Introduction
The prognosis of oral squamous cell carcinoma
(OSCC), which includes head and neck squamous cell carcinoma, is
greatly influenced by metastases, such as to the lymph node. The
5-year survival rate of patients without lymph node metastases is
between 63 and 86%, while the 5-year survival rate of patients with
lymph node metastases is between 20 and 36% (1,2).
Although traditional thinking suggests the relationship between
lymph node metastasis and prognosis is a consequence of the
malignancy (3), metastasizing
capacity (4) or acquisition of
resistance to chemotherapy (5) of
tumor cells, the involvement of the immune system in the metastatic
process remains poorly explored. To better understand this
relationship, we analyzed tumor immunological properties between
primary tumor cells and cells from a lymph node metastasis using
the L5-11 cell line that we established. The L5-11 cell line was
derived from a lymph node metastasis from the murine OSCC cell line
Sq-1979. Mice transplanted with the L5-11 cells had larger spleens
and tumors than those transplanted with Sq-1979. Therefore, we
hypothesized that a tumor immunosuppressive mechanism was acquired
by the lymph node metastasis cell line L5-11.
Recently, the presence of abnormally differentiated
bone marrow-derived cells has been appreciated as a major
immunological characteristic of cancer (6). Many studies have also demonstrated
that myeloid derived suppressor cells (MDSCs) downregulate the
immune response to tumors, infectious diseases and inflammation
(7,8). MDSCs are subdivided into two groups by
phenotype: i) monocytic MDSC (M-MDSC), which assume the form and
phenotypes of immature mononuclear cells and have the cell surface
markers CD11b+, Ly6Chi and Ly6G−;
and ii) polymorphonuclear MDSC (PMN-MDSC), which assume the form
and phenotype similar to immature polymorphonuclear cells and have
the cell surface markers CD11b+, Ly6Clo and
Ly6G+ (9). MDSCs inhibit
T cell function (10), induce
regulatory T cells (11) and
support tumor growth and metastasis (12), all of which serve a pro-tumor
function. MDSC from peripheral lymphoid organs suppress
antigen-specific CD8+ T cells, in contrast, MDSC from a
tumor microenvironment suppress both antigen-specific and
non-specific T cell activity (13).
However, the activity of MDSCs from a metastasized tumor have not
been studied, nor have basic comparisons of MDSCs derived from
primary and metastasized tumors. Moreover, there are few studies
regarding the role of MDSCs in OSCC. Therefore, we aimed to clarify
the characteristics of MDSCs derived from primary tumors and
metastases, and our data suggest that the poor prognosis associated
with metastasis in OSCC involves tumor immunosuppression.
Materials and methods
Mice
Male C3H/HeN mice, 6–8 weeks old, were purchased
from Chubu Kagaku Shizai, Co., Ltd. (Nagoya, Japan). The mice were
maintained ad libitum on Oriental MF solid chow (Oriental
Yeast, Co., Ltd. Tokyo, Japan). The present study was approved by
the Animal Ethics Committee of Asahi University (no. 14–018 and
16–017).
Cell culture and establishment of a
metastasized cell line
The C3H murine OSCC cell line Sq-1979 was obtained
from the RIKEN BioResource Center (Ibaraki, Japan). Tumor cells
were cultured in minimum essential medium (MEM; Wako Pure Chemical
Industries, Ltd., Osaka, Japan) containing 10% fetal bovine serum
(FBS; Biowest, Nuaille, France) and antibiotics (100 U/ml
penicillin and 100 µg/ml streptomycin) at 37°C in a humidified
atmosphere containing 5% CO2.
Sq-1979 cells (1×107) were suspended in
0.1 ml of phosphate-buffered saline (PBS), then subcutaneously
inoculated in the posterior neck area of mice. After 3 months,
metastasized regional lymph nodes were dissected into MEM
supplemented with 10% FBS and minced to isolate attached cells.
Then, the metastasized sub-clone, L5-11 was isolated by the serial
limiting dilution method. Cell images were taken using a Nikon
Eclipse Ti inverted microscope system (Nikon, Tokyo, Japan).
Evaluation of cell growth using the
MTT assay
Sq-1979 or L5-11 cells were seeded into 96-well
plates at a density of 1×103 cells/well and allowed to
adhere for 24 h. Cell viability was assessed daily by addition of 5
µl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT) using a Cell Proliferation kit I (Roche Diagnostics GmbH,
Mannheim, Germany) according to the manufacturers instructions. The
number of viable cells was assessed by measuring the absorbance of
the produced formazan crystals at 595 nm with a Tecan SpectraFluor
Plus XFluor4 software (Tecan Japan, Co., Ltd., Kawasaki, Japan).
The measurement was performed daily for 4 days. Cell growth was
calculated relative to the value on the first day, which was set at
100%.
In vivo tumor growth analysis
Sq-1979 or L5-11 cells (1×106) were
resuspended in 0.1 ml PBS and subcutaneously injected into the
lateroabdominal area of male C3H/HeN mice. Tumor size was observed
every 3 days; the tumor volume was calculated using the formula:
a×b2/2 where ‘a’ is the length and ‘b’ is the width of
tumor diameter. Mice were sacrificed after 30 days. There were no
significant differences in body weight of the mice.
Preparation of tumor infiltrating
cells and spleen cells
Mice were sacrificed 21 days after tumor
implantation for analysis. Tumor tissue was finely chopped,
incubated with the enzyme mix from the tumor dissociation kit (MACS
Miltenyi Biotec; Miltenyi Biotec GmbH, Berdish-Gladbach, Germany)
according to the manufacturers instruction, and then the
resuspended-sample was applied to a MACS SmartStrainers (70 µm)
placed on a 15-ml conical. Red blood cells in the sample were
removed using a red blood cell lysis solution (MACS Miltenyi
Biotec). For total spleen cell isolation, red blood cells were
removed using the same procedures, and the cells were applied to a
MACS SmartStrainers (70 µm) placed on a 15-ml conical. After
collection, total cell numbers were counted with a counting
chamber.
Flow cytometry
Fc receptors in the sample were blocked by
anti-mouse CD16/32 (eBioscience, Santa Clara, CA, USA) using 1 µg
per million cells for 10 min on ice. Then, cells were labeled with
Mouse MDSC Flow Cocktail 2 with isotype control, which contained PE
anti-mouse CD11b (clone M1/70), FITC anti-mouse Ly-6G (clone 1A8),
and APC anti-mouse Ly6C (clone HK1.4), or FITC anti-mouse CD8a
clone:53-6.7, PE anti-mouse CD4 clone:GK1.5 (all from Sony
Biotechnology, Inc., San Jose, CA, USA), and FITC mouse IgG2 and
PE-mouse IgG2b (BD Biosciences, San Diego, CA, USA) for 30 min in
the dark. Cells were then washed and resuspended in FACS buffer
(PBS with 0.5% BSA). Data acquisition and analysis were performed
using EC800 flow cytometry analyzer with EC800 analysis software
(Sony Biotechnology Inc.).
Real-time PCR
Total RNA was purified using the RNeasy Mini kit
(Qiagen, Heiden, Germany), and 600 ng total RNA was used for
reverse transcription with iScript™Advanced cDNA Synthesis kit
(Mediatech, Herndon, VA, USA). For real-time PCR analysis, 1 µl
cDNA samples diluted 20x, 1 µl each of the upper and lower primer
(Final 500 nM), 7 µl nuclease-free water, and 10 µl SsoAdvanced
SYBR-Green Supermix (Bio-Rad Laboratoties) were used. The following
primers were designed by Primer Express software (Biosystems,
Waltham, MA, USA): VEGFA forward, 5-GCTGTGCAGG CTGTAACGAT-3 and
reverse, 5-GGTCTGCATTCACATCT GCTGTGC-3; VEGFR1 forward,
5-ATCTATAAGGCAGC GGATTGACCG-3 and reverse, 5-CACGGAGGTGTTGAA
AGACTGGA-3; Npn1 forward, 5-AATCGAAAACCCAG GGTACCTCAC-3 and
reverse, 5-GCAGTCTCTGTCCTCC AAATGGAA-3; Npn2 forward,
5-ACTGTACCTTCACC ATCCTGGCC-3 and reverse, 5-GGTCCAACATGTGG
AATGCCATC-3; Sema3a forward, 5-AGATGCTCCATT CCAGTTTGTTCAC-3 and
reverse, 5-ACATAAGCCACCG CATCACTTGTA-3; Sema3b forward,
5-GGATGCATGT CTCTGAGCTCCG-3 and reverse, 5-GGCCCAGCCATAA
CTCATTTGTC-3; Sema3f forward, 5-ATGGCTGATAT CCGCATGGTCTT-3 and
reverse, 5-CCTTAGTGGACTT CATCGAGGGC-3; S100A9 forward,
5-CATCATGGAGG ACCTGGACACAA-3 and reverse, 5-GCAGCTTCTCA
TGACAGGCAAAGA-3; IL-6 forward, 5-CCTACCCCA ATTTCCAATGCTCTC-3 and
reverse, 5-GCATAACGCA CTAGGTTTGCCG-3; and HIF1α forward, 5-TGGATTT
TGGCAGCGATGACAC-3 and reverse, 5-AGTGGCTTTG GAGTTTCCGATGA-3. The
reaction conditions consisted of one 5-min cycle at 95°C, followed
by 45 cycles at 95°C for 10 sec, and 72°C for 10 sec. The reaction
and absolute quantitation analysis were performed with a Thermal
Cycler Dice® Real-Time System TP800 (Takara Bio, Inc.,
Shiga, Japan).
Statistical analysis
All quantitative data are presented as the mean ±
SD. Data were evaluated using one-way analysis of variance followed
by Tukey-Kramer test for Figs. 2C,
3B and C, 4B and D. Unpaired two-tailed Students
t-test was performed for the two-group comparisons in Figs. 3E and F and 4E and F. P<0.05 was
considered statistically significant.
Results
Characterization of the murine lymph
node metastasis cell line derived from OSCC
After establishing the L5-11 murine lymph node
metastasis cell line, derived from the murine OSCC cell line
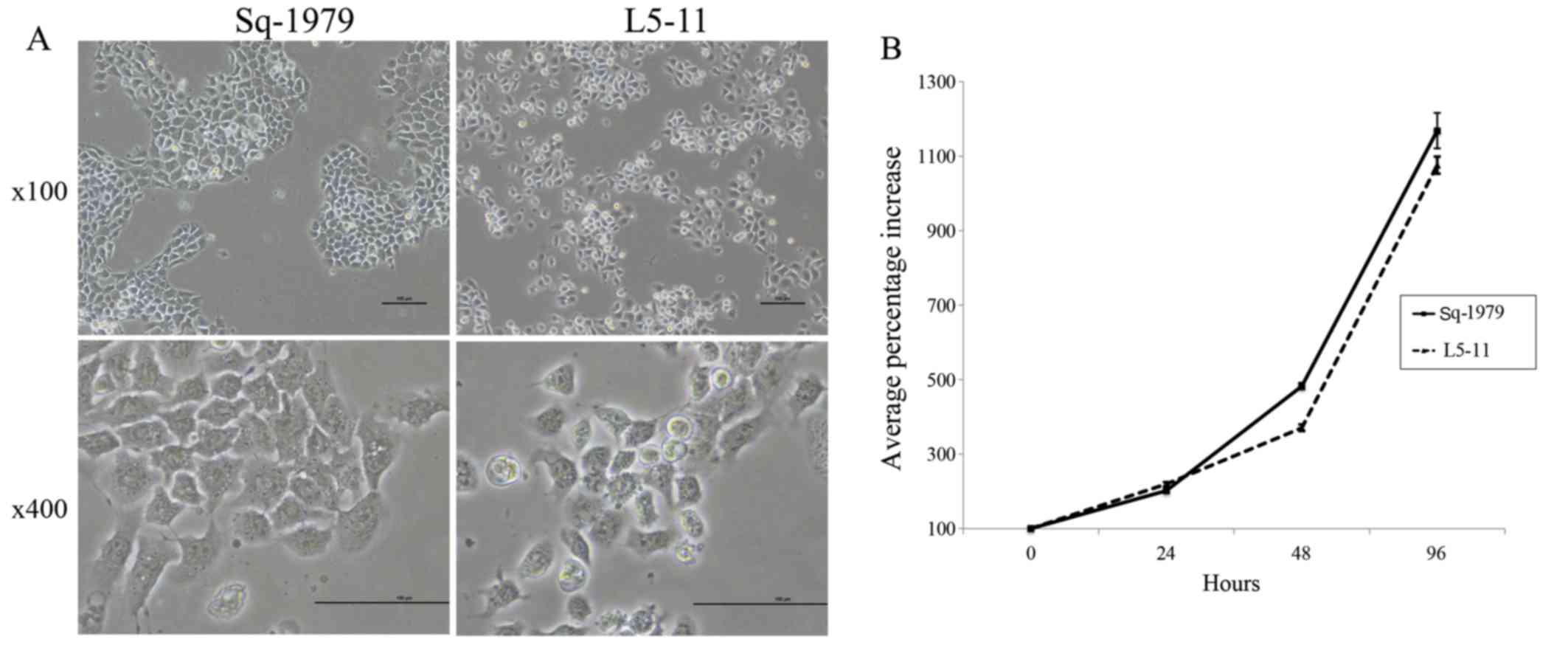
Sq-1979, we compared the morphology of the two lines. Sq-1979 cells
grew in island structures, similar to the original squamous
epithelium. However, L5-11 cells formed fewer of these structures,
and when they were observed they were small punctate arrangements
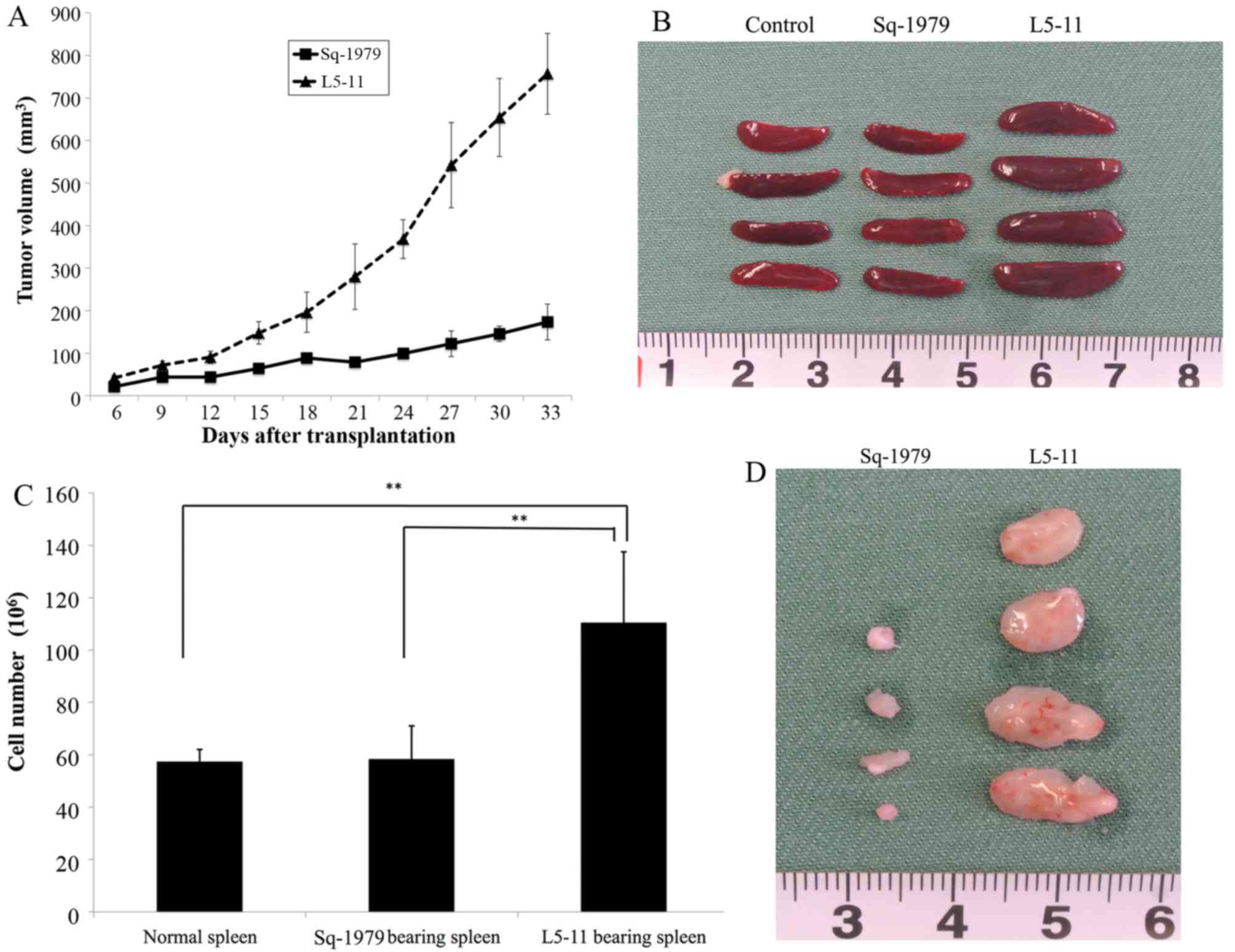
that had poor epithelial characteristics (Fig. 1A). Next, we compared the growth rate
of these cells. The in vitro growth rate was not
significantly different between Sq-1979 and L5-11 cells (Fig. 1B). To test in vivo growth
rates, we generated isogenic heterotopic transplantation models
from both tumor cell lines and measured tumor diameters. In
contrast to the in vitro growth rates, L5-11 tumors grew
faster than Sq-1979 tumors (Fig.
2A). We removed spleens and tumors at day 21
post-transplantation and both were larger in the L5-11 group
compared with the Sq-1979 group (Fig.
2B-D).
Metastatic OSCC cell syngeneic grafts
induce increased splenic PMN-MDSCs
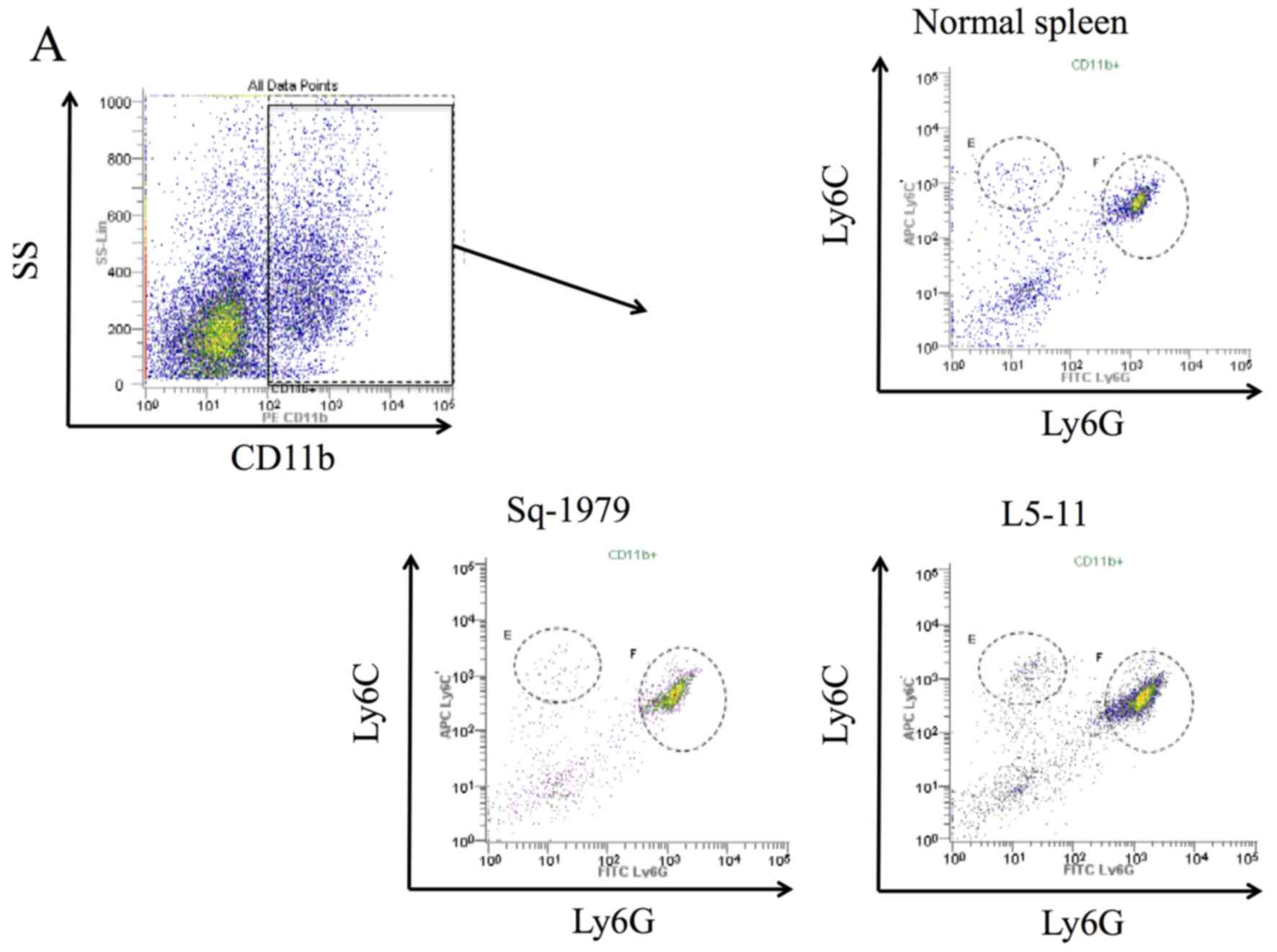
We examined the population of splenic MDSCs to
compare tumor immunogenicity between Sq-1979 and L5-11 syngeneic
grafts because, in addition to increased tumor growth in the L5-11
group, the recipient mice also showed significantly increased
spleen volume (Fig. 2B-D). The
proportion of M-MDSCs were ~2% from all groups, and there were no
significant differences observed. The absolute M-MDSC cell numbers
for each group were 1.3×106 cells in the control mice,
1.2×106 cells from Sq-1979-transplanted mice and
2.0×106 cells from L5-11-transplanted mice; these
results did not show significant differences (Fig. 3A and B). Conversely, the proportion
of PMN-MDSCs from L5-11-transplanted mice was significantly
increased (8.4%) compared with negative control mice (5.0%) and
Sq-1979-transplanted mice (5.9%). These changes were also reflected
in the absolute cell numbers of PMN-MDSCs, which were also
significantly increased in L5-11-transplanted mice compared with
control or Sq-1979-transplanted mice (8.8×106,
2.8×106 and 3.4×106 cells, respectively;
Fig. 3A and C).
Metastatic OSCC cell syngeneic grafts
induce increased intratumoral M- and PMN-MDSCs
We concurrently examined the population of
intratumoral MDSCs. The absolute cell number of both M-MDSCs and
PMN-MDSCs in the tumor were significantly increased in L5-11
compared with Sq-1979 tumors (Fig.
3D-F). Although the fraction of M-MDSCs in L5-11 syngeneic
grafts was increased to 0.89% compared with 0.09% in Sq-1979, the
fraction of PMN-MDSCs from L5-11 markedly increased to 38.1%
compared with 2.7% in Sq-1979 (Fig.
3D-F). Furthermore, we examined whether the fraction of
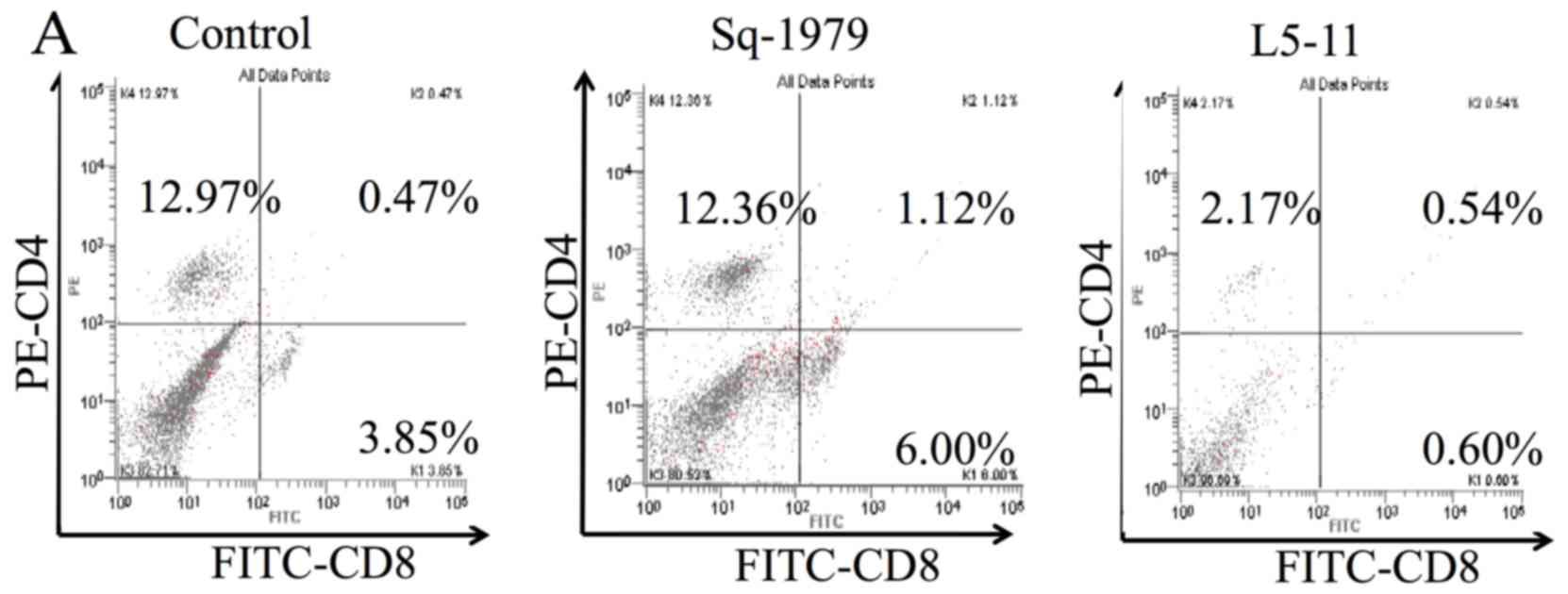
CD8+ or CD4+ cells changed. Splenic
CD4+ and CD8+ fractions and cell numbers from
L5-11-transplanted mice were lower than the control and
Sq-1979-transplanted mice (Fig.
4A-C). Most significantly, the fraction of splenic
CD8+ cells from Sq-1979-transplanted mice significantly
increased to 5.2% compared with 2.8% in the control and 0.3% in
L5-11 (Fig. 4B). The fraction and
number of intratumoral CD8+ cells in L5-11 significantly
increased compared with Sq-1979 (Fig.
4D and E). Although the number of intratumoral CD4+
cells in L5-11 significantly increased to 0.3×106 cells
compared with 0.05×106 cells in Sq-1979, there was no
significant difference in the fraction of intratumoral
CD4+ (Fig. 4F).
Analysis of tumor-derived factors that
induce MDSCs
We next explored which factors from tumor cells
induce MDSCs. Because the metastasis OSCC cells induced more
PMN-MDSCs than the primary OSCC cells in the spleen and tumor
(Fig. 3), we hypothesized that
tumor-derived factors may be related to the induction of MDSCs.
Using real-time PCR to assess the expression of several genes
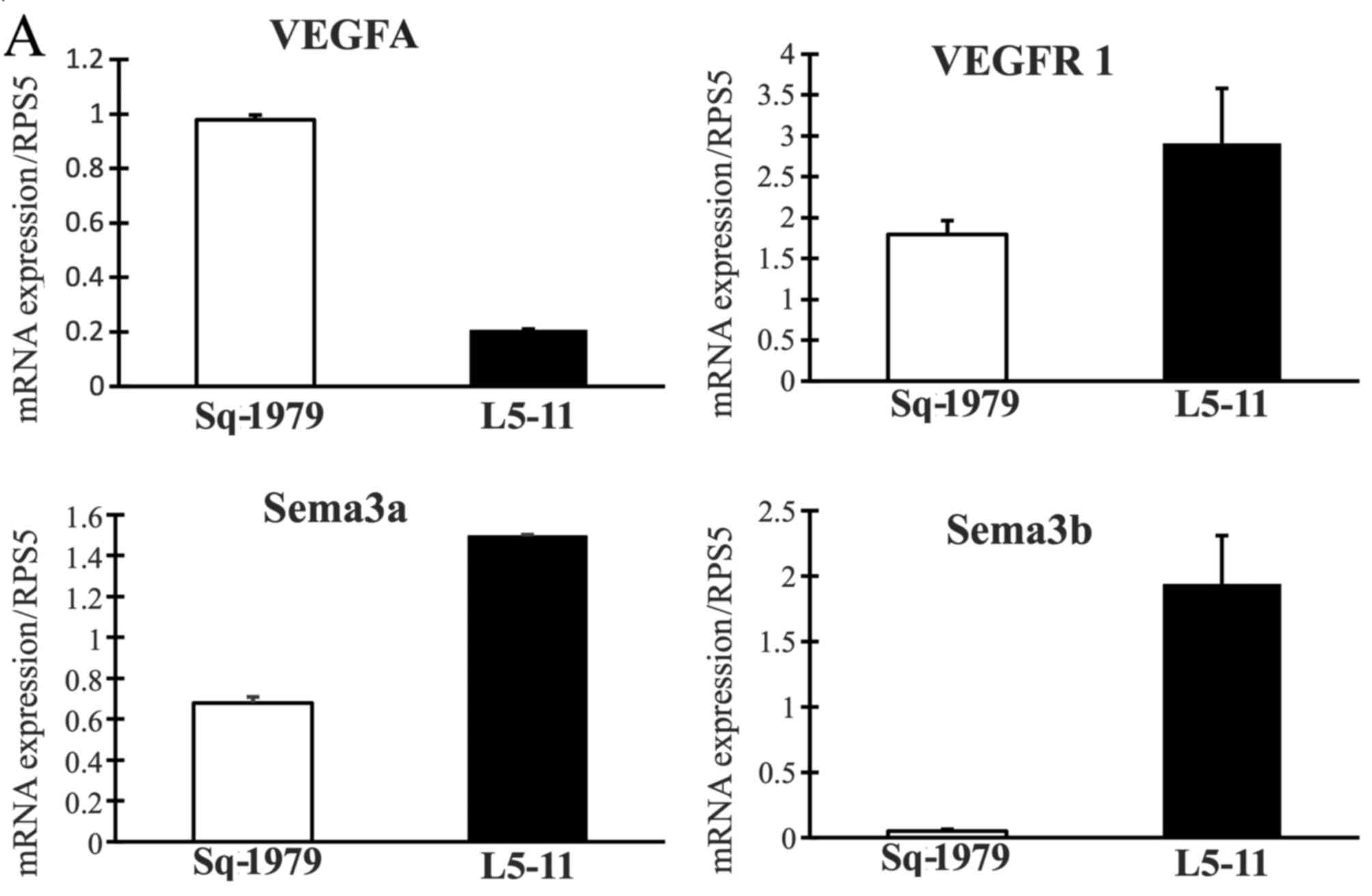
related to MDSCs, we found that VEGFR1, Sema3a, Sema3b, Sema3f,
IL-6 and S100A9 levels were significantly increased in L5-11 cells
compared with primary Sq-1979 cells (Fig. 5). Conversely, VEGFA, Npn2 and HIF1α
levels decreased despite previous reports suggesting these are
regulation factors for MDSCs.
Discussion
In the present study, we first examined tumor
immunogenicity in an isogenic heterotopic tumor transplantation
model of a primary OSCC cell line (Sq-1979) and an isogenic
heterotopic tumor transplantation model of a lymph node metastasis
cell line derived from the Sq-1979 cells. Our data showed that the
M-MDSC fraction of the tumor and PMN-MDSC number and fractions of
the spleen and tumor in the lymph node metastasis model were
significantly increased (Fig. 3).
These data indicated that the lymph node metastasis OSCC cells
developed the ability to induce PMN-MDSCs in both the spleen and
tumor. Therefore, the tumor immune suppression by PMN-MDSCs should
be considered a poor prognostic factor for metastasis. Some reports
have demonstrated that the primary tumor can induce MDSCs in the
spleen and tumor (14,15). The ability of MDSCs to support tumor
growth and metastasis can be divided into functions: i) protection
of tumor cells from immune-mediated killing; ii) remodeling the
tumor microenvironment; iii) establishment of a pre-metastatic
niche; and iv) interaction with tumor cells to induce ‘stemness’
and facilitate epithelial to mesenchymal transition (12). PMN-MDSCs have a similar phenotype as
neutrophils and are the largest population of MDSCs, making up
>75–80% of the total MDSCs in tumor-bearing mice (16). This study suggested that both
splenic and intratumoral PMN-MDSCs increased not only in the
primary tumor model but also in the lymph node metastasis model of
OSCC. Moreover, the number and fraction of PMN-MDSCs significantly
increased in both the spleen and tumor of the metastasis model
compared with the primary tumor model (Fig. 3). Notably, the fraction of
intratumoral PMN-MDSCs in the lymph node metastasis model markedly
increased to 38.1% compared with 2.7% in the primary tumor model
(Fig. 3F). Other reports have
demonstrated that M-MDSCs possess the main tumor immunosuppressive
functions of MDSCs (17,18), suggesting that the immunosuppressive
functions of PMN-MDSCs are less robust than M-MDSCs (19). Nevertheless, PMN-MDSCs play a
critical role in tumor immunity (12,20),
for example, PMN-MDSCs can inhibit antigen-nonspecific T cell
immune responses. The proportion of splenic PMN-MDSCs significantly
increased to 8.4% in L5-11-transplanted mice compared with 5% in
controls or 5.9% in Sq-1979-transplanted mice (Fig. 3C), while, there were no significant
differences in the percentage of splenic PMN-MDSC between controls
and Sq-1979-transplanted mice (Fig.
3C). Conversely, splenic CD8+ cells from
Sq-1979-transplanted mice significantly increased to 5.2% compared
with 2.8% in controls; however, in L5-11-transplanted mice there
was a significant decrease to 0.3% compared with controls. These
data suggested that CD8+ cells had been induced in
hematopoietic organs of the OSCC primary model, but had been
inhibited in the OSCC metastasis model. At first, we anticipated
that both the number and fraction of intratumoral CD4+
and CD8+ cells would be decreased in the metastasis
model as in the spleen. However, contrary to our expectations, both
the absolute cell numbers and the fraction of intratumoral
CD8+ cells in the metastasis model significantly
increased, and the number of intratumoral CD4+ cells
significantly increased but the fraction was unchanged (Fig. 4D and F). The augmentation of
intratumoral CD4+ cells in the metastasis model may be
due to increased numbers of total intratumoral cells compared with
the primary tumor model. Although, the fraction of intratumoral
CD8+ cells in the metastasis model significantly
increased to 2.3% compared with 1.3% in the primary model, the
augmentation of CD8+ cells may not be as effective for
tumor growth suppression because intratumoral PMN-MDSC in the
metastasis model, which have immune suppressive activity against
antigen-specific T cells, increased markedly.
It is often thought that differences in the
properties of cancer cells themselves affect the rate of primary
tumor growth and metastasis. However, our analysis of syngeneic
graft models suggested that the ability of the tumor to induce
specific immune cell infiltration may be more important than simple
proliferation rates. Therefore, we then analyzed potential
tumor-derived factors that may be related to the induction of
MDSCs. VEGF is an angiogenic factor secreted from tumor cells, and
VEGF signaling through VEGFR1 and Npn1 has been shown to contribute
to tumor growth and invasion (21).
VEGF production is promoted by HIF-1α (22,23),
and VEGF, IL-6 and S100A9 have been shown to induce the expansion
of MDSCs (24,25). The semaphorin family of secreted and
membrane-bound ligands that signal through Npn1, 2 enhance
angiogenesis and lymphangiogenesis and attract tumor-associated
macrophages to the tumor microenvironment (26–28).
Therefore, we analyzed the expression of these genes in Sq-1979 and
L5-11 cells. The results showed that sema3a, sema3b, sema3f,
VEGFR1, Npn1 and S100A9 mRNA levels were increased in L5-11 cells
compared with Sq-1979 cells (Fig.
5). These data indicate that acquired expression of these
factors in the L5-11 cell line promote tumor growth and metastasis
through the construction of pro-tumor microenvironment, inducing
angiogenesis and/or inducing the expansion of MDSCs.
In conclusion, our results clearly demonstrate that
splenic and intratumoral MDSCs are enhanced in the lymph node
metastasis model of OSCC. When assaying for changes in gene
expression that could account for the increased expansion of MDSCs,
neither VEGF nor HIF-1α were significantly changed, but VEGFR,
S100A9 and IL-6 were significantly enhanced in the lymph node
metastasis OSCC cells. These factors may affect the induction of
MDSCs, which promote tumor growth and metastasis.
Acknowledgements
The present study was supported in part by JSPS
KAKENHI (grant nos. 24791982, 26462854 and 26861748).
References
|
1
|
Kalnins IK, Leonard AG, Sako K, Razack MS
and Shedd DP: Correlation between prognosis and degree of lymph
node involvement in carcinoma of the oral cavity. Am J Surg.
134:450–454. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grandi C, Alloisio M, Moglia D, Podrecca
S, Sala L, Salvatori P and Molinari R: Prognostic significance of
lymphatic spread in head and neck carcinomas: Therapeutic
implications. Head Neck Surg. 8:67–73. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu CC, Chen PN, Peng CY, Yu CH and Chou
MY: Suppression of miR-204 enables oral squamous cell carcinomas to
promote cancer stemness, EMT traits, and lymph node metastasis.
Oncotarget. 7:20180–20192. 2016.PubMed/NCBI
|
|
4
|
Ohnishi K, Yamaguchi M, Erdenebaatar C,
Saito F, Tashiro H, Katabuchi H, Takeya M and Komohara Y:
Prognostic significance of CD169-positive lymph node sinus
macrophages in patients with endometrial carcinoma. Cancer Sci.
107:846–852. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jung EJ, Santarpia L, Kim J, Esteva FJ,
Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST, et al:
Plasma microRNA 210 levels correlate with sensitivity to
trastuzumab and tumor presence in breast cancer patients. Cancer.
118:2603–2614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Condamine T, Ramachandran I, Youn JI and
Gabrilovich DI: Regulation of tumor metastasis by myeloid-derived
suppressor cells. Annu Rev Med. 66:97–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heim CE, Vidlak D, Scherr TD, Kozel JA,
Holzapfel M, Muirhead DE and Kielian T: Myeloid-derived suppressor
cells contribute to Staphylococcus aureus orthopedic biofilm
infection. J Immunol. 192:3778–3792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gabrilovich DI and Nagaraj S:
Myeloid-derived suppressor cells as regulators of the immune
system. Nat Rev Immunol. 9:162–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Youn JI, Kumar V, Collazo M, Nefedova Y,
Condamine T, Cheng P, Villagra A, Antonia S, McCaffrey JC, Fishman
M, et al: Epigenetic silencing of retinoblastoma gene regulates
pathologic differentiation of myeloid cells in cancer. Nat Immunol.
14:211–220. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagaraj S, Schrum AG, Cho HI, Celis E and
Gabrilovich DI: Mechanism of T cell tolerance induced by
myeloid-derived suppressor cells. J Immunol. 184:3106–3116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weiss JM, Subleski JJ, Back T, Chen X,
Watkins SK, Yagita H, Sayers TJ, Murphy WJ and Wiltrout RH:
Regulatory T cells and myeloid-derived suppressor cells in the
tumor microenvironment undergo Fas-dependent cell death during
IL-2/alphaCD40 therapy. J Immunol. 192:5821–5829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marvel D and Gabrilovich DI:
Myeloid-derived suppressor cells in the tumor microenvironment:
Expect the unexpected. J Clin Invest. 125:3356–3364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Corzo CA, Condamine T, Lu L, Cotter MJ,
Youn JI, Cheng P, Cho HI, Celis E, Quiceno DG, Padhya T, et al:
HIF-1α regulates function and differentiation of myeloid-derived
suppressor cells in the tumor microenvironment. J Exp Med.
207:2439–2453. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khaled YS, Ammori BJ and Elkord E:
Increased levels of granulocytic myeloid-derived suppressor cells
in peripheral blood and tumour tissue of pancreatic cancer
patients. J Immunol Res. 2014:8798972014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walter S, Weinschenk T, Stenzl A, Zdrojowy
R, Pluzanska A, Szczylik C, Staehler M, Brugger W, Dietrich PY,
Mendrzyk R, et al: Multipeptide immune response to cancer vaccine
IMA901 after single-dose cyclophosphamide associates with longer
patient survival. Nat Med. 18:1254–1261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Youn JI, Collazo M, Shalova IN, Biswas SK
and Gabrilovich DI: Characterization of the nature of granulocytic
myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc
Biol. 91:167–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wynn TA: Myeloid-cell differentiation
redefined in cancer. Nat Immunol. 14:197–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haverkamp JM, Smith AM, Weinlich R, Dillon
CP, Qualls JE, Neale G, Koss B, Kim Y, Bronte V, Herold MJ, et al:
Myeloid-derived suppressor activity is mediated by monocytic
lineages maintained by continuous inhibition of extrinsic and
intrinsic death pathways. Immunity. 41:947–959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bronte V, Brandau S, Chen SH, Colombo MP,
Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A,
Ostrand-Rosenberg S, et al: Recommendations for myeloid-derived
suppressor cell nomenclature and characterization standards. Nat
Commun. 7:121502016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schouppe E, Mommer C, Movahedi K, Laoui D,
Morias Y, Gysemans C, Luyckx A, De Baetselier P and Van
Ginderachter JA: Tumor-induced myeloid-derived suppressor cell
subsets exert either inhibitory or stimulatory effects on distinct
CD8+ T-cell activation events. Eur J Immunol.
43:2930–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo M, Hou L, Li J, Shao S, Huang S, Meng
D, Liu L, Feng L, Xia P, Qin T, et al: VEGF/NRP-1axis promotes
progression of breast cancer via enhancement of
epithelial-mesenchymal transition and activation of NF-κB and
β-catenin. Cancer Lett. 373:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goel HL and Mercurio AM: VEGF targets the
tumour cell. Nat Rev Cancer. 13:871–882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Francesco EM, Lappano R, Santolla MF,
Marsico S, Caruso A and Maggiolini M: HIF-1α/GPER signaling
mediates the expression of VEGF induced by hypoxia in breast cancer
associated fibroblasts (CAFs). Breast Cancer Res. 15:R642013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Noman MZ, Desantis G, Janji B, Hasmim M,
Karray S, Dessen P, Bronte V and Chouaib S: PD-L1 is a novel direct
target of HIF-1α, and its blockade under hypoxia enhanced
MDSC-mediated T cell activation. J Exp Med. 211:781–790. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X, Eksioglu EA, Zhou J, Zhang L, Djeu
J, Fortenbery N, Epling-Burnette P, Van Bijnen S, Dolstra H, Cannon
J, et al: Induction of myelodysplasia by myeloid-derived suppressor
cells. J Clin Invest. 123:4595–4611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miao HQ, Lee P, Lin H, Soker S and
Klagsbrun M: Neuropilin-1 expression by tumor cells promotes tumor
angiogenesis and progression. FASEB J. 14:2532–2539. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Casazza A, Laoui D, Wenes M, Rizzolio S,
Bassani N, Mambretti M, Deschoemaeker S, Van Ginderachter JA,
Tamagnone L and Mazzone M: Impeding macrophage entry into hypoxic
tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis
and restores antitumor immunity. Cancer Cell. 24:695–709. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu ZQ, Zhou SL, Zhou ZJ, Luo CB, Chen EB,
Zhan H, Wang PC, Dai Z, Zhou J, Fan J, et al: Overexpression of
semaphorin 3A promotes tumor progression and predicts poor
prognosis in hepatocellular carcinoma after curative resection.
Oncotarget. 7:51733–51746. 2016.PubMed/NCBI
|