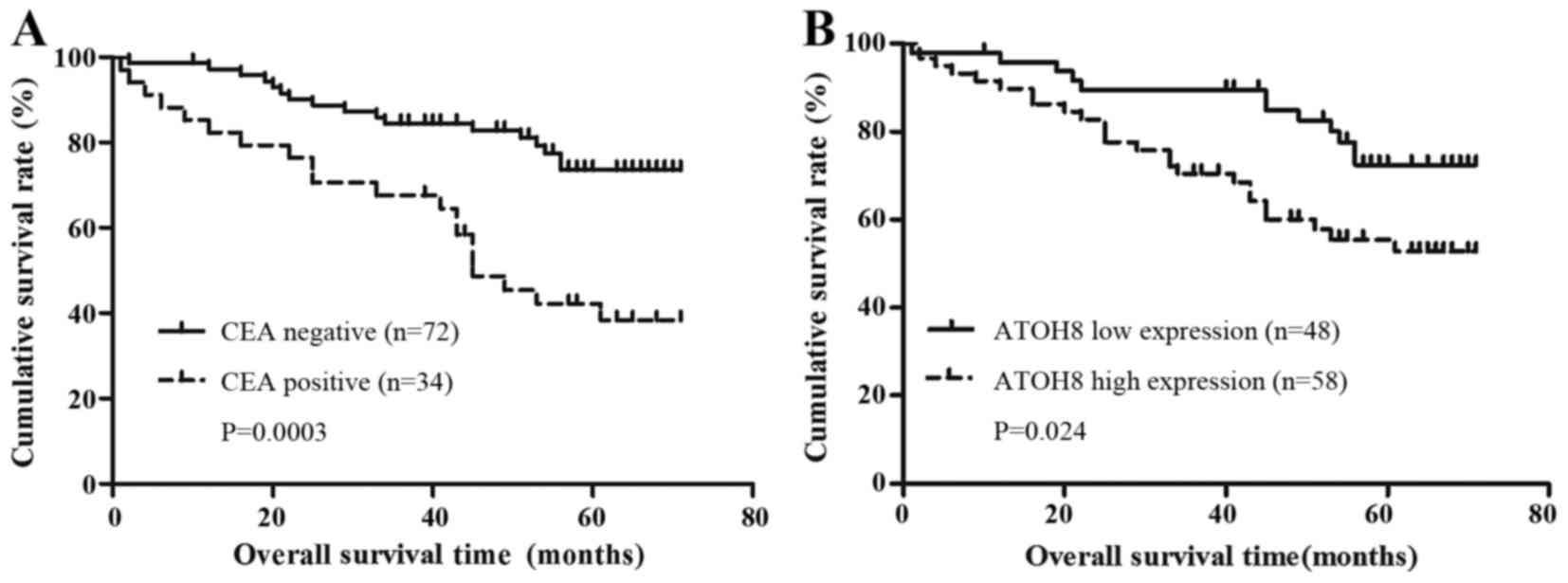
the serum CEA level with the prognosis of CRC patients by
Kaplan-Meier survival analysis. It suggested that the patients with
a high level of pre-surgery CEA had a shorter OS time (95% CI,
37.3–54.1 months vs. 95% CI, 56.9–65.6 months, P=0.0003; Fig. 2A). Moreover, when comparing the
expression of ATOH8 with OS, the high expression group was observed
to have statistically worse OS (95% CI, 45.2–57.7 months vs. 95%
CI, 56.7–67.0 months; P=0.024, Fig.
2B).
Lentivirus-mediated ATOH8 knockdown in
CRC cells
The expression of ATOH8 mRNA was analysed in the
four colon cancer cell lines HCT116, SW620, LoVo and RKO with
qRT-PCR. The relative expression ratio of ATOH8 mRNA in the four
cancer cell lines was normalized to LoVo cells. The relative
expression levels of ATOH8 mRNA in the four cancer cell lines were
26.58±1.75, 23.85±1.74, 0.81±0.19 and 0.23±0.06 in the HCT116,
SW620, LoVo and RKO cells, respectively (Fig. 3A). Expression in the SW620 and
HCT116 cells was relatively high, so we chose these two cell lines
in the following experiment. Then, we designed a negative control
siRNA (NC) and three different ATOH8 siRNAs (KDs) (siRNA#1,
siRNA#2, siRNA#2) and cloned them into GFP-lentiviral vectors. The
groups transfected with NC and KDs were visible in the dark under
fluorescence microscopy, while the non-transfected control group
(CON) was invisible, which means the lentiviral vectors were
successfully transfected into the SW620 (Fig. 3B) and HCT116 cells (data not shown).
Then, we screened the expression of ATOH8 mRNA in SW620 and HCT116
cells using qRT-PCR to ascertain the knockdown efficiency. The
results suggested that ATOH8 expression was effectively
downregulated by siRNA#1 (KD1, decreased to 71.6±11.9% and
81.6±7.3%; P<0.05) and siRNA#2 (KD2, decreased to 50.3±10.0% and
65.0±5.0%; P<0.001) in the SW620 and HCT116 cells, respectively,
compared to the controls. However, siRNA#3 (KD3) did not decrease
ATOH8 expression in either the SW620 (109.7±8.2%) or HCT116
(104.4±7.3%) cells (Fig. 3C). As
the knockdown efficiency of KD2 in SW620 cells was the highest, we
chose the siRNA#2-SW620 cells in the following experiments. Before
additional in vitro experiments, the expression of ATOH8 of
the CON, NC and KD groups was confirmed by qRT-PCR (Fig. 3D; P<0.001) and western blotting
(Fig. 3E; P<0.01).
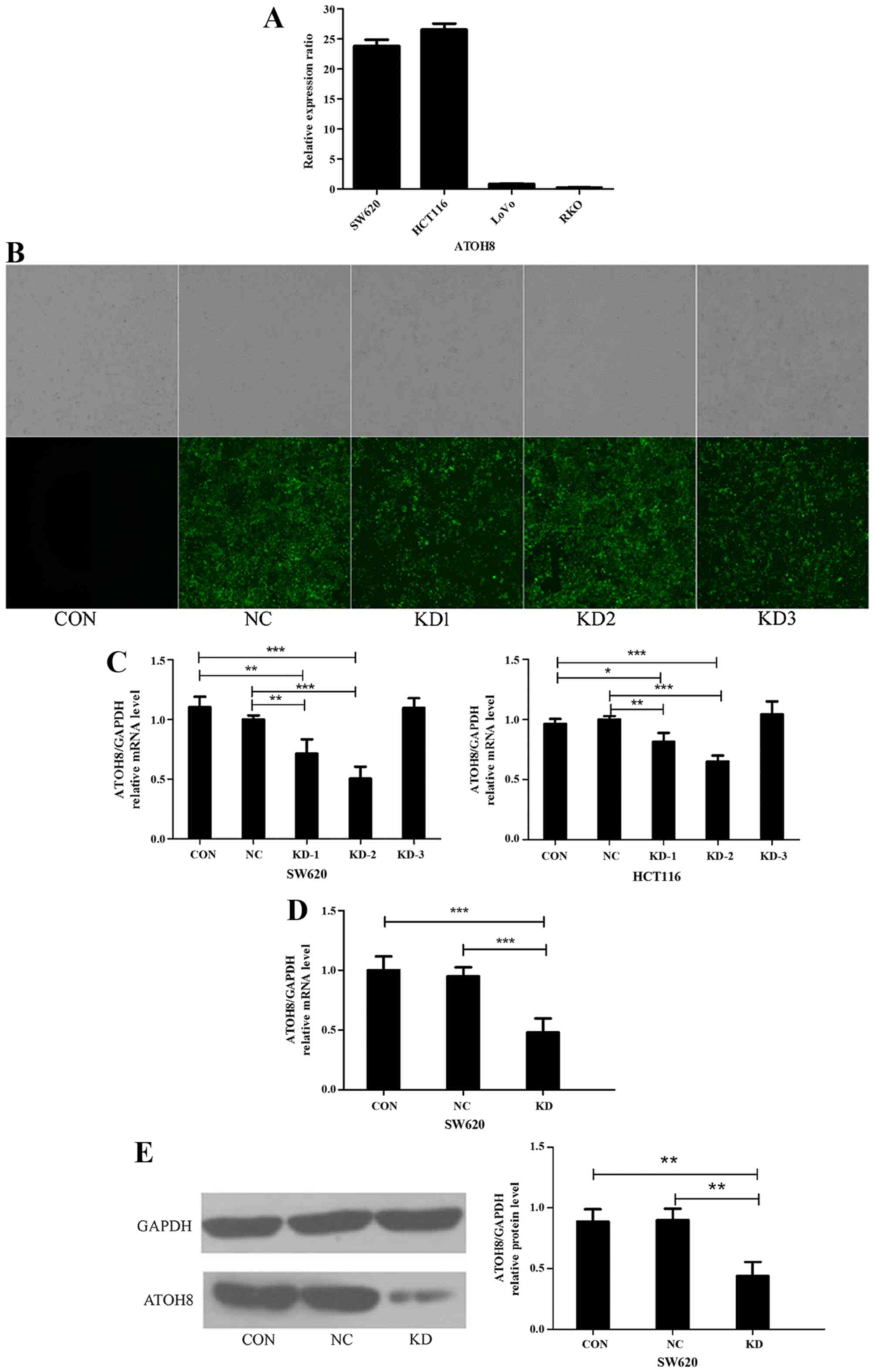 | Figure 3.Lentiviral-mediated ATOH8 knockdown in
CRC cells. (A) Relative expression ratio of ATOH8 in SW620, HCT116,
LoVo and RKO colon cancer cell lines as normalized to LoVo cells.
The expression levels of ATOH8 in SW620 and HCT116 cells were
relatively high. (B) The transfection of different GFP-lentiviral
siRNA vectors (NC, KD1, KD2 and KD3) in SW620 cells was evaluated
by fluorescence microscopy. (C) The knockdown efficiency of each
lentiviral siRNA vector in SW620 and HCT116 cells was confirmed by
qRT-PCR, and it revealed that ATOH8 expression was effectively
downregulated by both KD1 and KD2 compared with CON and NC. The
expression of ATOH8 in the CON, NC and KD groups was confirmed by
(D) qRT-PCR and (E) western blotting. CON, non-transfected control
group; NC, negative control group; KD, ATOH8-siRNA transfection
group; *P<0.05, **P<0.01, ***P<0.001. ATOH8, atonal
homolog 8; CRC, colorectal cancer; GFP, green fluorescent
protein. |
Knockdown of ATOH8 suppresses CRC cell
proliferation
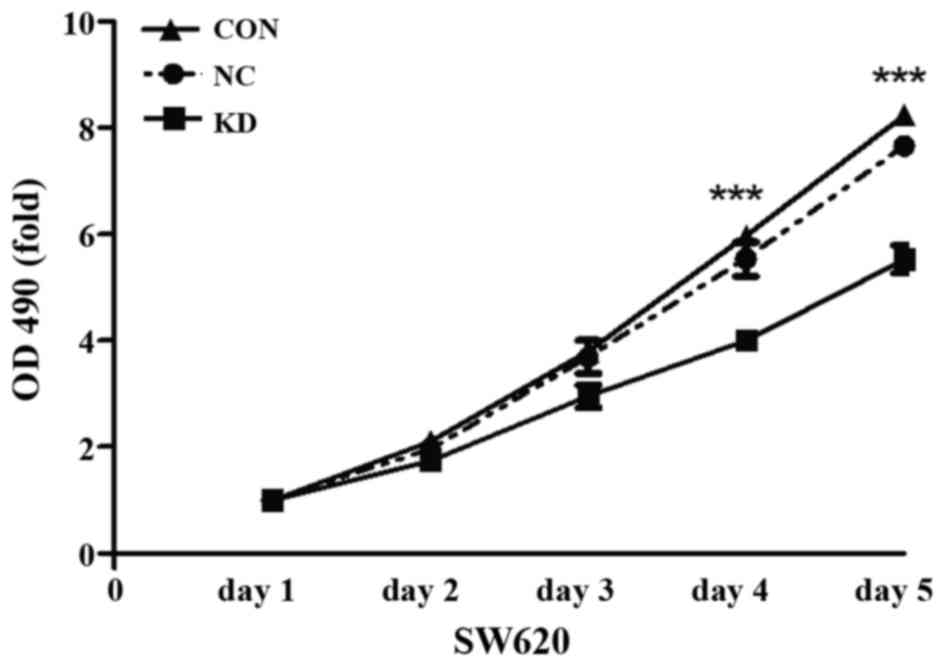
We performed an MTT assay to investigate the effect
of ATOH8 knockdown on the proliferation ability of SW620 cells. As
shown in Fig. 4, the proliferation
of the KD group on day 4 and 5 was markedly decreased compared with
the CON and NC group (P<0.001).
Knockdown of ATOH8 increases cell
apoptosis
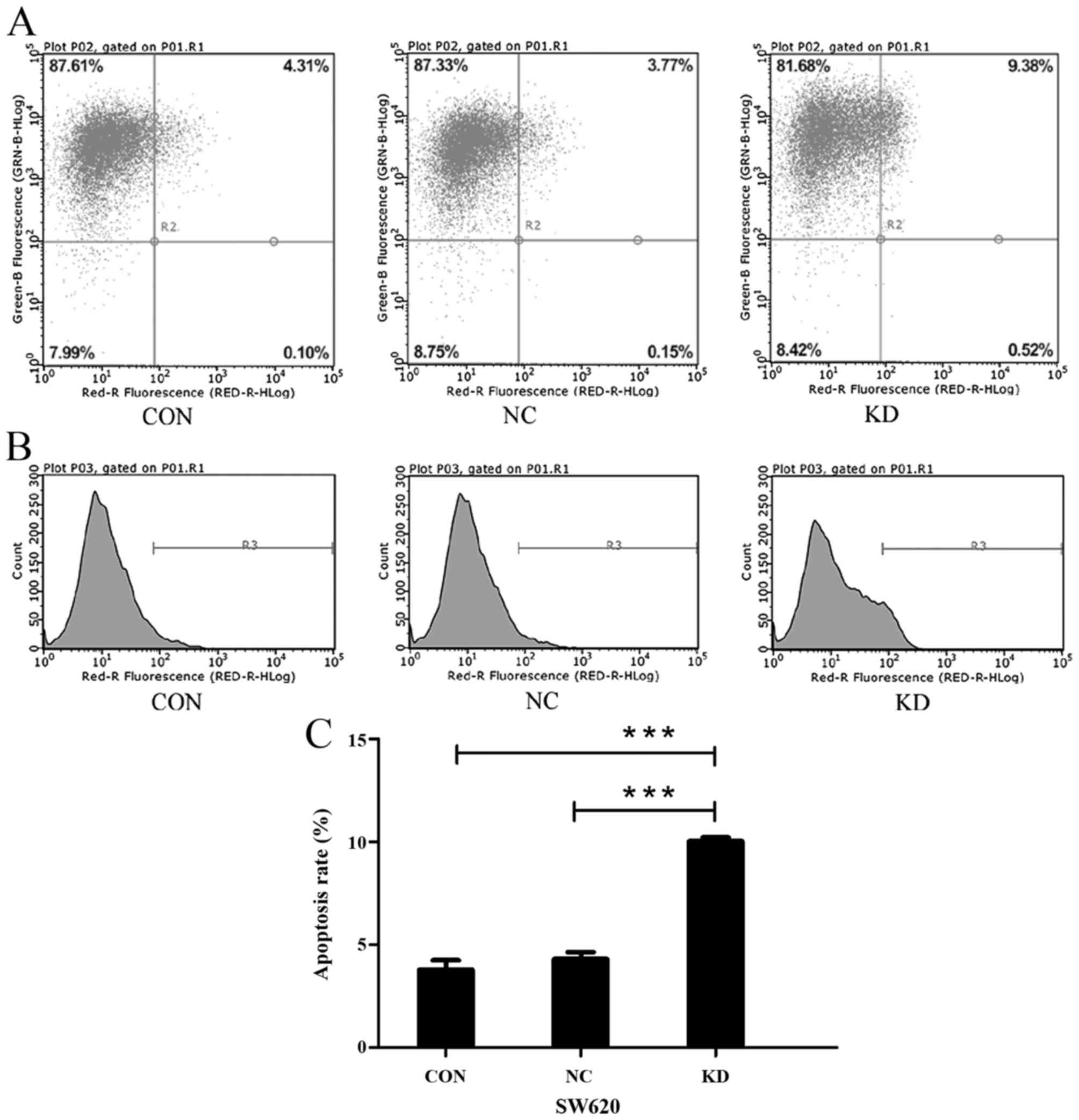
Following five days of transfection, the confluence
of the cells reached ~85% in the CON, NC and KD groups, and then we
performed Annexin V-APC single staining assay (Fig. 5A and B). As shown in Fig. 5C, the percentage of apoptotic cells
in the KD group (10.04±0.178%) was significantly increased compared
to the CON group (3.8±0.445%; P<0.001) and NC group (4.3±0.342%;
P<0.001).
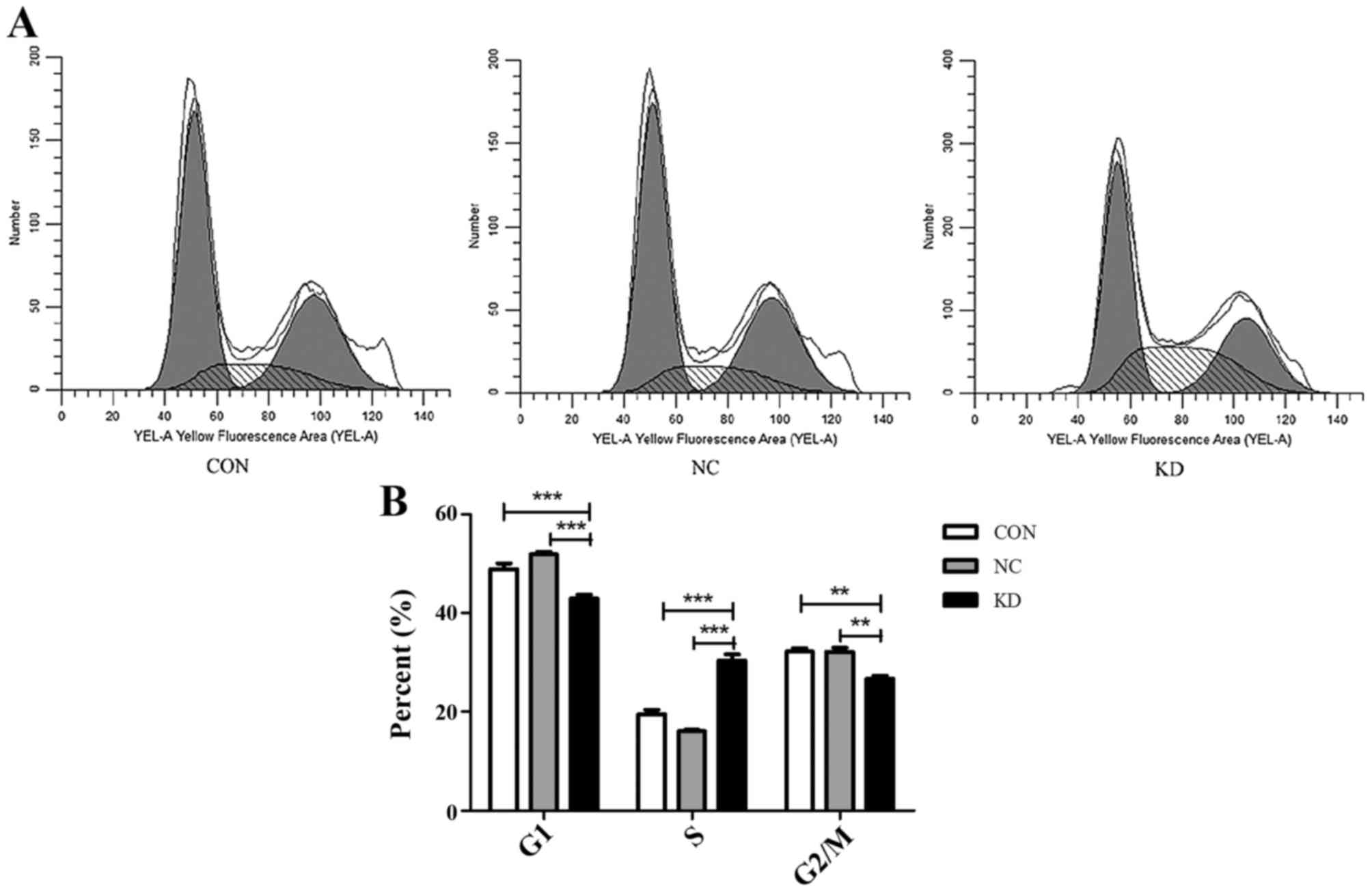
Knockdown of ATOH8 arrests the cell
cycle in the S phase
To investigate the role of ATOH8 knockdown in cell
cycle progression, flow cytometry was performed, which revealed
that ATOH8 knockdown arrested the cell cycle in SW620 cells. As
shown in Fig. 6, the percentage of
the G1 phase cells (CON vs. NC vs. KD, 48.87 vs. 51.87 vs. 42.93%,
respectively; P<0.001) and the G2/M phase cells (32.15 vs. 32.12
vs. 26.69%; P<0.01) was significantly decreased in the
ATOH8-knockdown group while the percentage of cells in the S phase
(19.49 vs. 16.02 vs. 30.38%; P<0.001) was increased.
Collectively, these data demonstrated that ATOH8 knockdown led to
cell cycle arrest during the S phase.
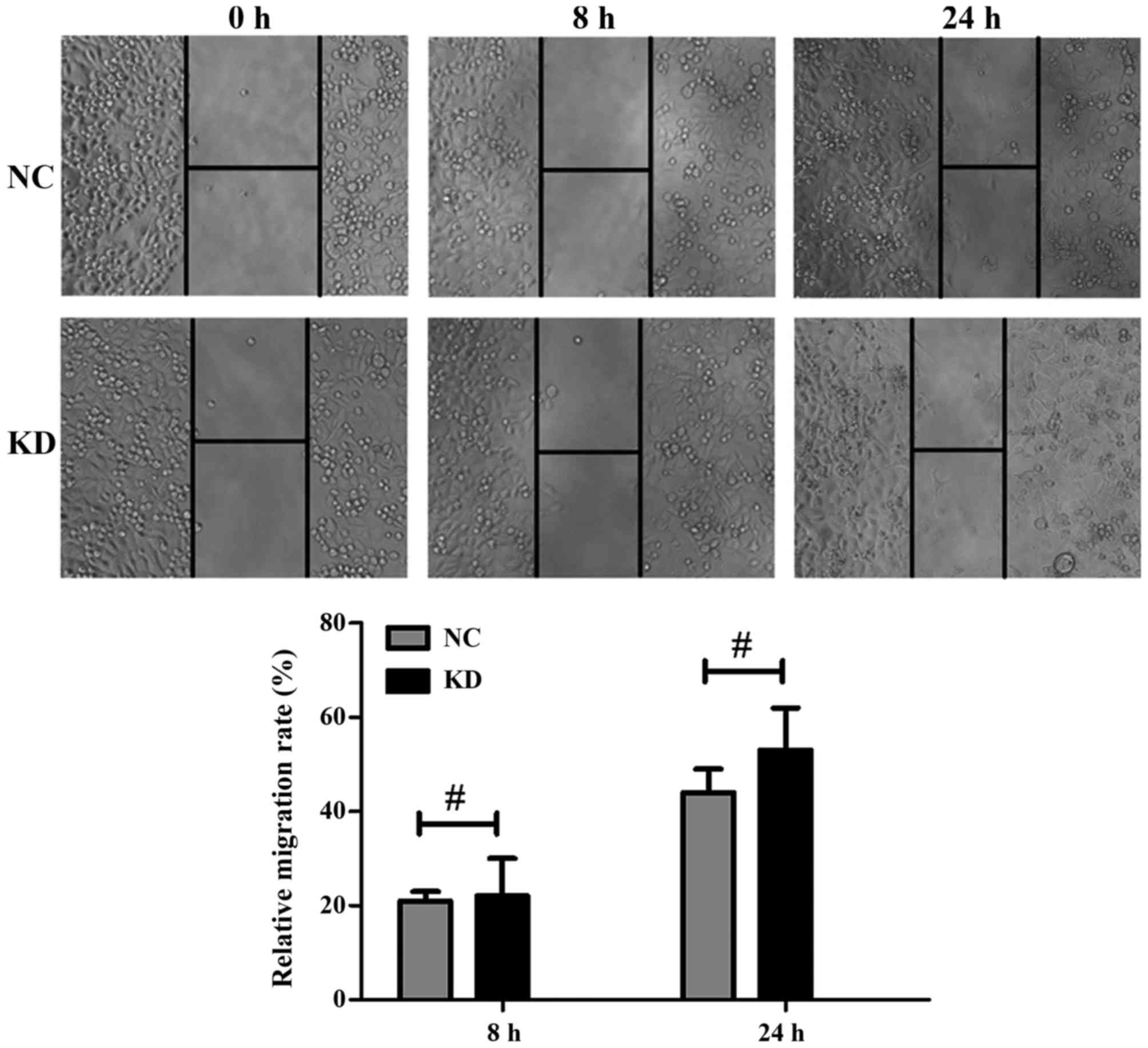
ATOH8 knockdown has no effect on the
cell motility of CRC cells
The wound healing assay was used to determine the
effects of ATOH8 downregulation on cell motility. As shown in
Fig. 7, the wound healing assay
indicated ATOH8 knockdown had no effect on the cell migration rate
of SW620 cells. The relative migration rate at 8 h of NC and KD
group was 21±2 and 22±8%, respectively, P>0.05; and migration
rate at 24 h was 44±5 and 53±9%, respectively, P>0.05.
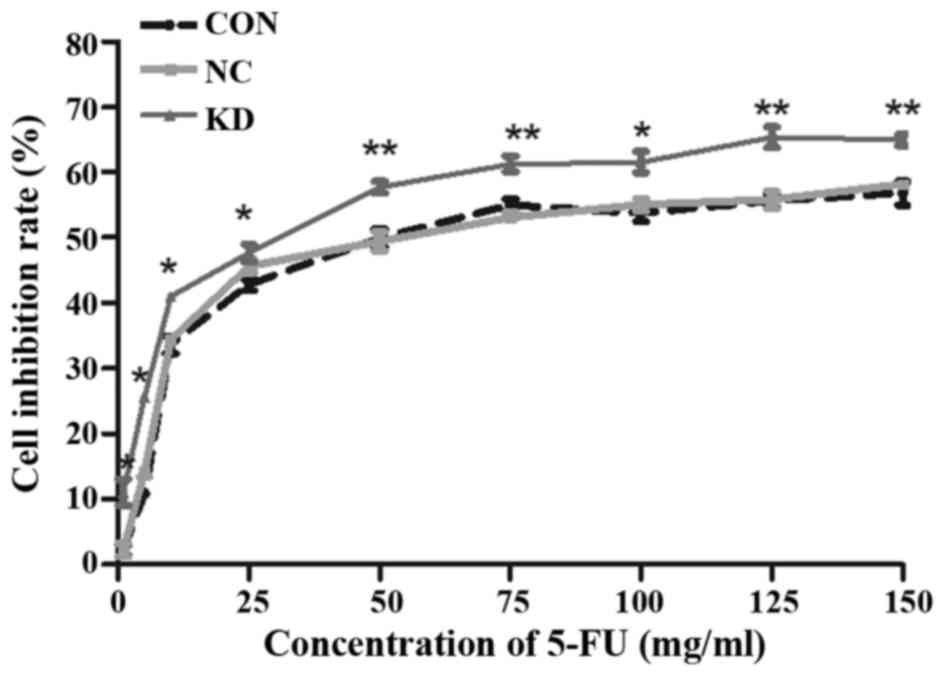
ATOH8 knockdown increases the
sensitivity of CRC cells to 5-FU
The effect of ATOH8 knockdown on drug sensitivity
was performed by evaluating the cytotoxicity of 5-FU in SW620
cells. The inhibitory effect of various concentrations of 5-FU was
assessed by MTT assay following 24 h of treatment. The inhibitory
effect of 5-FU was dose-dependent in the CON, NC and KD groups
(Fig. 8). In addition, the
inhibitory rate of the KD group was increased more significantly
than the CON and NC groups as the drug concentration increased,
which meant that ATOH8 knockdown increased the sensitivity of SW620
to 5-FU.
Discussion
ATOH8 belongs to the atonal family and consists of
321 amino acids with a bHLH domain. It has been shown that ATOH8
has important regulatory functions during the differentiation of
pancreatic precursor cells, neurons, podocytes, skeletal muscle,
retina and endothelial cells (6,8–11,19,20).
With the improvement of array techniques, it was found that ATOH8
exhibited abnormal expression in various cancers such as prostate
and breast cancer, glioblastoma multiforme and hepatocellular
carcinoma (12,13,15–17).
Song et al (12) focused on
the function of ATOH8 in hepatocellular cancer. They demonstrated
that ATOH8 expression was decreased in hepatocellular cancer, and
that the low expression of ATOH8 was associated with poor
differentiation, high serum AFP level and worse overall survival.
The same researchers constructed ATOH8 overexpression and
low-expression models in HCC cell lines and demonstrated that
upregulation of ATOH8 decreased tumor proliferation, invasion, and
migratory abilities as well as increased the sensitivity of HCC
cell lines to chemotherapy, while downregulation of ATOH8 showed
the opposite effects. Moreover, Wang et al (13) designed a series of experiments in
nasopharyngeal carcinoma (NPC) and demonstrated that the inhibition
of ATOH8 expression promoted a malignant phenotype of NPC and that
this malignant phenotype could be reversed by ATOH8 restoration.
These two studies suggested that ATOH8 acted as a tumor-suppressor
gene in HCC and NPC. However, a study in breast cancer (17) suggested that ATOH8 was a downstream
effector of IL-6/STAT3 signaling, which increased the stemness
potential of breast cancer cell lines. Thus, the function of ATOH8
in cancer is controversial. The role of ATOH8 in colorectal cancer
(CRC) is currently unknown, thus, we investigated the expression of
ATOH8 in CRC tissues and the function of this gene in
vitro.
By comparing 106 biopsies of carcinoma tissues with
their corresponding peritumoral tissues, immunohistochemical
results demonstrated that the expression of ATOH8 was clearly
increased in the carcinoma tissues. Bowden et al (21) demonstrated that in familial
adenomatous polyposis adenomas, a high risk subset of CRC, ATOH8
expression was increased compared with normal colon epithelial
cells, which supports our results. When we compared the expression
of ATOH8 with clinicopathological features, we found that high
expression of ATOH8 was associated with a high serum CEA level.
There are several studies that have shown that a high serum CEA
level is a poor prognosis predictor for CRC patients (22–24).
Thus, we also investigated the relationship between the serum CEA
level and the OS of the patients, and the result was consistent
with former studies (22–24). Furthermore, our results based on the
Kaplan-Meier analysis revealed that patients with high levels of
ATOH8 expression had a shorter overall survival time than those
with low expression. Considering all these results, we proposed
that the expression of ATOH8 is a potential prognostic factor of
colorectal carcinoma.
To explore the function of ATOH8 in colon cancer, we
suppressed ATOH8 expression in SW620 cells through ATOH8-siRNA
transfection. We successfully constructed a downregulated ATOH8
model confirmed by western blotting and qRT-PCR. The MTT assay
suggested that the growth rate of CRC cells was significantly
decreased in the downregulated ATOH8 group compared to the control
and negative groups, which suggests that ATOH8 promotes cell
proliferation in CRC cell lines. Studies have demonstrated that
different types of bHLH transcriptional factors have marked
similarity in many functional regions (25), and numerous bHLH transcriptional
factors play important roles in tumor growth and proliferation. For
example, Ascle2, a bHLH transcriptional factors (TF), promoted
cellular proliferation in CRC (26), and E2A (i.e., isoforms E12 and E47)
and accelerated the cellular growth rate in prostate cancer
(27).
Flow cytometric analysis suggested that the
proportion of cells in the G0/G1 phase increased while the
percentage of cells in the S and G2/M phase decreased, which means
that ATOH8 depletion induced S phase arrest accompanied by a
decrease in cell proliferation. Additionally, ATOH8 depletion
increased the proportion of apoptotic CRC cells. There are no other
studies that have been conducted concerning the effect of ATOH8 on
the cell cycle and apoptosis in malignancy, thus far. However, it
has been reported that some bHLH transcriptional factors, such as
Hes1, through its inhibitor induced apoptosis in cancer cells by
inhibiting the Notch signaling pathway. Currently, some Hes1
inhibitors are in clinical trials (28,29).
The effect of ATOH8 on the cell motility of CRC
cells was also investigated by wound healing assay. The results
indicated that ATOH8 depletion did not affect the cell motility of
colon cancer cells. Although, in HCC and NPC (12,13),
the expression of ATOH8 was associated with the invasive and
migratory abilities of the cancer, in these two tumor types ATOH8
functioned as a cancer suppressor gene.
The potential therapeutic value of ATOH8 in CRC
treatment was also investigated. From our results, the inhibitory
rate of the ATOH8-downregulated cells was higher than the controls
at different concentrations of 5-FU, which implies that
downregulation of ATOH8 increases the sensitivity of colon cancer
cells to chemotherapy. It also suggests that ATOH8 may be a
potential therapeutic target for CRC.
The first major limitation of the present study was
the fact that we did not explore the molecular mechanism of ATOH8
in CRC. Secondly, we only explored the function of ATOH8 in an
in vitro cell model without in vivo experiments. Our
next goal is to find the downstream effector of ATOH8 and the
corresponding signaling pathways.
In summary, our data demonstrated that ATOH8
expression was upregulated in CRC patients, and that it predicted
poor prognosis. The upregulation of ATOH8 may increase the
malignancy potential of CRC. Finally, ATOH8 is a potential
therapeutic target of CRC.
Acknowledgements
The present study received research funding from the
Zhejiang Provincial Natural Science Foundation of China
(LY15H160059).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Zuo T, Zeng H, Zhang S
and He J: National cancer incidence and mortality in China, 2012.
Chin J Cancer Res. 28:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weir B, Zhao X and Meyerson M: Somatic
alterations in the human cancer genome. Cancer Cell. 6:433–438.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang B, Balakrishnan-Renuka A, Napirei M,
Theiss C and Brand-Saberi B: Spatiotemporal expression of Math6
during mouse embryonic development. Histochem Cell Biol.
143:575–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inoue C, Bae SK, Takatsuka K, Inoue T,
Bessho Y and Kageyama R: Math6, a bHLH gene expressed in the
developing nervous system, regulates neuronal versus glial
differentiation. Genes Cells. 6:977–986. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lynn FC, Sanchez L, Gomis R, German MS and
Gasa R: Identification of the bHLH factor Math6 as a novel
component of the embryonic pancreas transcriptional network. PLoS
One. 3:e24302008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ross MD, Martinka S, Mukherjee A, Sedor
JR, Vinson C and Bruggeman LA: Math6 expression during kidney
development and altered expression in a mouse model of
glomerulosclerosis. Dev Dyn. 235:3102–3109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang F, Wasserman SM, Torres-Vazquez J,
Weinstein B, Cao F, Li Z, Wilson KD, Yue W, Wu JC, Xie X, et al:
The role of Hath6, a newly identified shear-stress-responsive
transcription factor, in endothelial cell differentiation and
function. J Cell Sci. 127:1428–1440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Güttsches AK, Balakrishnan-Renuka A, Kley
RA, Tegenthoff M, Brand-Saberi B and Vorgerd M: ATOH8: A novel
marker in human muscle fiber regeneration. Histochem Cell Biol.
143:443–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yao J, Zhou J, Liu Q, Lu D, Wang L, Qiao X
and Jia W: Atoh8, a bHLH transcription factor, is required for the
development of retina and skeletal muscle in zebrafish. PLoS One.
5:e109452010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song Y, Pan G, Chen L, Ma S, Zeng T, Chan
Man TH, Li L, Lian Q, Chow R, Cai X, et al: Loss of ATOH8 increases
stem cell features of hepatocellular carcinoma cells.
Gastroenterology. 149:1068–1081.e5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Xie J, Yan M, Wang J, Wang X,
Zhang J, Zhang Y, Li P, Lei X, Huang Q, et al: Downregulation of
ATOH8 induced by EBV-encoded LMP1 contributes to the malignant
phenotype of nasopharyngeal carcinoma. Oncotarget. 7:26765–26779.
2016.PubMed/NCBI
|
|
14
|
Zaravinos A, Lambrou GI, Boulalas I,
Delakas D and Spandidos DA: Identification of common differentially
expressed genes in urinary bladder cancer. PLoS One. 6:e181352011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Freire P, Vilela M, Deus H, Kim YW, Koul
D, Colman H, Aldape KD, Bogler O, Yung WK, Coombes K, et al:
Exploratory analysis of the copy number alterations in glioblastoma
multiforme. PLoS One. 3:e40762008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hazelett DJ, Rhie SK, Gaddis M, Yan C,
Lakeland DL, Coetzee SG, Henderson BE, Noushmehr H, Cozen W,
Kote-Jarai Z, et al: Ellipse/GAME-ON consortium; Practical
consortium: Comprehensive functional annotation of 77 prostate
cancer risk loci. PLoS Genet. 10:e10041022014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang A, Chen Y, Shen W, Gao R, Zhou W,
Luo Y, Luo N, Stupack D and Xiang R: Abstract 1956: The basic
helix-loop-helix (bHLH) transcriptional factor ATOH8 promotes the
stemness of breast cancer cells via Oct4 and Nanog. Cancer Res.
74:(Suppl 19): Abstract nr 1956.
2014.doi:10.1158/1538-7445.AM2014-1956.
|
|
18
|
Chu X, Zhao P, Lv Y and Liu L: Decreased
expression of TFPI-2 correlated with increased expression of CD133
in cholangiocarcinoma. Int J Clin Exp Pathol. 8:328–336.
2015.PubMed/NCBI
|
|
19
|
Ejarque M, Mir-Coll J, Gomis R, German MS,
Lynn FC and Gasa R: Generation of a conditional allele of the
transcription factor atonal homolog 8 (Atoh8). PLoS One.
11:e01462732016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balakrishnan-Renuka A, Morosan-Puopolo G,
Yusuf F, Abduelmula A, Chen J, Zoidl G, Philippi S, Dai F and
Brand-Saberi B: ATOH8, a regulator of skeletal myogenesis in the
hypaxial myotome of the trunk. Histochem Cell Biol. 141:289–300.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bowden NA, Croft A and Scott RJ: Gene
expression profiling in familial adenomatous polyposis adenomas and
desmoid disease. Hered Cancer Clin Pract. 5:79–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tampellini M, Ottone A, Alabiso I,
Baratelli C, Forti L, Berruti A, Aroasio E and Scagliotti GV: The
prognostic role of baseline CEA and CA 19–9 values and their
time-dependent variations in advanced colorectal cancer patients
submitted to first-line therapy. Tumor Biol. 36:1519–1527. 2015.
View Article : Google Scholar
|
|
23
|
Yang KL, Yang SH, Liang WY, Kuo YJ, Lin
JK, Lin TC, Chen WS, Jiang JK, Wang HS, Chang SC, et al:
Carcinoembryonic antigen (CEA) level, CEA ratio, and treatment
outcome of rectal cancer patients receiving pre-operative
chemoradiation and surgery. Radiat Oncol. 8:432013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen L, Jiang B, Di J, Zhang C, Wang Z,
Zhang N, Xing J, Cui M, Yang H, Yao Z, et al: Predictive value of
preoperative detection of CEA and CA199 for prognosis in patients
with stage II–III colorectal cancer. Zhonghua Wei Chang Wai Ke Za
Zhi. 18:914–919. 2015.(In Chinese). PubMed/NCBI
|
|
25
|
Tsigelny IF, Kouznetsova VL, Pingle SC and
Kesari S: bHLH Transcription factors inhibitors for cancer therapy:
General features for in silico drug design. Curr Med Chem.
21:3227–3243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu R, Yang Y, Tian Y, Bai J, Zhang X, Li
X, Peng Z, He Y, Chen L, Pan Q, et al: Ascl2 knockdown results in
tumor growth arrest by miRNA-302b-related inhibition of colon
cancer progenitor cells. PLoS One. 7:e321702012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Patel D and Chaudhary J: Increased
expression of bHLH transcription factor E2A (TCF3) in prostate
cancer promotes proliferation and confers resistance to doxorubicin
induced apoptosis. Biochem Biophys Res Commun. 422:146–151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Axelson H: The Notch signaling cascade in
neuroblastoma: Role of the basic helix-loop-helix proteins HASH-1
and HES-1. Cancer Lett. 204:171–178. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sang L, Roberts JM and Coller HA:
Hijacking HES1: How tumors co-opt the anti-differentiation
strategies of quiescent cells. Trends Mol Med. 16:17–26. 2010.
View Article : Google Scholar : PubMed/NCBI
|