Introduction
Oral squamous cell carcinoma (OSCC) is a common
malignancy that affects ~300,000 individuals per year worldwide
(1). OSCC is often associated with
loss of eating and speech functions, disfigurement, and
psychological distress. The primary treatment for OSCC is surgical
intervention. Despite considerable advances in the treatment of
OSCC over the past two decades, the overall disease outcomes have
improved only modestly (2). Local
tumor recurrence affects ~60% of patients, and metastasis develops
in ~15–25% of patients (3). The
prevention and management of OSCC will greatly benefit from the
identification of molecular markers and targets indicative of the
disease (4,5).
Over the course of the last 20 years, saliva has
been used to evaluate periodontal disease and the risk of dental
caries. It has recently been reported that biomarkers for various
diseases, including cancer, may be identified in the saliva,
indicating the potential value of saliva as a test sample instead
of blood. Recently, salivary diagnosis using various biochemical
analytical techniques for the detection of breast and pancreatic
cancers has been developed (6).
Using two-dimensional electrophoresis for whole
saliva, which can be easily sampled in a non-invasive manner,
Katakura et al (7)
successfully identified an enolase 1 that is characteristically
expressed in the whole saliva of patients with oral cancer.
Therefore, the research program of the present authors has
continued to focus on salivary metabolomics in our conducting a
metabolome analysis and attempting a simultaneous exhaustive search
for low-molecular-weight markers for the identification of a
plethora of metabolites. Sugimoto et al (6) reported 24 candidate metabolites from
saliva samples that were able to serve as biomarkers for cancer
patients of various races, geographic regions, and tumor types.
This previous study used capillary electrophoresis-mass
spectrometry (CE-MS), which is a combined method that has been
adapted for the high-resolution separation of ionic compounds, and
may be used for metabolome analysis.
The purpose of the present study was to identify
metabolic biomarkers in Japanese patients with OSCC using CE-MS
metabolome analysis of saliva.
Materials and methods
Subjects
Saliva was obtained from Japanese patients with OSCC
(n=22) and from healthy controls (n=21) who visited the Department
of Dentistry, Oral and Maxillofacial Surgery, Tokyo Dental Collage
Ichikawa General Hospital, Tokyo, Japan between September 2013 and
March 2015. None of the patients had received any prior treatment
in the form of chemotherapy or radiotherapy, and no patient had a
history of prior malignancy; information regarding the samples is
summarized in Table I. Healthy
controls were selected amongst individuals that did not have a
history of mucosal diseases in the oral cavity, immunodeficiency,
autoimmune disorders, hepatitis, or human immunodeficiency virus
(HIV) infection. Written informed consent was obtained from all the
subjects. The present study was approved by the ethics committee of
Tokyo Dental College (Tokyo, Japan; no. 105).
 | Table I.Clinical characteristics of the
patients with OSCC and healthy controls. |
Table I.
Clinical characteristics of the
patients with OSCC and healthy controls.
| Characteristics | OSCC patients
(n=22) | Healthy controls
(n=21) |
|---|
| Age (yrs.; mean ±
SD) | 68±13 | 56±8 |
| Gender |
| Male | 13 | 8 |
|
Female | 9 | 13 |
| Tumor site |
|
Tongue | 15 |
|
Gingiva | 6 |
| Oral
floor | 1 |
| T classification |
| T1 | 7 |
| T2 | 7 |
| T3 | 1 |
| T4 | 7 |
| N classification |
| N0 | 19 |
| N1 | 2 |
| N2 | 1 |
| N3 | 0 |
| Stage |
| I | 7 |
| II | 7 |
|
III | 1 |
| IV | 7 |
Sample collection and preparation
All subjects received professional mechanical tooth
cleaning by a dental hygienist the day prior to sample collection,
and saliva was collected at 8:00 a.m. the following morning under
fasting conditions after sufficient gargling and other oral hygiene
steps. The subjects were instructed to spit into 50-cc tubes, which
were placed in a Styrofoam cup filled with crushed ice. The
subjects were reminded not to cough up mucus. It usually took 5–10
min to collect 5 ml of unstimulated saliva. Saliva collection was
performed in a restful private room. The saliva samples were
centrifuged at 2,600 × g for 15 min at 4°C, and spun for a further
20 min in cases where incomplete separation was observed. After the
impurities in the saliva were percolated with a centrifugal filter
(Nanosep®; Pall Corporation, Port Washington, NY, USA),
equal amounts of the supernatant were transferred to two fresh
tubes, and the samples were processed and frozen within 30 min.
Frozen saliva was thawed and dissolved at room temperature. Prior
to the metabolome analyses, each saliva sample (45 µl) was added to
5 µl Milli-Q water (Merck Millipore, Billerica, MA, USA) containing
internal standards and 20 mM each of methionine sulfone,
D-camphor-10-sulfonic acid (Wako Pure Chemical Industries, Ltd.
Osaka, Japan), 2-(n-morpholino)ethanesulfonic acid (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan), 3-aminopyrrolidine
(Sigma-Aldrich Japan K.K., Tokyo, Japan), and trimesate (Wako Pure
Chemical Industries, Ltd.).
CE-MS metabolome analysis
Cation analysis was performed using a CE capillary
electrophoresis system (G1600AX), a G6220A LC/MSD time-of-flight
(TOF) system, a 1100-series isocratic high-performance liquid
chromatography (HPLC) pump, a CE-MS adapter kit, and a
CE-electrospray ionization (ESI)-MS sprayer kit (Agilent
Technologies GmbH, Waldbronn, Germany). Anion analysis was
performed using a CE capillary electrophoresis system (G1600AX), a
G1969A LC/MSD TOF system, a 1100-series isocratic HPLC pump, a
CE-MS adapter kit, and a CE-ESI source-MS sprayer kit (Agilent
Technologies GmbH). For the cation and anion analyses, the CE-MS
adapter kit included a capillary cassette that facilitates
thermostatic control of the capillary. The CE-ESI-MS sprayer kit
simplifies coupling of the CE system with the MS system, and is
equipped with an electrospray source. For system control and data
acquisition, 3D-CE ChemStation software (rev. A.09.03.SR1 and
A.10.02) and Agilent MassHunter software were used for CE and
TOF-MS (B.04.00 and B.02.00) analyses, respectively. The original
Agilent SST316Ti stainless steel ESI needle was replaced with a
passivated SST316Ti stainless steel and platinum needle (passivated
with 1% formic acid and a 20% aqueous solution of isopropanol at
80°C for 30 min) for anion analysis.
Processing of CE-TOF-MS data and
statistical analysis
The metabolite standards, instrumentation, and
CE-TOF-MS conditions used in the present study were identical to
those previously described (8),
with slight modifications in the lock mass system setting. The
metabolites were analyzed using a fused silica capillary (50 µm
i.d.×80 cm total length) with a commercial electrophoresis buffer
(Solution ID: H3301-1001 for cation analysis and H3302-1021 for
anion analysis; Human Metabolome Technologies, Inc., Yamagata,
Japan) as the electrolyte.
Hierarchical cluster analysis was performed using
the proprietary software packages, PeakStat and SampleStat,
respectively. Detected metabolites were plotted on metabolic
pathway maps using Visualization and Analysis of Networks
Containing Experimental Data (VANTED) software. Statistical
analyses were performed with the Wilcoxon rank sum test to compare
the two groups. P<0.05 was considered to indicate a
statistically significant value.
Metabolite identification
Although CE-TOF-MS provides accurate molecular mass
information at the milli-m/z level, the m/z value alone is seldom
sufficient to identify a metabolite (9,10).
Therefore, in the present analysis a combination of the m/z values
and the migration times predicted by the artificial neural networks
(ANNs) (11) were used to identify
the metabolites. In brief, the ANN model was first trained using
the measured migration times and molecular descriptors of standard
compounds with the net charge calculated from the pKa values. The
trained ANN model then predicted the migration times of the
candidate metabolites. Herein, the compounds selected as candidates
were available in the Kyoto Encyclopedia of Gene and Genomics
(KEGG) database (12) and the Human
Metabolome Database (HMDB) (13).
The composition formulae obtained using the MS data and the matched
candidates were confirmed by their isotope distribution
patterns.
Results
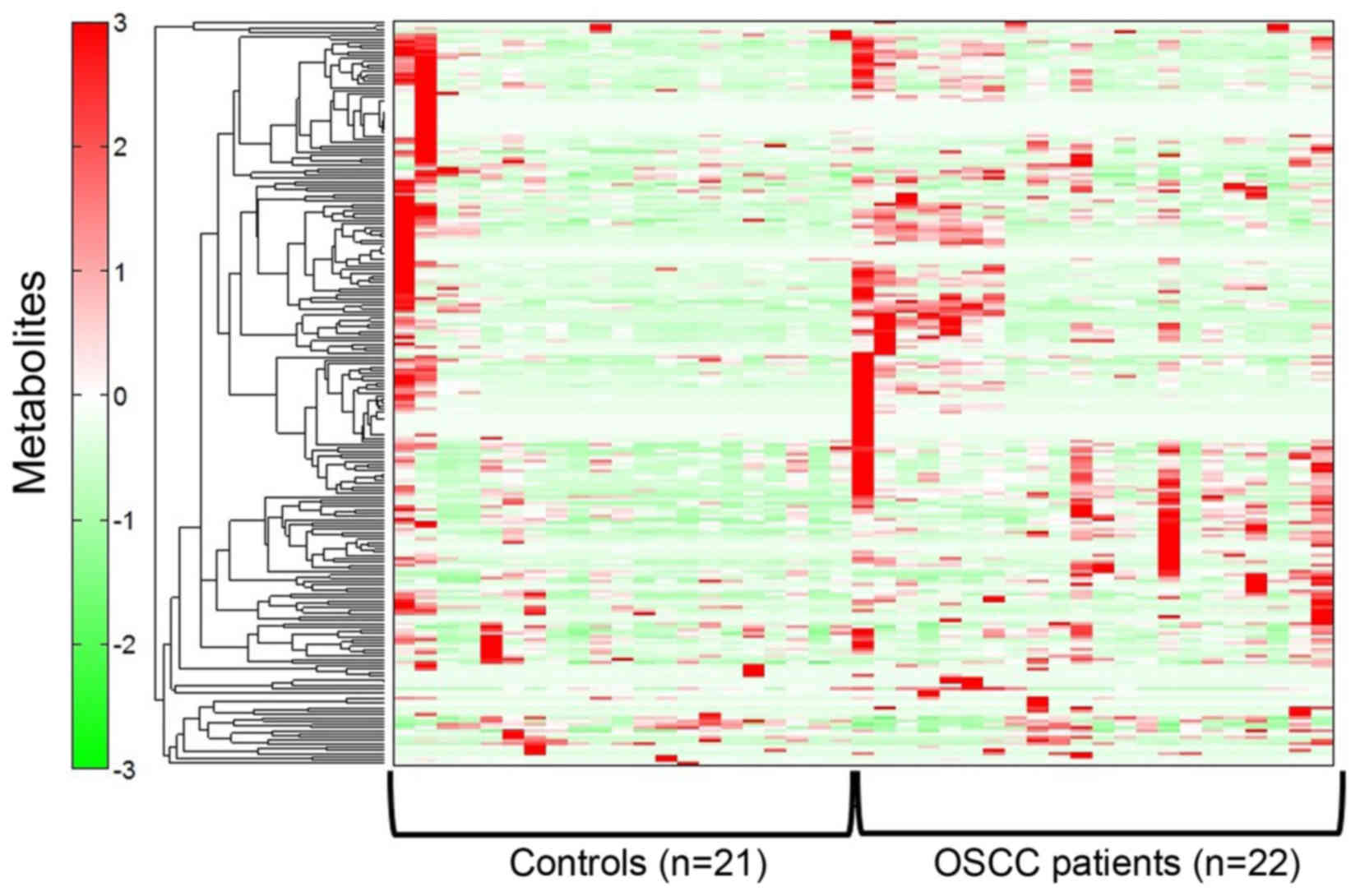
Heat map representation of metabolome
analysis from patients with OSCC and healthy controls
The saliva samples from 22 patients with OSCC and 21
healthy controls were collected, and metabolites were extracted for
CE-MS metabolome analysis.
After having eliminated the extra peaks, such as the
isotopic and fragment peaks, the CE-TOF-MS analysis resulted in the
detection of 499 peaks in all the saliva samples. Of these, 251
peaks were attributable to known standard metabolites (135 cation
peaks and 116 anion peaks). These were identified and quantified
with the metabolite standards by matching the m/z values with the
normalized migration times. The remaining 248 peaks detected
belonged to unknown metabolites (45 cation peaks and 203 anion
peaks). The score results are presented as a heat map (Fig. 1).
Metabolome pathway in patients with
OSCC and healthy controls
A total of 499 metabolites were detected as peaks in
patients with OSCC and healthy controls using CE-MS. Of the total
number of metabolites, 25 were identified as potential markers that
could be used to discriminate between individuals with OSCC and
healthy controls (Table II):
Choline, p-hydroxyphenylacetic acid, and 2-hydroxy-4-methylvaleric
acid (P<0.001); valine, 3-phenyllactic acid, leucine, hexanoic
acid, octanoic acid, terephthalic acid, γ-butyrobetaine, and
3-(4-hydroxyphenyl)propionic acid (P<0.01); and isoleucine,
tryptophan, 3-phenylpropionic acid, 2-hydroxyvaleric acid, butyric
acid, cadaverine, 2-oxoisovaleric acid,
N6,N6,N6-trimethyllysine, taurine, glycolic acid,
3-hydroxybutyric acid, heptanoic acid, alanine, and urea
(P<0.05). Among these, seven salivary metabolites in patients
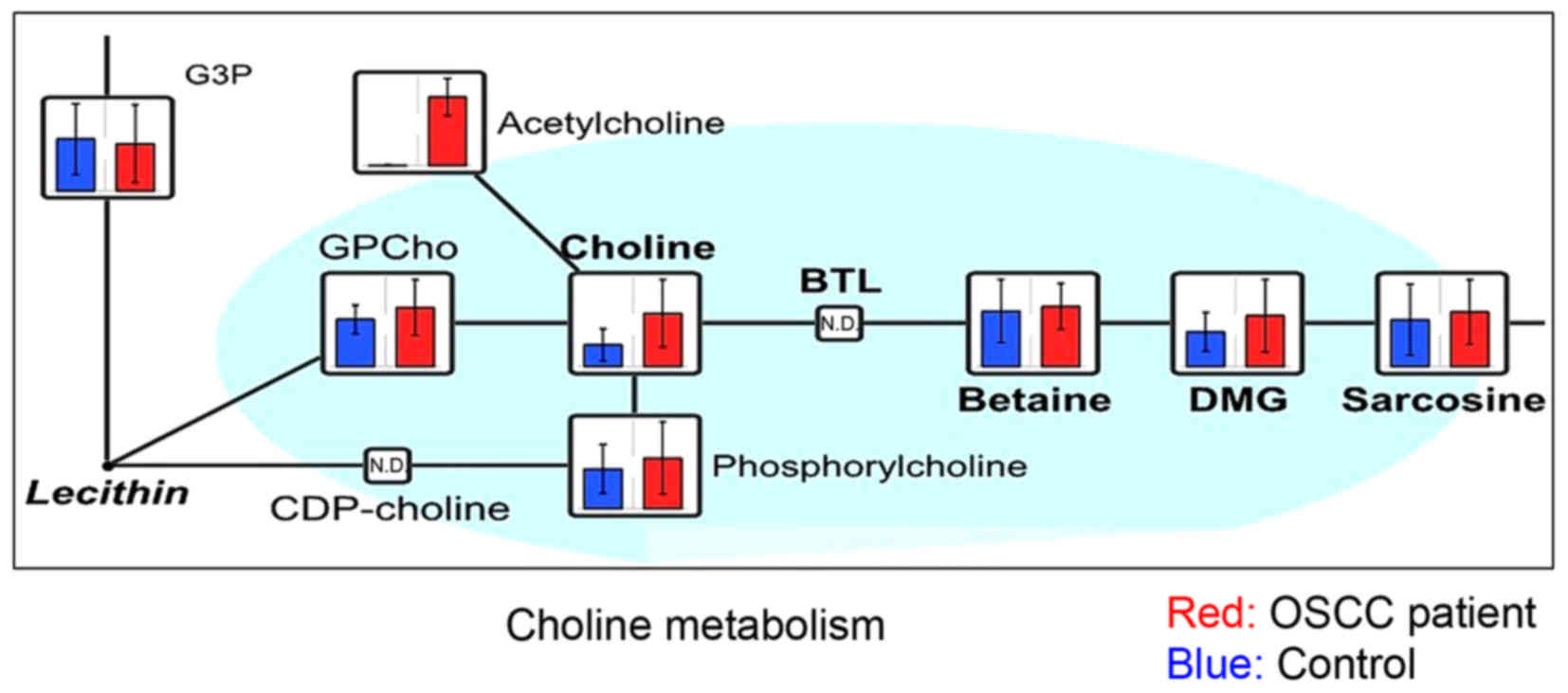
with OSCC were further characterized: Choline (Fig. 2), metabolites of the branched-chain
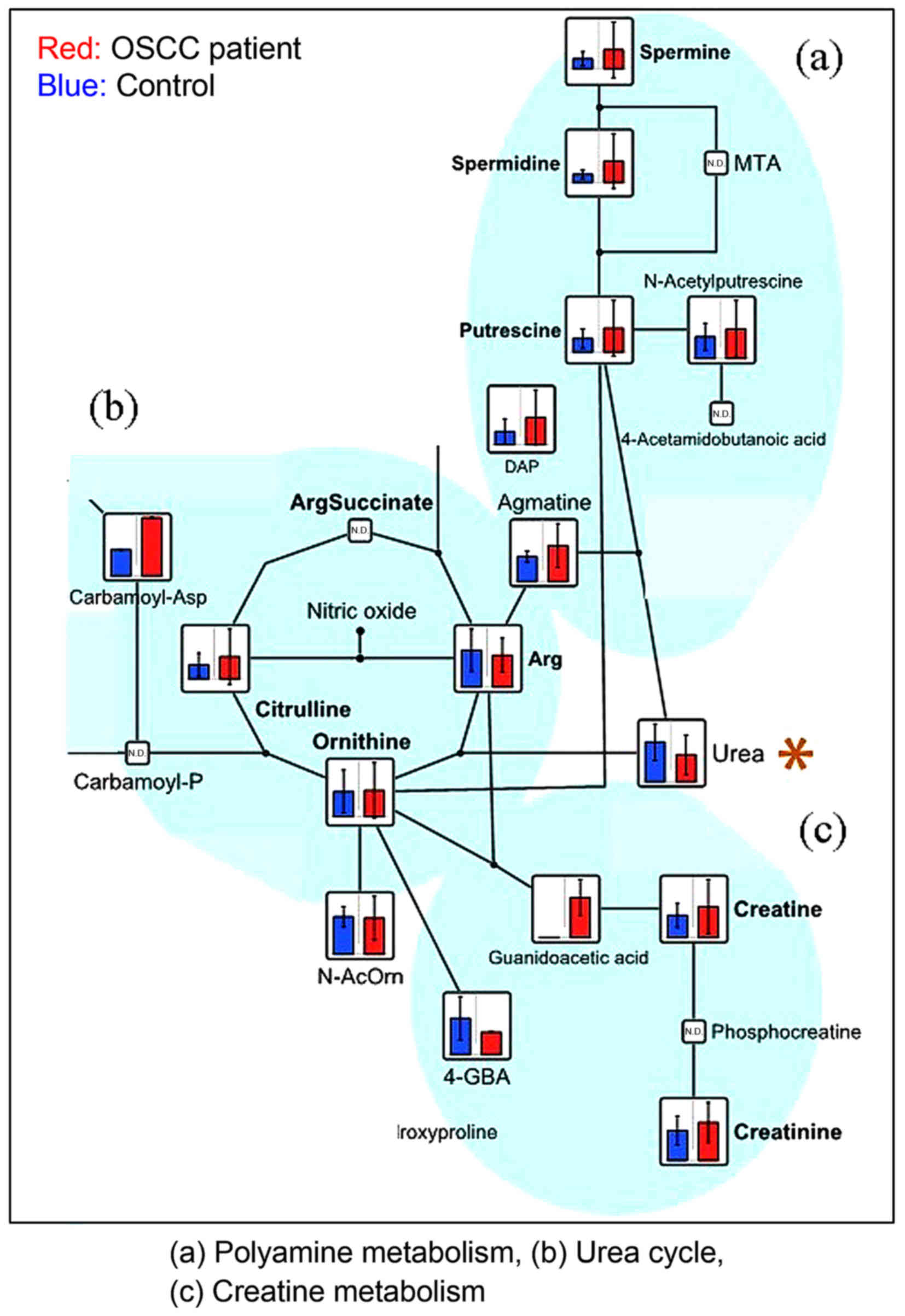
amino acids (BCAA) cycle (valine, isoleucine, leucine, and
2-oxoisovaleric acid) (Fig. 3),
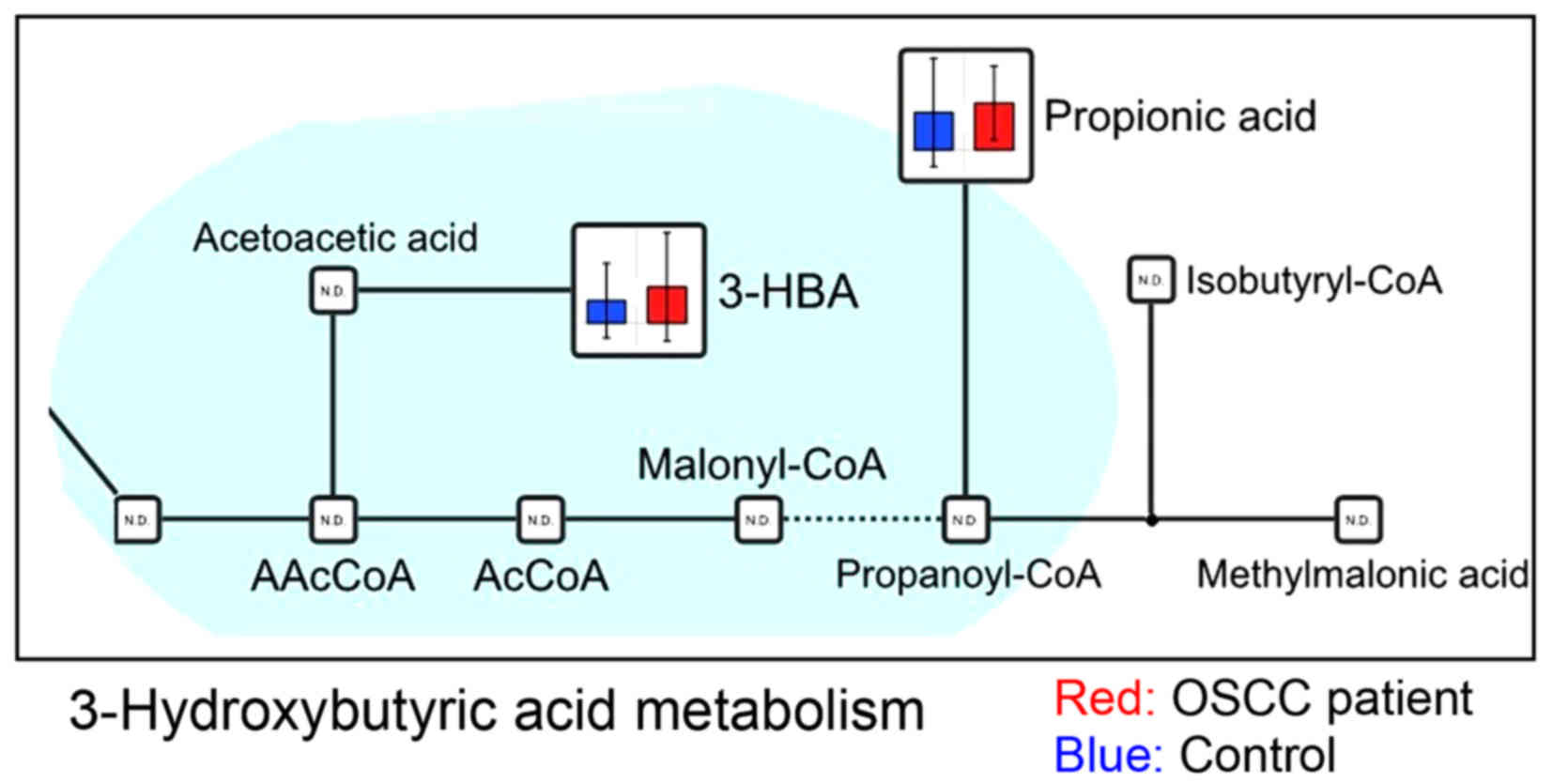
urea (Fig. 4), and 3-hydroxybutyric
acid (Fig. 5). Choline showed the
greatest statistically significant difference between patients with
OSCC and healthy controls in the present study. Sugimoto et
al (6) previously reported that
choline and the metabolites of the BCAA cycle could be salivary
biomarkers for OSCC, but 2-oxoisovaleric acid was not detected in
the previous study. Urea was the only metabolite that exhibited a
lower level in patients with OSCC compared with healthy controls
(Fig. 4). There has been no
previous report of 3-hydroxybutyric acid in OSCC (Fig. 5).
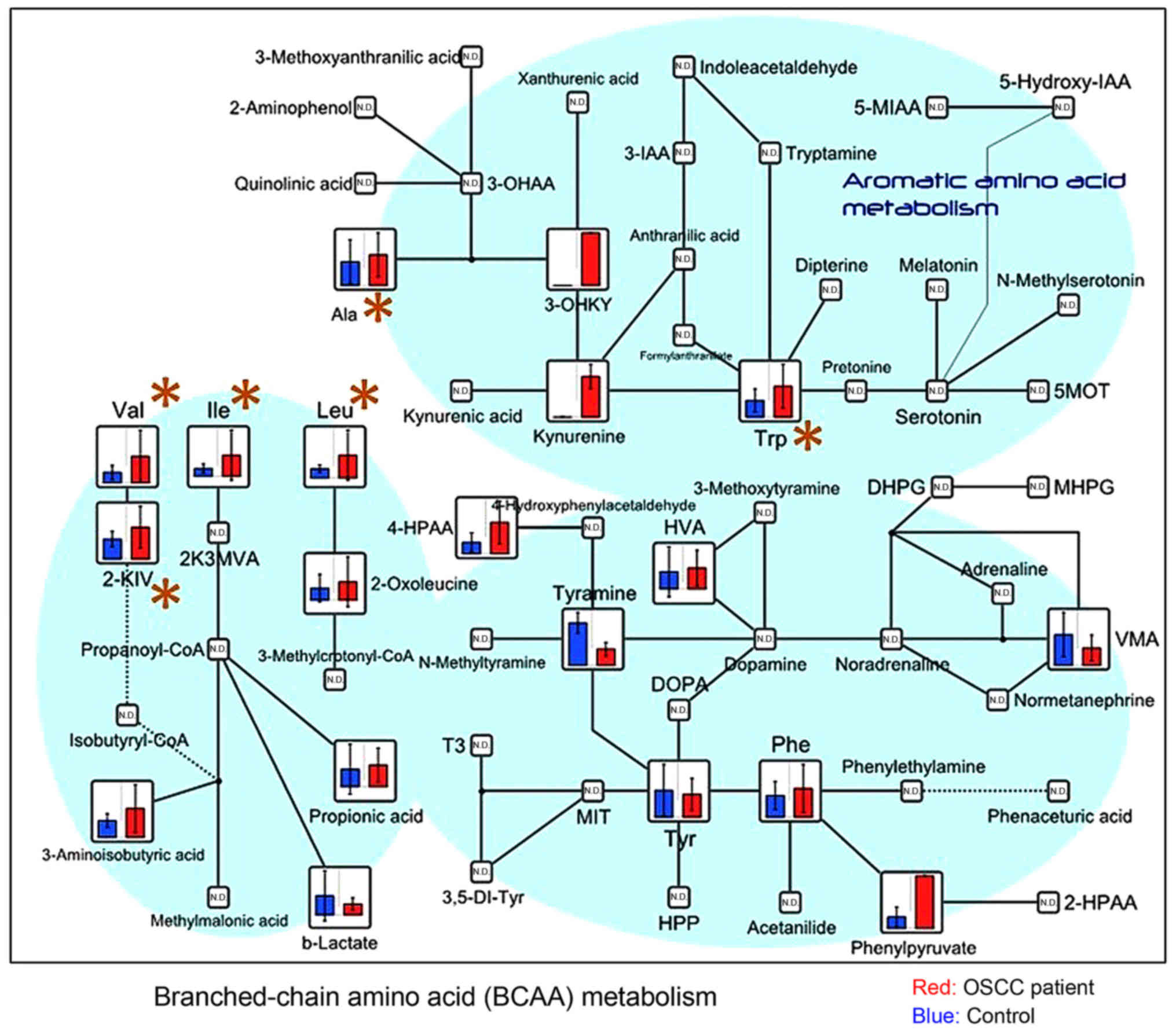 | Figure 3.Metabolome data map of metabolites
involved in the BCAA and aromatic amino acid pathways, detected in
the saliva obtained from patients with OSCC and controls. Levels of
valine (P=0.002), isoleucine (P=0.011), leucine (P=0.004), and
2-oxoisovaleric acid (P=0.030), which are components of the BCAA
cycle, exhibited a significant difference between groups. BCAA,
branched-chain amino acids; OSCC, oral squamous cell carcinoma;
3-IAA, indole-3-acetic acid; 5-MIAA, 5-methoxyindoleacetic acid;
2-HPAA, 2-hydroxyphosphonocarboxylic acid; MVA, mevalonic acid;
KIV, α-ketoisovaleric acid; VMA, zanillylmandelic acid; DOPA,
dihydroxyphenylalanine; T3, triiodothyronine; MIT,
monoiodotyrosine; HPP, hydroxyphenylpyruvate; DHPG,
dihydroxyphenylglycine; MHPG, 3-methoxy-4-hydroxyphenylglycol;
5MOT, 5 methoxytryptamine. |
 | Table II.Significance of candidate metabolomes
as biomarkers of OSCC. |
Table II.
Significance of candidate metabolomes
as biomarkers of OSCC.
| Peak no. | Compound name | Ratioa |
P-valueb |
|---|
| 1 | Choline | 2.5 | 6.E-05 |
| 2 |
p-Hydroxyphenylacetic acid | 2.7 | 0.001 |
| 3 |
2-Hydroxy-4-methylvaleric acid | 2.7 | 0.001 |
| 4 | Valine | 2.6 | 0.002 |
| 5 | 3-Phenyllactic
acid | 1.8 | 0.003 |
| 6 | Leucine | 2.5 | 0.004 |
| 7 | Hexanoic acid | 3.3 | 0.005 |
| 8 | Octanoic acid | 1.8 | 0.007 |
| 9 | Terephthalic
acid | 2.1 | 0.007 |
| 10 |
γ-Butyrobetaine | 1.9 | 0.010 |
| 11 | 3-(4-Hydroxyphenyl)
propionic acid | 3.0 | 0.010 |
| 12 | Isoleucine | 2.7 | 0.011 |
| 13 | Tryptophan | 1.9 | 0.014 |
| 14 | 3-Phenylpropionic
acid | 2.9 | 0.016 |
| 15 | 2-Hydroxyvaleric
acid | 1.0 | 0.017 |
| 16 | Butyric acid | 2.6 | 0.019 |
| 17 | Cadaverine | 3.4 | 0.026 |
| 18 | 2-Oxoisovaleric
acid | 1.6 | 0.030 |
| 19 |
N6,N6,N6-Trimethyllysine | 2.5 | 0.035 |
| 20 | Taurine | 1.9 | 0.035 |
| 21 | Glycolic acid | 1.1 | 0.036 |
| 22 | 3-Hydroxybutyric
acid | 1.6 | 0.037 |
| 23 | Heptanoic acid | 1.2 | 0.037 |
| 24 | Alanine | 1.3 | 0.046 |
| 25 | Urea | 0.7 | 0.026 |
Discussion
Oral cancer, one of the six most common human
cancers, is often not diagnosed until it has reached an advanced
stage and has a low overall 5-year survival rate of <50%, which
has not essentially changed over the past few decades (14,15).
Patients with OSCC often present with symptoms at a late stage, and
there is a high recurrence rate following treatment, particularly
in patients with neck lymph node metastasis.
The use of saliva as the sample for biomarker
detection is a big merit for both the patient and the dentist,
because saliva may be collected repeatedly in a non-invasive manner
for oral cancer screening. In addition to the straightforward
sample collection, other advantages of using a saliva sample
compared with other body fluids, such as serum or urine, include
the ability to obtain sufficient quantities for analysis and the
lower costs of storage and shipping (16). Various changes in a patient's
condition are reflected in the blood, but specific markers are
observed in the serum for malignant tumors. These changes also have
a high possibility of being reflected in the saliva, since serum
components are detectable in the saliva (6). In the present study, oral
cancer-specific markers with a high discrimination ability were
identified and characterized, demonstrating the potential use of
salivary metabolomics in OSCC diagnosis. A previous study
successfully identified an enolase 1 that is characteristically
expressed in the saliva of patients with OSCC using a
two-dimensional electrophoretic method (7). Therefore, the focus of the present
study was on developing a convenient method for metabolome analysis
that may be used to analyze whole saliva samples, which are able to
be collected non-invasively and repeatedly. However, it is also
important to consider the bacteria present in the oral cavity when
using saliva as a biomarker sample. For example,
P-hydroxyphenylacetic acid was significantly increased in the OSCC
patient saliva compared with healthy control saliva in the present
study. P-Hydroxyphenylacetic acid is a metabolic enzyme of
tyrosine, which is produced by Porphyromonas gingivalis as a
metabolic end-product (17). In the
present study, strict criteria for saliva collection were
implemented in order to standardize the conditions. All the
subjects received professional mechanical tooth cleaning by a
dental hygienist the day prior to sample collection, and saliva was
collected at 8:00 a.m. the following morning under fasting
conditions after sufficient gargling and other oral hygiene
steps.
It is widely acknowledged that cancer cells
predominantly use glycolysis rather than the oxidative
phosphorylation circuit called the tricarboxylic acid (TCA) cycle
(Warburg effect) (18). This effect
has been detected in stomach cancer and colon cancer tissues based
on metabolome analysis with CE-MS (19). Similar metabolic pathway activity
has also been reported for oral cancer tissue (6). Since the metabolism observed in the
saliva of patients with OSCC is significantly different from the
Warburg effect, this suggested that other metabolic pathways should
be considered.
Sugimoto et al (6) reported 24 candidate metabolites that
were able to serve as biomarkers for OSCC from saliva samples of
patients of various races, and 7 of these metabolites (taurine,
valine, leucine, isoleucine, choline, cadaverine, and tryptophan)
were also detected as potential biomarkers in the present study.
The concentration of choline in the saliva of patients with OSCC
was significantly higher compared with that in the saliva of
healthy controls; this metabolite exhibited the most significant
difference between the two groups. Choline, a quaternary amine, is
an essential nutrient that is predominantly supplied through the
diet, and choline-containing metabolites are important constituents
of the phospholipid metabolism of cell membranes and are associated
with malignant transformation, including in breast, brain, and
prostate cancer (20). In tumors,
choline is highly metabolized to phosphocholine and is oxidized to
betaine; hence, a low concentration of choline and high
concentrations of phosphocholine and betaine have been observed
(21). Furthermore, previous
studies have shown that the levels of choline metabolites were
higher in tumors compared with benign lesions or normal tissues
(22). An excessive increase in
plasma choline levels in the tumor cells of patients with breast
cancer was also reported (23).
Aberrant choline metabolism may be due to enhanced membrane
synthesis and degradation, which reflect the excessive
proliferation of cancer cells. The saliva of patients with oral
cancer displayed a profile showing increased levels of
phosphocholine and glycerophosphocholine (6). In the present study,
glycerophosphocholine was detected in certain of the samples, but
no statistically significant difference was observed between the
two groups. However, this result should be interpreted with
caution, since choline is included in various foods. This might not
have been an issue in the present study, however, given that the
saliva of all the subjects was collected in the morning under
fasting conditions after sufficient gargling and oral hygiene.
However, to clarify this effect, choline should be detected in
paired OSCC and normal tissues simultaneously using CE-MS and
another analysis method, such as real-time polymerase chain
reaction (RT-PCR) or immunohistochemistry.
BCAAs, such as leucine, isoleucine, and valine, are
implicated in various diseases. Branched-chain aminotransferase,
which produces a branched chain α-keto acid, is an enzyme that
catalyzes a reversible amino group transfer reaction in the BCAA
degradation system; branched-chain α-keto acid dehydrogenase is an
enzyme that catalyzes the second step. Finally, acetyl-CoA is
formed from leucine, succinyl-CoA and acetoacetate are formed from
isoleucine, and succinyl-CoA is formed from valine. The metabolic
pathway produces numerous intermediates to be consumed in the TCA
cycle. For example, maple syrup urine disease is characterized by
dysfunction in BCAA metabolism (24). Valine has been reported as a
metabolite that differs significantly in the saliva of patients
with uterine (25), colon (26), renal (27), and oral (6) cancer. Leucine levels have been shown
to be markedly elevated in women with rectal cancer (28). Cancer cells require excess nutrients
and energy to adapt to increased biosynthetic activity, which is
correlated with glutamine activity. Several anticancer agents
(e.g., l-asparaginase) used in clinical practice utilize mechanisms
that inhibit glutamine. Glutamine contributes to the cellular
import of leucine, which controls the amino acid/Rag/mammalian
target of rapamycin complex 1 (mTORC1) signaling pathway (29). α-Keto-carboxylic acid, consisting of
amino groups, receives isoleucine from glutamic acid. A significant
difference in isoleucine was detected in serum samples of patients
with uterine cancer when analyzed by nuclear magnetic resonance
(NMR) spectroscopy (30). The
levels of isoleucine were also reported to differ markedly in the
serum of patients with lung cancer (31), and in patients with schizophrenic
disease (32). It was reported that
the levels of BCAAs, including leucine, isoleucine, and valine,
were significantly higher in cancer patients compared with control
subjects (6), and the same results
were identified with the patients with OSCC in the present study.
The metabolic pathway produces numerous intermediates to be
consumed in the TCA cycle. Therefore, metabolism of BCAAs may also
serve an important role in the energy production of oral cancer. In
the present study, the levels of valine, isoleucine, leucine, and
2-oxoisovaleric acid, which are all involved in the BCAA cycle,
significantly differed between the two groups. However, relatively
little is known concerning the function of 2-oxoisovaleric acid in
general, and it has not been reported in any previous cancer
metabolomic study, including those for OSCC.
Urea is a highly soluble organic compound formed in
the liver from ammonia produced by the deamination of amino acids.
It is the principal end-product of protein catabolism, and accounts
for approximately half of the total urinary solids. Urea is formed
in a cyclic pathway known as the urea cycle. Urea was shown to be
significantly increased in the urine samples of patients with
gastric cancer compared with healthy controls (33); however, there has been no previous
report of variations in the urea level in patients with OSCC. In
the present study, urea was the only metabolite that was
significantly higher in the healthy controls compared with patients
with OSCC. Urea production is impaired under conditions of poor
nutrition and Helicobacter pylori infection. As dietary
intake becomes difficult for patients with OSCC due to pain, their
protein intake is likely to become insufficient. H. pylori
produces a urease (34) that
catalyzes the conversion of urea into ammonia and carbon dioxide
contained in the stomach mucus. Urea produced under the influence
of H. pylori metabolism is used in the production of
ammonia. No statistically significant differences in urea levels
were identified between patients with tongue and gingiva cancer
(unpublished data). To the best of our knowledge, no previous
metabolomic study of OSCC tumor and saliva samples has identified
urea as a marker for OSCC. However, the present findings indicate
that decreased urea in the saliva is possibly a biomarker for OSCC.
Nevertheless, as mentioned above, there is a requirement for
further evaluation of the association between this finding and the
metabolism of oral bacteria.
3-Hydroxybutyric acid has an asymmetric carbon atom,
and is one of the ketone bodies. It is synthesized in the liver
from acetyl-CoA, when the blood glucose concentration, which is
used as an energy source, is low. Regarding the increased level of
3-hydroxybutyric acid in the saliva of patients with OSCC, the
higher lipid levels in the OSCC saliva may be associated with a
higher metabolic turnover and the demand for membrane biosynthesis
for cell proliferation, leading to a higher utilization rate of
lipids. 3-Hydroxybutyric acid levels are increased in ketosis.
3-Hydroxyisovalerate, which is derived from isovaleryl-CoA, a
catabolic intermediate of leucine, has been attracting attention
recently as a potential biomarker of ovarian (35), liver (36), pancreatic (37), and gastric (38) cancer, with markedly increased
differences observed in the serum or urine. As oral cancer cells
use ketone bodies to generate energy, it could possibly be detected
in the saliva. Although there is no report of this metabolite in
OSCC, 3-hydroxybutyric acid levels in tongue cancer were higher
than those in gingiva cancer (unpublished data). In a further
study, we will investigate a role and function of 3-hydroxybutyrate
in OSCC patient.
In summary, the present analysis of the saliva of
Japanese patients with OSCC revealed similar choline and BCAA cycle
levels, as identified in the study by Sugimoto et al
(6). However, the levels of
3-hydroxybutyric acid and 2-oxoisovaleric acid were higher in the
saliva of patients with OSCC compared with the saliva of healthy
control subjects, whereas urea levels were lower in Japanese OSCC
samples compared with those of healthy controls. The findings in
the present study regarding metabolism specific to OSCC may provide
a novel strategy for the detection of OSCC, and thereby improve the
treatment efficacy. However, in future studies, CE-MS should be
combined with other analytical techniques, such as HPLC, RT-PCR and
immunochemical staining, and other metabolome analytical
techniques, such as NMR and LC-MS spectroscopy. Investigating the
association between metabolites and the gene copy number, and gene
and protein expression levels in the metabolic pathway of samples
from patients with OSCC, may help to elucidate the mechanisms
underlying carcinogenesis.
Acknowledgements
We thank Professor Nobuo Takano (Oral Cancer Center,
Tokyo Dental College), Dr Masahiro Sugimoto (Keio University,
Tokyo, Japan), and Dr Kenjiro Kami (Human Metabolome Technologies
Inc., Yamagata, Japan) for their technical advice.
References
|
1
|
Sudbø J: Novel management of oral cancer:
A paradigm of predictive oncology. Clin Med Res. 2:233–242. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eheman C, Henley SJ, Ballard-Barbash R,
Jacobs EJ, Schymura MJ, Noone AM, Pan L, Anderson RN, Fulton JE,
Kohler BA, et al: Annual Report to the Nation on the status of
cancer, 1975–2008, featuring cancers associated with excess weight
and lack of sufficient physical activity. Cancer. 118:2338–2366.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Genden EM, Ferlito A, Bradley PJ, Rinaldo
A and Scully C: Neck disease and distant metastases. Oral Oncol.
39:207–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sabichi AL, Demierre MF, Hawk ET, Lerman
CE and Lippman SM: Frontiers in cancer prevention research. Cancer
Res. 63:5649–5655. 2003.PubMed/NCBI
|
|
5
|
Spafford MF, Koch WM, Reed AL, Califano
JA, Xu LH, Eisenberger CF, Yip L, Leong PL, Wu L, Liu SX, et al:
Detection of head and neck squamous cell carcinoma among exfoliated
oral mucosal cells by microsatellite analysis. Clin Cancer Res.
7:607–612. 2001.PubMed/NCBI
|
|
6
|
Sugimoto M, Wong DT, Hirayama A, Soga T
and Tomita M: Capillary electrophoresis mass spectrometry-based
saliva metabolomics identified oral, breast and pancreatic
cancer-specific profiles. Metabolomics. 6:78–95. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katakura A, Yamamoto N, Sakuma T, Sugahara
K, Onda T, Noguchi S and Shibahara T: A screening test for oral
cancer using saliva samples: Proteomic analysis of biomarkers in
whole saliva. J Oral Maxillofac Surg. 27:1–5. 2015.
|
|
8
|
Soga T, Baran R, Suematsu M, Ueno Y, Ikeda
S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, et al:
Differential metabolomics reveals ophthalmic acid as an oxidative
stress biomarker indicating hepatic glutathione consumption. J Biol
Chem. 281:16768–16776. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kind T and Fiehn O: Metabolomic database
annotations via query of elemental compositions: Mass accuracy is
insufficient even at less than 1 ppm. BMC Bioinformatics.
7:2342006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kind T and Fiehn O: Seven Golden Rules for
heuristic filtering of molecular formulas obtained by accurate mass
spectrometry. BMC Bioinformatics. 8:1052007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sugimoto M, Kikuchi S, Arita M, Soga T,
Nishioka T and Tomita M: Large-scale prediction of cationic
metabolite identity and migration time in capillary electrophoresis
mass spectrometry using artificial neural networks. Anal Chem.
77:78–84. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goto S, Okuno Y, Hattori M, Nishioka T and
Kanehisa M: LIGAND: Database of chemical compounds and reactions in
biological pathways. Nucleic Acids Res. 30:402–404. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wishart DS, Tzur D, Knox C, Eisner R, Guo
AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al: HMDB:
The human metabolome database. Nucleic Acids Res. 35:D521–D526.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vokes EE, Weichselbaum RR, Lippman SM and
Hong WK: Head and neck cancer. N Engl J Med. 328:184–194. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Banks RE, Dunn MJ, Hochstrasser DF,
Sanchez JC, Blackstock W, Pappin DJ and Selby PJ: Proteomics: New
perspectives, new biomedical opportunities. Lancet. 356:1749–1756.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, St John MA, Zhou X, Kim Y, Sinha U,
Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH, et al:
Salivary transcriptome diagnostics for oral cancer detection. Clin
Cancer Res. 10:8442–8450. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takahama U, Imamura H and Hirota S:
Nitration of the salivary component 4-hydroxyphenylacetic acid in
the human oral cavity: Enhancement of nitration under acidic
conditions. Eur J Oral Sci. 117:555–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Warburg OH: The metabolism of tumours:
investigations from the Kaiser Wilhelm Institute for Biology.
Berlin-Dahlem. Richard R. Smith Inc.; New York: pp. 129–169.
1931
|
|
19
|
Hirayama A, Kami K, Sugimoto M, Sugawara
M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et
al: Quantitative metabolome profiling of colon and stomach cancer
microenvironment by capillary electrophoresis time-of-flight mass
spectrometry. Cancer Res. 69:4918–4925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ackerstaff E, Glunde K and Bhujwalla ZM:
Choline phospholipid metabolism: A target in cancer cells? J Cell
Biochem. 90:525–533. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katz-Brull R, Seger D, Rivenson-Segal D,
Rushkin E and Degani H: Metabolic markers of breast cancer:
Enhanced choline metabolism and reduced choline-ether-phospholipid
synthesis. Cancer Res. 62:1966–1970. 2002.PubMed/NCBI
|
|
22
|
Haddadin IS, McIntosh A, Meisamy S, Corum
C, Snyder Styczynski AL, Powell NJ, Nelson MT, Yee D, Garwood M and
Bolan PJ: Metabolite quantification and high-field MRS in breast
cancer. NMR Biomed. 22:65–76. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Katz-Brull R, Margalit R and Degani H:
Differential routing of choline in implanted breast cancer and
normal organs. Magn Reson Med. 46:31–38. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Harris RA, Zhang B, Goodwin GW, Kuntz MJ,
Shimomura Y, Rougraff P, Dexter P, Zhao Y, Gibson R and Crabb DW:
Regulation of the branched-chain alpha-ketoacid dehydrogenase and
elucidation of a molecular basis for maple syrup urine disease. Adv
Enzyme Regul. 30:245–263. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mustafa A, Gupta S, Hudes GR, Egleston BL,
Uzzo RG and Kruger WD: Serum amino acid levels as a biomarker for
renal cell carcinoma. J Urol. 186:1206–1212. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma Y, Zhang P, Wang F, Liu W, Yang J and
Qin H: An integrated proteomics and metabolomics approach for
defining oncofetal biomarkers in the colorectal cancer. Ann Surg.
255:720–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gaudet MM, Falk RT, Stevens RD, Gunter MJ,
Bain JR, Pfeiffer RM, Potischman N, Lissowska J, Peplonska B,
Brinton LA, et al: Analysis of serum metabolic profiles in women
with endometrial cancer and controls in a population-based
case-control study. J Clin Endocrinol Metab. 97:3216–3223. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cross AJ, Moore SC, Boca S, Huang WY,
Xiong X, Stolzenberg-Solomon R, Sinha R and Sampson JN: A
prospective study of serum metabolites and colorectal cancer risk.
Cancer. 120:3049–3057. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Willems L, Jacque N, Jacquel A, Neveux N,
Maciel TT, Lambert M, Schmitt A, Poulain L, Green AS, Uzunov M, et
al: Inhibiting glutamine uptake represents an attractive new
strategy for treating acute myeloid leukemia. Blood. 122:3521–3532.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye N, Liu C and Shi P: Metabolomics
analysis of cervical cancer, cervical intraepithelial neoplasia and
chronic cervicitis by 1H NMR spectroscopy. Eur J Gynaecol Oncol.
36:174–180. 2015.PubMed/NCBI
|
|
31
|
Deja S, Porebska I, Kowal A, Zabek A, Barg
W, Pawelczyk K, Stanimirova I, Daszykowski M, Korzeniewska A,
Jankowska R, et al: Metabolomics provide new insights on lung
cancer staging and discrimination from chronic obstructive
pulmonary disease. J Pharm Biomed Anal. 100:369–380. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Luca V, Viggiano E, Messina G, Viggiano
A, Borlido C, Viggiano A and Monda M: Peripheral amino acid levels
in schizophrenia and antipsychotic treatment. Psychiatry Investig.
5:203–208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang Q, Wang C and Li B: Metabolomic
analysis using liquid chromatography/mass spectrometry for gastric
cancer. Appl Biochem Biotechnol. 176:2170–2184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shirai M: Transcription regulation of the
urease operon in Helicobacter pyiori in response to pH and
mechanisms of stable colonizaion in the stomach. Yamaguchi Med.
50:593–601. 2001.
|
|
35
|
Hilvo M, de Santiago I, Gopalacharyulu P,
Schmitt WD, Budczies J, Kuhberg M, Dietel M, Aittokallio T,
Markowetz F, Denkert C, et al: Accumulated metabolites of
hydroxybutyric acid serve as diagnostic and prognostic biomarkers
of ovarian high-grade serous carcinomas. Cancer Res. 76:796–804.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zeng J, Yin P, Tan Y, Dong L, Hu C, Huang
Q, Lu X, Wang H and Xu G: Metabolomics study of hepatocellular
carcinoma: Discovery and validation of serum potential biomarkers
by using capillary electrophoresis-mass spectrometry. J Proteome
Res. 13:3420–3431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
OuYang D, Xu J, Huang H and Chen Z:
Metabolomic profiling of serum from human pancreatic cancer
patients using 1H NMR spectroscopy and principal component
analysis. Appl Biochem Biotechnol. 165:148–154. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hur H, Paik MJ, Xuan Y, Nguyen DT, Ham IH,
Yun J, Cho YK, Lee G and Han SU: Quantitative measurement of
organic acids in tissues from gastric cancer patients indicates
increased glucose metabolism in gastric cancer. PLoS One.
9:e985812014. View Article : Google Scholar : PubMed/NCBI
|