Introduction
Breast cancer, with a high mortality around the
world, is one of the most frequent cancers among women. There are
more than 1.7 million new cases of breast cancer worldwide in one
year, with more than 522,000 deaths (1). Despite the fact that both basic
scientists and clinical researchers have carried out many
investigations on breast cancer, the prognosis remains unfavorable,
the main reasons are attributed to the immature diagnosis and
therapy and the adverse effects of surgeries (2,3).
MicroRNAs (miRNAs) are short non-coding RNA
molecules which are 20-25-nt long highly conserved and bind
primarily to sequences within the 3 untranslated region (3UTR) of
mRNAs. The primary function of miRNAs is to regulate its target
gene expression by degradation or post-transcriptional regulation
(4,5). miRNAs target mRNA with a
semi-complimentary seed sequence (6–9 bp) which guides binding to
the response elements. One miRNA may have many potential targets
because each seed sequence may match many mRNA molecules (5). Hundreds of miRNAs are found in
well-defined transcriptional units, and can be in either intronic
or exonic regions in non-coding transcriptional regions, or as
intronic miRNAs in coding regions (6). Several previous studies have proposed
that microRNAs are involved in a broad range of biological
processes through their regulation of complex signal transduction
pathways (6,7). Half of human miRNAs can lead to the
dysregulation of oncogenes and tumor suppressors in cancers by
locating at fragile sites (8).
Recent studies have revealed that miRNA signatures can stratify
molecular subgroups of breast cancer (9,10).
Moreover, some miRNAs are able to predict the tumor progression and
there are some reports on prognosis of patients with breast cancer
(11,12). There is only one study on miR-597 in
pediatric patients with immune thrombocytopenic purpura (13), but there have been no studies on the
function of miR-597 in breast cancer.
Fos-related antigen 2 (FOSL2), also named FRA-2,
belongs to the AP-1 transcription factor family which includes the
various isoforms of Fos and Jun (14). FOSL2 has been reported as a specific
function gene in bone development (15) and appears to associate with diverse
physiological and pathological processes, such as photoperiodic
regulation (16) fibrosis (17) and even in cancer (18). In addition, FOSL2 has been
documented as a key determinant of cellular plasticity during CD4 T
cell differentiation (19). Some
studies have reported that FOSL2 plays a key role in the regulation
in some pathways, such as TGF-β pathway, for example, FOSL2 induce
TGF-β expression as a transcriptional regulator in cardiac
fibroblasts (20). In human breast
cancer, FOSL2 expression has been reported to enhance invasive
properties (20,21), but there have been few studies on
the other functions.
In the present study, we first found that miR-597
was significantly downregulated in breast cancer tissues compared
with the adjacent normal tissues. We then studied the expression
and function of miR-597 in breast cancer cells. We also verified
that FOSL2 was a direct target of miR-597, and that miR-597
negatively regulated FOSL2 expression to inhibit cell
proliferation, migration and invasion in breast cancer.
Materials and methods
Cell culture and clinical
specimens
Five breast carcinoma cell lines (T-47D, SK-BR-3,
MDA-MB-231, MCF7 and BT-474) were purchased from the Chinese
Academy of Science Cell Bank (Shanghai, China). Esophageal squamous
cell carcinoma (EC-109), human cervical adenocarcinoma (HeLa),
prostate cancer (PC1) and MCF10A cells were kindly provided by the
Central Laboratory, Tongji University Affiliated People's 10th
Hospital (Shanghai, China). All cells except MCF10A were cultured
with 10% fetal bovine serum (FBS; Gibco-BRL/Life Technologies,
Paisley, UK) in high-glucose Dulbeccos modified Eagles medium
(DMEM; HyClone Laboratories, Inc., Logan, UT, USA) and supplemented
with antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin;
Gibco-BRL, Grand Island, NY, USA) at 37°C with 5% CO2 in
a humidified incubator. The MCF10A was maintained in complete media
[DMEM/F12 (50:50 mix) supplemented with 5% horse serum, 10 mg/ml 10
mM HEPES, insulin, 20 ng/ml epidermal growth factor, 0.5 mg/ml
hydrocortisone and 100 ng/ml cholera toxin]. Thirty-eight
pair-matched breast tissue samples were obtained from the
Department of Breast and Thyroid Surgery, Tongji University
Affiliated People's 10th Hospital (Shanghai, China). All samples
were obtained surgically and immediately snap- frozen and stored in
liquid nitrogen until use. The Institutional Review Board approved
the tissue procurement protocol used in the study, and the
appropriate informed consent was obtained from the patients.
RNA isolation and quantitative
real-time PCR
Total RNA was isolated by TRIzol reagent according
to the manufacturers protocol (Invitrogen, Carlsbad, CA, USA). cDNA
was performed using 1000 ng of total RNA as the template with
looped primers and using a PrimeScript RT reagent kit (RR037A;
Takara Bio, Inc., Shiga, Japan). Quantitative real-time PCR was
performed in a 96-well plate according to the protocol of the KAPA
SYBR FAST Universal qPCR kit (KK4601; Kapa Biosystems, Wilmington,
MA, USA). An ABI 7900HT PCR sequencer (Applied Biosystems, Foster
City, CA, USA) was used for the data measured, and gene expression
was calculated using the ∆∆Ct method for relative quantitation
using 18S or U6 as endogenous reference genes. The sequences of
specific primers are 5-GAGAGGAACA AGCTGGCTGC-3 and
5-GCTTCTCCTTCTCCTTCTGC-3 for FOSL2; 5-CGTACAGCATCCTGGAGATA-3
and 5-CC TTCAGCTTACAGTCATTG-3 for FGF10; 5-CTGAGCTC
TCTGAGTGCAAC-3 and 5-GGTCTGAGCTGTATCGC TGC-3 for EGFR;
5-GCAAGACTCCAGCGCCTTCT-3 and 5-CTCATCTTCTTGTTCCTCCTC-3 for
MYC; 5-CCTGG ATACCGCAGCTAGGA-3 and 5-GCGGCGCAATACGAA
TGCCCC-3 for 18S RNA; 5-ACACTCCAGCTGGGTGTG TCACTCGATGAC-3
and 5-TGGTGTCGTGGAGTCG-3 for miR-597; 5-CTCGCTTCGGCAGCACA-3
and 5-AACGCT TCACGAATTTGCGT-3 for U6.
Cell proliferation, migration and
invasion assays
Cell proliferation was detected using MTT assay,
1,000 cells transfected with synthesized oligonucleotides were
seeded in 96-well plates. At 0, 48, 72 and 96 h, 100 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution (0.5 mg/ml) (Sigma-Aldrich, St. Louis, MO, USA) was added
to each well, after incubated for another 4 h the medium was
removed, and 150 µl of dimethyl sulfoxide (DMSO) was added. The
plates were read at a wavelength of 490 nm. Cell migration was
detected by the wound-healing assay, an artificial wound was
created by a 200 µl pipette tip. To visualize migrated cells and
wound healing, images were taken at 0 and 36 h. Cell invasion was
detected using a Transwell chamber (Millipore, Billerica, MA, USA)
with Matrigel (BD Biosciences, San Jose, CA, USA). Cells
(1×105) were cultured in the top chamber of Transwell
coated with Matrigel. Cells were cultured in medium with no fetal
bovine serum (FBS) in the upper chamber and added with FBS-DMEM in
the lower chamber. After 36 h, invasive cells on the lower chamber
were fixed in 95% ethanol and stained with 0.5% crystal violet, the
numbers were determined using ImageJ software (National Institutes
of Health, Bethesda, MD, USA).
Cell cycle assay
A total of 5×105 transfected cells were
seeded into 6-well plates and incubated in 0.2% serum medium for
cell synchronization. Subsequently, the cells were incubated with
complete medium for 48 h. The cells were stained with propidium
iodide (PI) (550825; BD Biosciences, San Diego, CA, USA) and
~1×105 cells were examined with a FACS flow cytometer
and analyzed with ModFit software (BD Biosciences, San Jose, CA,
USA).
EdU assay
A total of 1.5×105 transfected cells were
plated into 6-well plates, and the assay performed according to the
manufacturers protocol of a Cell-Light™ EdU Apollo® 488
In Vitro Imaging kit (C10310-3; Guangzhou RiboBio, Co.,
Ltd., Guangzhou, China). Finally, cells were visualized and counted
using a fluorescence microscope (Leica Microsystems, Wetzlar,
German).
Luciferase reporter assays
We used the pGL3-reporter luciferase vector to
construct the pGL3-FOSL2 3UTR or pGL3-FOSL2 3UTR-mut vectors. The
pGL3-FOSL2 3UTR-mut vector was obtained using a
KOD-Plus-Mutagenesis kit (F0936K; Toyobo, Co., Ltd., Osaka, Japan).
The Luciferase activity of NC and miR-597 were measured whit the
Dual-luciferase reporter assay system (Promega Corp., Fitchburg,
WI, USA) after transfection for 36 h. Firefly luciferase activity
was normalized to Renilla luciferase activity for each
transfected well. The sequences of specific primers are 5-AT
ACTCGAGAGCACCTTCAAGCGCTCCAG-3 and 5-GG AAGCTTCTGCTGCTTGGATTCATCTC-3
for FOSL2-3UTR, 5-ATACTCGAGATGTACCAGGATTATCCCGG-3′ and
5-GCGGAATTCTTACAGAGCCAGCAGAGTGG-3 for FOSL2-CDS.
Western blot analysis
Protein lysates were separated by sodium dodecyl
sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to
nitrocellulose membranes (Millipore, Billercia, MA, USA). The
primary antibodies were incubated overnight after blocking with 5%
fat-free milk for 1 h and the primary antibodies were β-actin
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), FOSL2
(ab124830; Abcam, Cambridge, UK), MMP-2 and MMP-9 (13132, 13667;
Cell Signaling Technologies, Beverly, MA, USA) and then incubated
with their corresponding secondary antibodies (LI-COR Biosciences,
Lincoln, NE, USA). Odyssey LI-CDR scanner (BD Biosciences) were
used for the membranes.
Statistical analysis
All statistical analyses were performed using the
SPSS version 20.0 (SPSS, Inc., Chicago, IL, USA). The data are
presented as the mean ± SD. The statistical significance is shown
as P<0.05, P<0.01.
Results
miR-597 is downregulated in breast
cancer tissues and cells
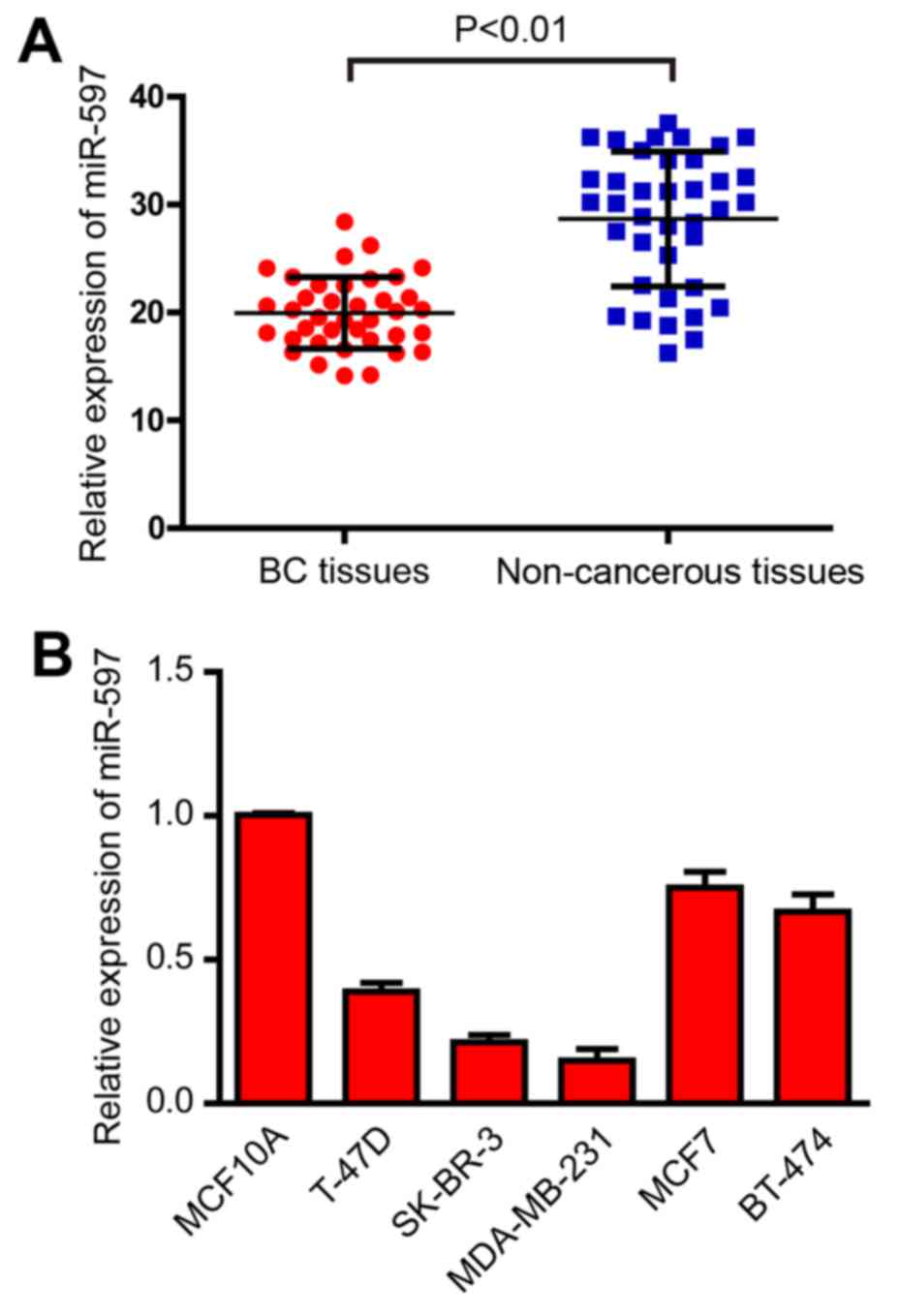
In order to ascertain the expression of miR-597 in
breast cancer, qRT-PCR was performed in 38 pairs of breast cancer
and non-cancerous tissues. As shown in Fig. 1A, the results demonstrated that
miR-597 was significantly downregulated in breast cancer tissues
(Fig. 1A). Human breast cancer cell
lines T-47D, SK-BR-3, MDA-MB-231, MCF7 and BT-474, and normal
breast cell line MCF10A were also detected by qRT-PCR. As shown in
Fig. 1B, the expression levels of
miR-597 were also downregulated in the five breast cancer cell
lines (Fig. 1B). Since MDA-MB-231
and SK-BR-3 cells exhibited the lowest miR-597 expression among the
five breast cancer cell lines, these two cell lines were chosen for
mature miR-597 mimic transfection and for further studies.
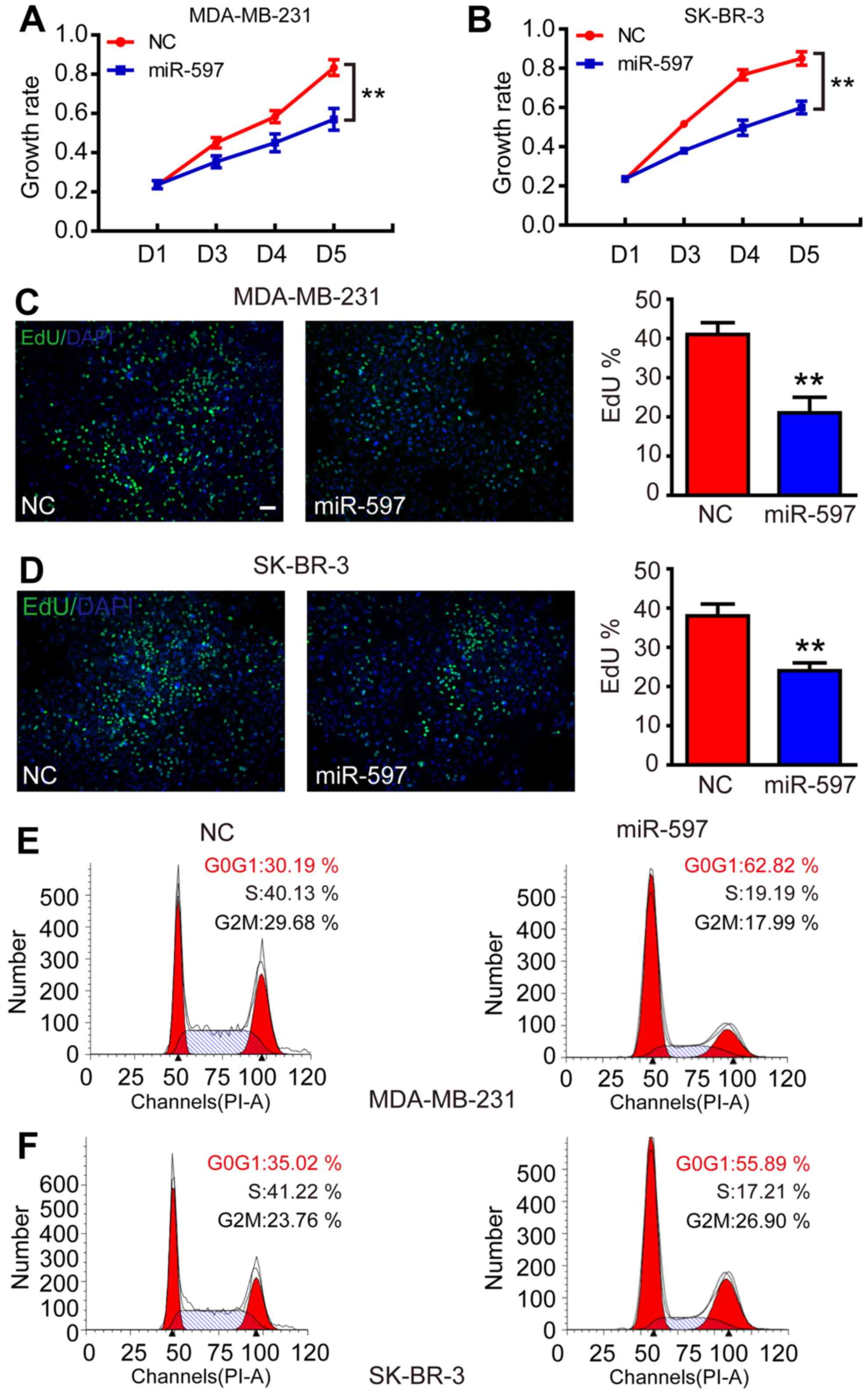
Overexpression of miR-597 inhibits
cell proliferation
Subsequently, to evaluate the function of miR-597 in
tumorigenesis, we transfected miR-597 mimics into MDA-MB-231 and
SK-BR-3 to increase the levels of ectopic miR-597. As a result, the
elevation of miR-597 significantly inhibited the growth rate of
MDA-MB-231 and SK-BR-3 from the third day (Fig. 2A and B; P<0.01). The cell
proliferation process was modulated by the cell cycle, thus, an EdU
assay was performed. The result showed that the miR-597 subclone
incorporated less EdU compared with the NC subclone (Fig. 2C and D; P<0.01), indicating that
miR-597 reduced cells into S-phase. In addition, flow cytometry
revealed that cells with miR-597 overexpression had a 20–30% higher
proportion at the G0/G1 phase and a 20% lower proportion at the S
phase compared with NC groups (Fig. 2E
and F). All results suggested that miR-597 inhibited cell
proliferation by modulating the G1-S phase of the cell cycle.
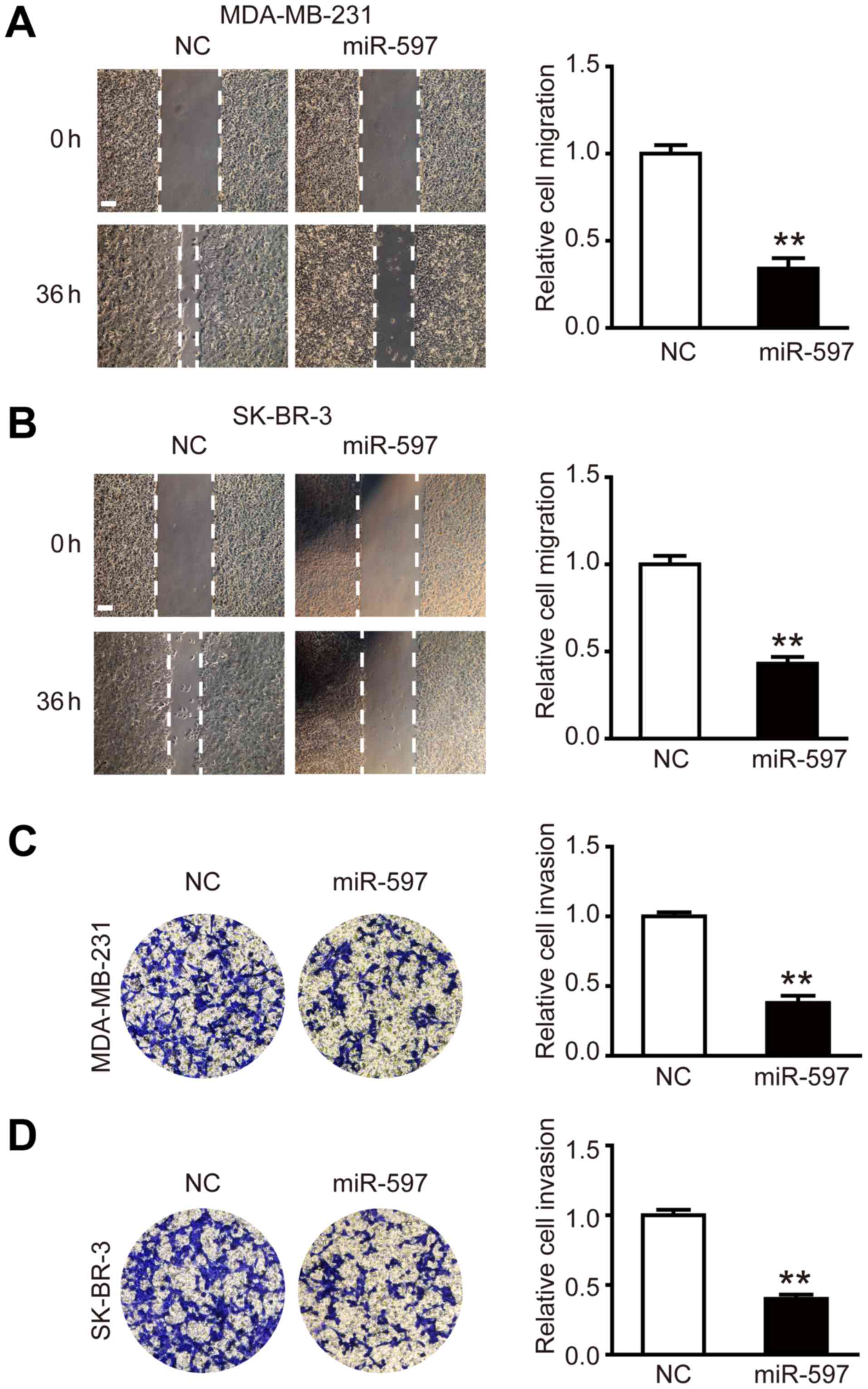
Overexpression of miR-597 inhibits
cell migration and invasion
To investigate the effect of miR-597 on the
migration and invasion of breast cancer cells, miR-597 mimic or its
negative control were transfected into MDA-MB-231 and SK-BR-3
cells, respectively. The wound healing assay was performed, the
results showed that the migration ability of MDA-MB-231 and SK-BR-3
cells in the miR-597 group was lower than NC groups at 36 h
post-wounding (Fig. 3A and B;
P<0.01). The cell invasion assay showed similar results, in the
miR-597 group, there was ~50% reduction in the average number of
cells penetrating the Transwell membrane and Matrigel compared with
NC group (Fig. 3C and D;
P<0.01). By detecting and analyzing with western blot analysis,
we found that the expression levels of invasion-related proteins
MMP-2 and MMP-9 were reduced (data not shown). Thus, the above
results suggest that miR-597 plays an important part in the cell
migration and invasion.
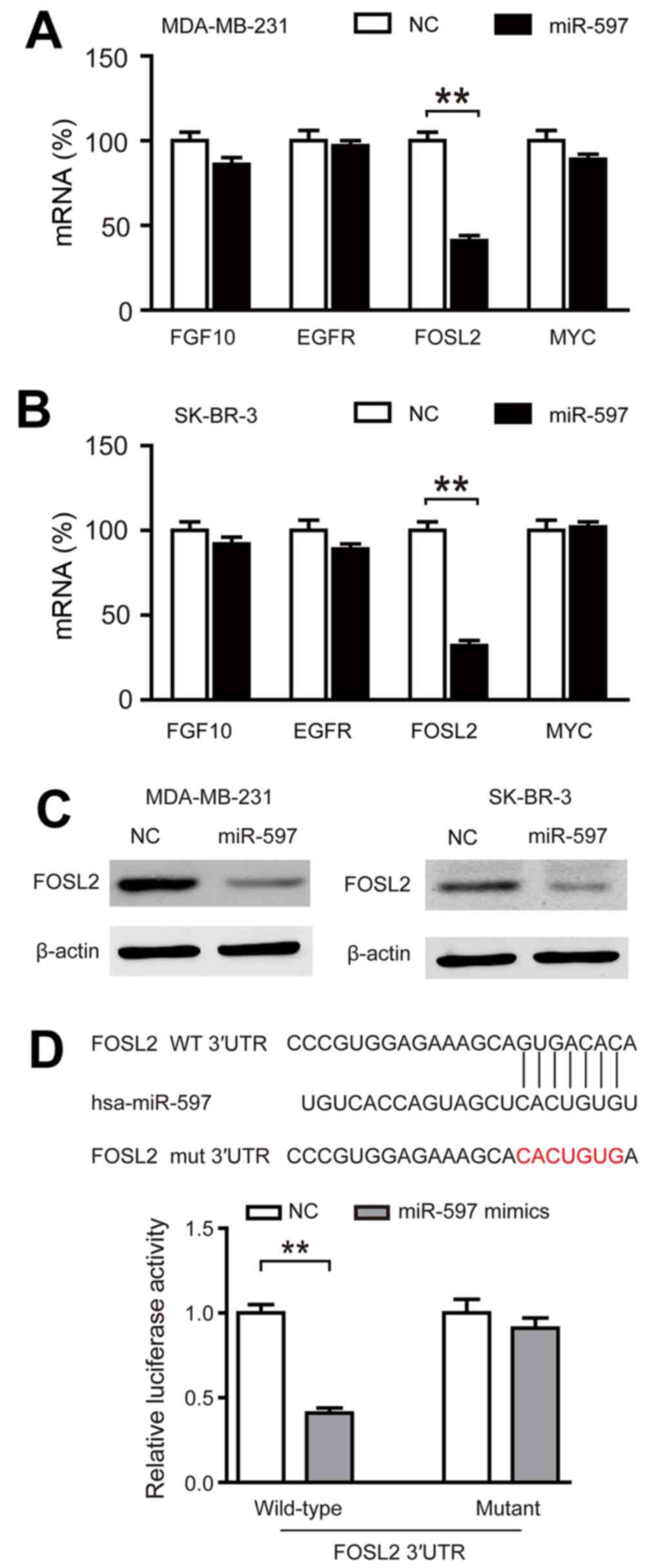
FOSL2 is a direct target of
miR-597
In order to find the targets of miR-597, we used
TargetScanHuman 6.2 and GeneCoDis3 to predict the targets of
miR-597 and we found that four genes (FGF10, EGFR, FOSL2 and MYC)
had functions that oppose miR-597 in breast cancer. By performing
qRT-PCR, we only found FOSL2 markedly inhibited by miR-597
(Fig. 4A and B; P<0.01). In
addition, the expression of FOSL2 protein in the miR-597 subclone
was lower than that in the NC subclone (Fig. 4C). The luciferase reporter assays
also demonstrated that miR-597 mimics significantly reduced the
activity of FOSL2 3-UTR, while there was no change in the mutant
group (Fig. 4D). These data
suggested that FOSL2 is a direct target of miR-597.
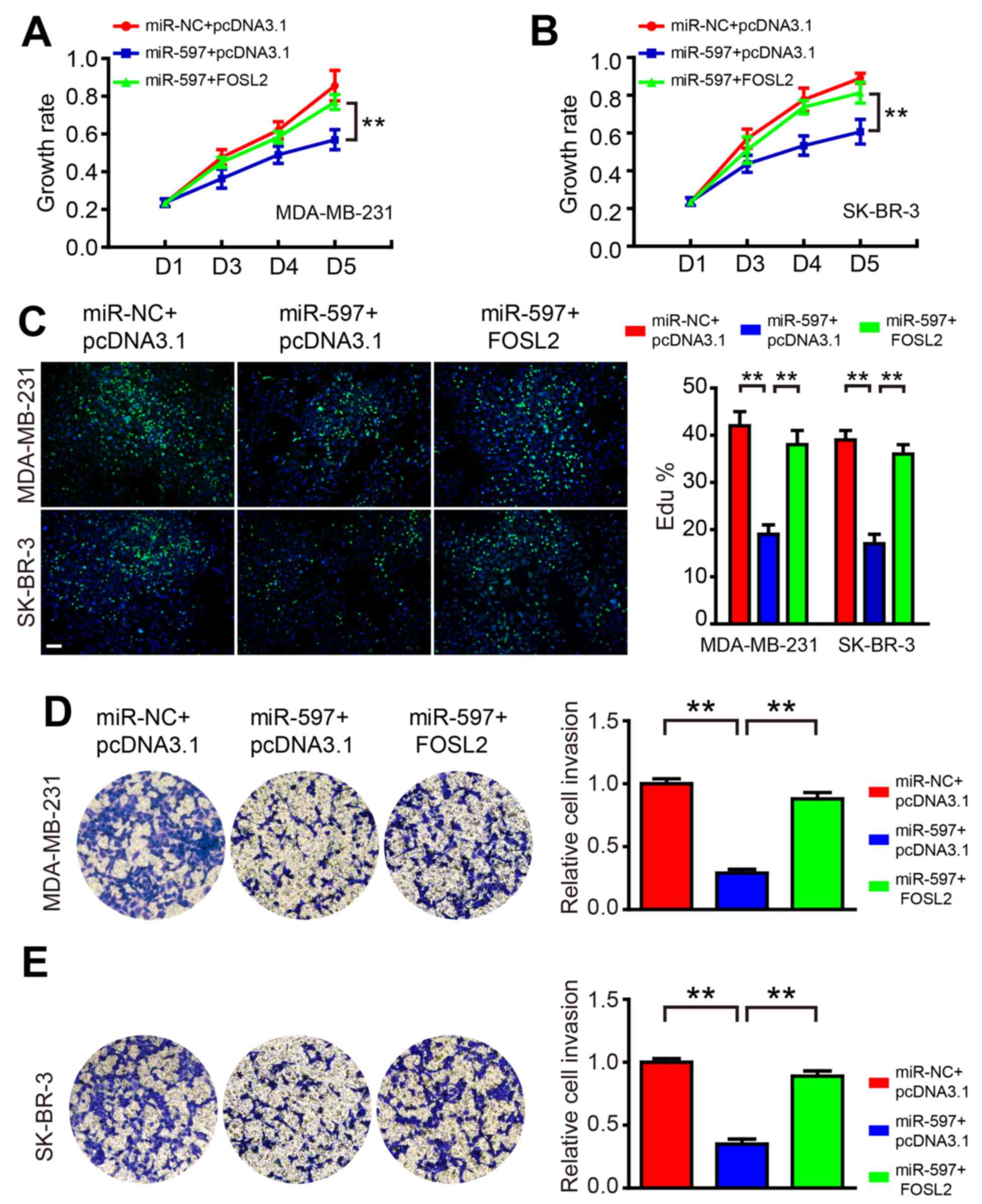
Overexpression of FOSL2 blocks the
roles of miR-597
To test the hypothesis that miR-597 regulates cell
proliferation and invasion by targeting FOSL2, we co-transfected
miR-597 mimics and FOSL2-expressing vector into MDA-MB-231 and
SK-BR-3 cells. The western blot result showed that the level of
FOSL2 decreased by miR-597 could be rescued by overexpression of
FOSL2 in cells (data not shown). As shown in Fig. 5, FOSL2 reintroduction reversed the
anti-proliferation and anti-invasion roles of miR-597, suggesting
that the effect of miR-597 on cell growth and invasion were largely
mediated by its downregulation of FOSL2.
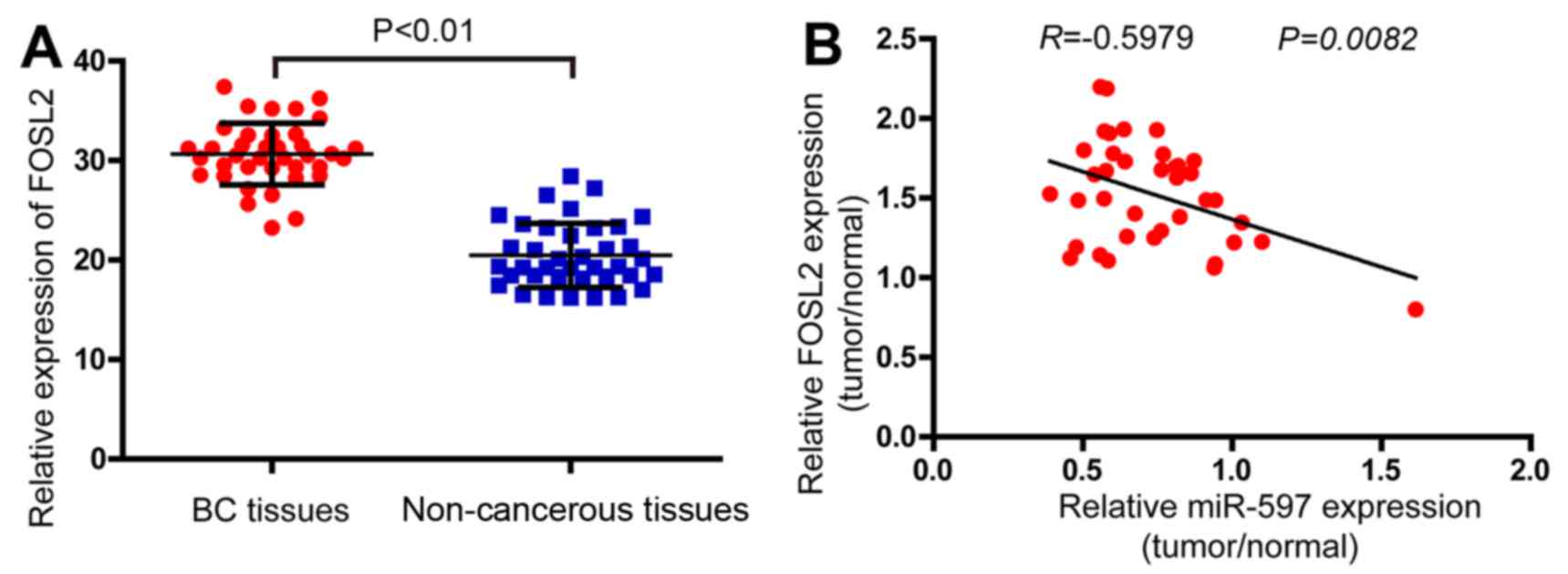
Effect of FOSL2 and miR-597 on the
clinicopathological features of breast cancer
In order to determine the expression levels of FOSL2
in breast cancer, the real-time PCR analysis was performed in 38
pairs of breast cancer and adjacent non-cancerous tissues. The
results demonstrated that the FOSL2 mRNA were higher in tumor
samples compared with the adjacent normal tissues (Fig. 6A). Finally, analyzing with Pearson
correlation the expression of FOSL2 and miR-597 (Fig. 1A) in these 38 tumor samples, we
found a negative correlation between the two (Fig. 6B, R=−0.5979; P=0.0082).
Discussion
Increasing number of miRNAs are reported in
different cancers. The altered expression of miRNAs lead to changes
in cell proliferation, migration, invasion, survival and apoptosis
underlying many diseases, included cancers. After the first report
on deregulation of miRNA in human lymphocytic leukemia (22) many other studies showed aberrant
expression profiles of miRNA in breast cancer. However, to the best
of our knowledge, this is the first report on the expression level
and biological function of miR-597 in breast cancer. In the present
study, we did not only clearly demonstrate that miR-597 functioned
as a tumor suppressor that inhibited breast cell proliferation,
migration and invasion, but we also identified that FOSL2 was one
of its target genes.
As the first investigation on miR-597 in breast
cancer, we speculated whether miR-597 had similar influence on
other types of cancers. Thus, the esophageal squamous cell line
EC-109, the cervical cancer cell line HeLa and prostate cancer cell
line PC1 were used for similar functional research (data not
shown), and their proliferation was also inhibited by miR-597, the
result suggested that miR-597 has similar function in regulating
cell proliferation on other types of cancer cells. However, we
detected the expression of FOLS2 in these types of cancers while
overexpression of miR-597, we found that the prostate cancer cell
line PC1 had an insignificant change of FOSL2 while others had a
significant down-expression. We considered that the function of
miR-597 in PC1 was not through FOSL2 but other targets, this
finding suggested that FOSL2 is one of the targets of miR-597.
There may be a number of other targets of miR-597 not yet
found.
The AP-1 family is known to be involved in cellular
proliferation and oncogenesis, depending on the combination of AP-1
proteins and the cellular context (23,24).
Fra-2 is one component of a functional AP-1 complex that can also
comprise of Jun-Jun, Jun-Fos or Jun-ATF2 dimers (25). FOSL2 was originally identified as a
gene encoding FOS related antigen gene-2 (26,27)
and shown to be oncogenic for chicken embryonic fibroblasts (CEFs)
(27). In our studies, we found
that FOSL2 was significantly upregulated in breast cancer tissues
compared with the adjacent normal tissues. FOSL2 overexpression in
the miR-597-overexpression cancer cells block the vast majority of
miR-597 roles, which is consistent with previous studies (20,21).
FOSL2 may be one of the targets of miR-597 in breast cancer cells,
it can partly rescue the effect in cell proliferation (Fig. 5A and B). Possibly there is another
target of miR-597 on this function which should be studied
further.
Various synthetic miRNAs have been reported to exert
powerful tumor-suppressive effects in many different cancer types,
such as bladder cancer, melanoma and gastric cancer (28–30),
even in breast cancer, artificial miRNA which targeted CXCR4 or
MTDH blocked the proliferation, invasion and metastasis (31,32).
All these findings suggested that artificially designed miRNA may
be a valuable approach in the treatment of breast cancer. Although
the synthetic miR-597 had powerful tumor-suppressive effects in
breast cancer, the mechanism remains still unknown. Two main
reasons were accepted for the abnormal versions of microRNAs: one
is the promoter modification (28)
and the other is the regulation of interacting proteins (29,30).
Regardless of the cause of the underexpression of miR-597, or which
pathway was affected by miR-597further studied are required.
Acknowledgements
The authors wish to thank Jiayi Wang, Shanshan Xu,
Ji Ma, Yue Zhang and Fenyong Sun of Shanghai Tongji University for
their technical assistance. The present study was supported by the
National Natural Science Foundation of China (grant no.
81371913).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 36:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cuzick J, DeCensi A, Arun B, Brown PH,
Castiglione M, Dunn B, Forbes JF, Glaus A, Howell A, von Minckwitz
G, et al: Preventive therapy for breast cancer: A consensus
statement. Lancet Oncol. 12:496–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quintavalle M, Elia L, Condorelli G and
Courtneidge SA: MicroRNA control of podosome formation in vascular
smooth muscle cells in vivo and in vitro. J Cell Biol. 189:13–22.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dang X, Ma A, Yang L, Hu H, Zhu B, Shang
D, Chen T and Luo Y: MicroRNA-26a regulates tumorigenic properties
of EZH2 in human lung carcinoma cells. Cancer Genet. 205:113–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009.PubMed/NCBI
|
|
8
|
Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang
Z, Tan F, Tan X, Zhou F, Sun J, et al: MicroRNA-25 promotes cell
migration and invasion in esophageal squamous cell carcinoma.
Biochem Biophys Res Commun. 421:640–645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G, et
al: MicroRNA signatures predict oestrogen receptor, progesterone
receptor and HER2/neu receptor status in breast cancer. Breast
Cancer Res. 11:R272009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Foekens JA, Sieuwerts AM, Smid M, Look MP,
de Weerd V, Boersma AW, Klijn JG, Wiemer EA and Martens JW: Four
miRNAs associated with aggressiveness of lymph node-negative,
estrogen receptor-positive human breast cancer. Proc Natl Acad Sci
USA. 105:13021–13026. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rothé F, Ignatiadis M, Chaboteaux C,
Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H,
Desmedt C, Harris AL, et al: Global microRNA expression profiling
identifies MiR-210 associated with tumor proliferation, invasion
and poor clinical outcome in breast cancer. PLoS One. 6:e209802011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bay A, Coskun E, Oztuzcu S, Ergun S,
Yilmaz F and Aktekin E: Plasma microRNA profiling of pediatric
patients with immune thrombocytopenic purpura. Blood Coagul
Fibrinolysis. 25:379–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tulchinsky E: Fos family members:
Regulation, structure and role in oncogenic transformation. Histol
Histopathol. 15:921–928. 2000.PubMed/NCBI
|
|
15
|
Bozec A, Bakiri L, Jimenez M, Rosen ED,
Catalá-Lehnen P, Schinke T, Schett G, Amling M and Wagner EF:
Osteoblast-specific expression of Fra-2/AP-1 controls adiponectin
and osteocalcin expression and affects metabolism. J Cell Sci.
126:5432–5440. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Engel L, Gupta BB, Lorenzkowski V,
Heinrich B, Schwerdtle I, Gerhold S, Holthues H, Vollrath L and
Spessert R: Fos-related antigen 2 (Fra-2) memorizes photoperiod in
the rat pineal gland. Neuroscience. 132:511–518. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roy S, Khanna S, Azad A, Schnitt R, He G,
Weigert C, Ichijo H and Sen CK: Fra-2 mediates oxygen-sensitive
induction of transforming growth factor beta in cardiac
fibroblasts. Cardiovasc Res. 87:647–655. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou L, Graves M, MacDonald G, Cipollone
J, Mueller CR and Roskelley CD: Microenvironmental regulation of
BRCA1 gene expression by c-Jun and Fra2 in premalignant human
ovarian surface epithelial cells. Mol Cancer Res. 11:272–281. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ciofani M, Madar A, Galan C, Sellars M,
Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et
al: A validated regulatory network for Th17 cell specification.
Cell. 151:289–303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schröder C, Schumacher U, Müller V, Wirtz
RM, Streichert T, Richter U, Wicklein D and Milde-Langosch K: The
transcription factor Fra-2 promotes mammary tumour progression by
changing the adhesive properties of breast cancer cells. Eur J
Cancer. 46:1650–1660. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Milde-Langosch K, Röder H, Andritzky B,
Aslan B, Hemminger G, Brinkmann A, Bamberger CM, Löning T and
Bamberger AM: The role of the AP-1 transcription factors c-Fos,
FosB, Fra-1 and Fra-2 in the invasion process of mammary
carcinomas. Breast Cancer Res Treat. 86:139–152. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eferl R and Wagner EF: AP-1: A
double-edged sword in tumorigenesis. Nat Rev Cancer. 3:859–868.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Daury L, Busson M, Tourkine N, Casas F,
Cassar-Malek I, Wrutniak-Cabello C, Castellazzi M and Cabello G:
Opposing functions of ATF2 and Fos-like transcription factors in
c-Jun-mediated myogenin expression and terminal differentiation of
avian myoblasts. Oncogene. 20:7998–8008. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsui M, Tokuhara M, Konuma Y, Nomura N
and Ishizaki R: Isolation of human fos-related genes and their
expression during monocyte-macrophage differentiation. Oncogene.
5:249–255. 1990.PubMed/NCBI
|
|
27
|
Nishina H, Sato H, Suzuki T, Sato M and
Iba H: Isolation and characterization of fra-2, an additional
member of the fos gene family. Proc Natl Acad Sci USA.
87:3619–3623. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Z, Zhan W, Wang Z, Zhu B, He Y, Peng J,
Cai S and Ma J: Inhibition of PRL-3 gene expression in gastric
cancer cell line SGC7901 via microRNA suppressed reduces peritoneal
metastasis. Biochem Biophys Res Commun. 348:229–237. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa
P, Gui Y and Cai Z: Synthetic miRNA-mowers targeting miR-183-96-182
cluster or miR-210 inhibit growth and migration and induce
apoptosis in bladder cancer cells. PLoS One. 7:e522802012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu X, Fang H, Chen H, Jiang X, Fang D,
Wang Y and Zhu D: An artificial miRNA against HPSE suppresses
melanoma invasion properties, correlating with a down-regulation of
chemokines and MAPK phosphorylation. PLoS One. 7:e386592012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang Z, Wu H, Reddy S, Zhu A, Wang S,
Blevins D, Yoon Y, Zhang Y and Shim H: Blockade of invasion and
metastasis of breast cancer cells via targeting CXCR4 with an
artificial microRNA. Biochem Biophys Res Commun. 363:542–546. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang S, Shu JZ, Cai Y, Bao Z and Liang QM:
Establishment and characterization of MTDH knockdown by artificial
MicroRNA interference - functions as a potential tumor suppressor
in breast cancer. Asian Pac J Cancer Prev. 13:2813–2818. 2012.
View Article : Google Scholar : PubMed/NCBI
|