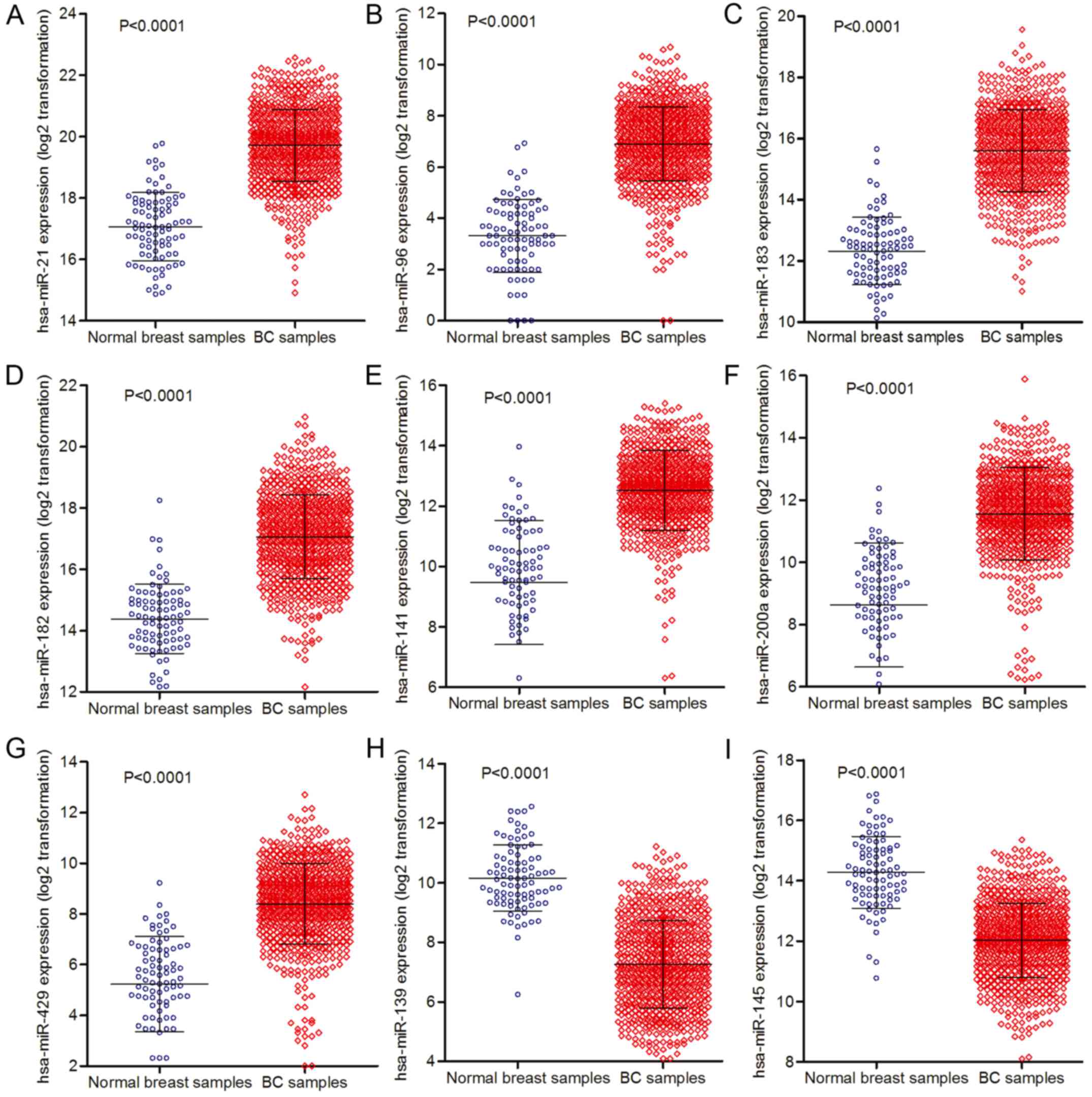
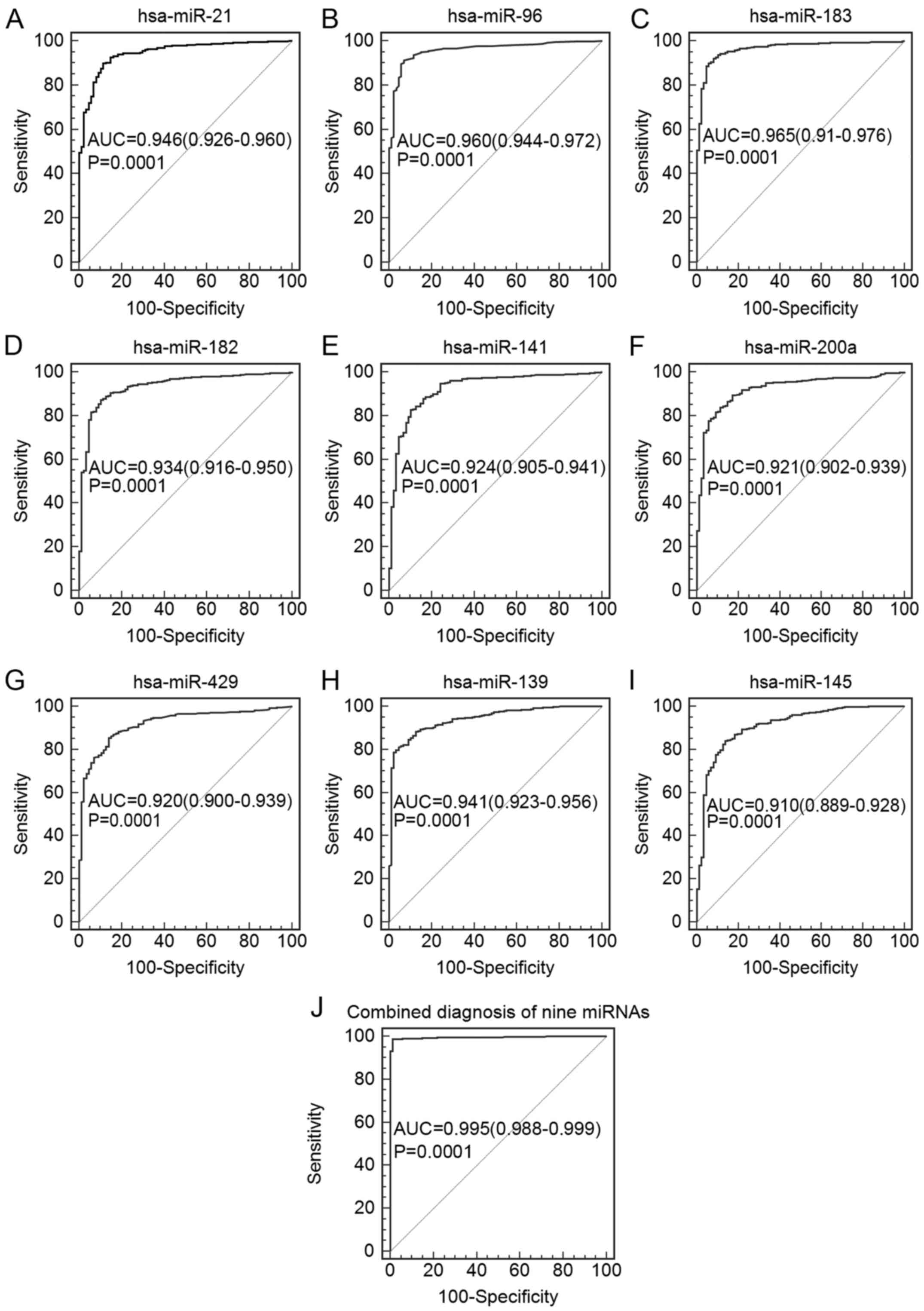
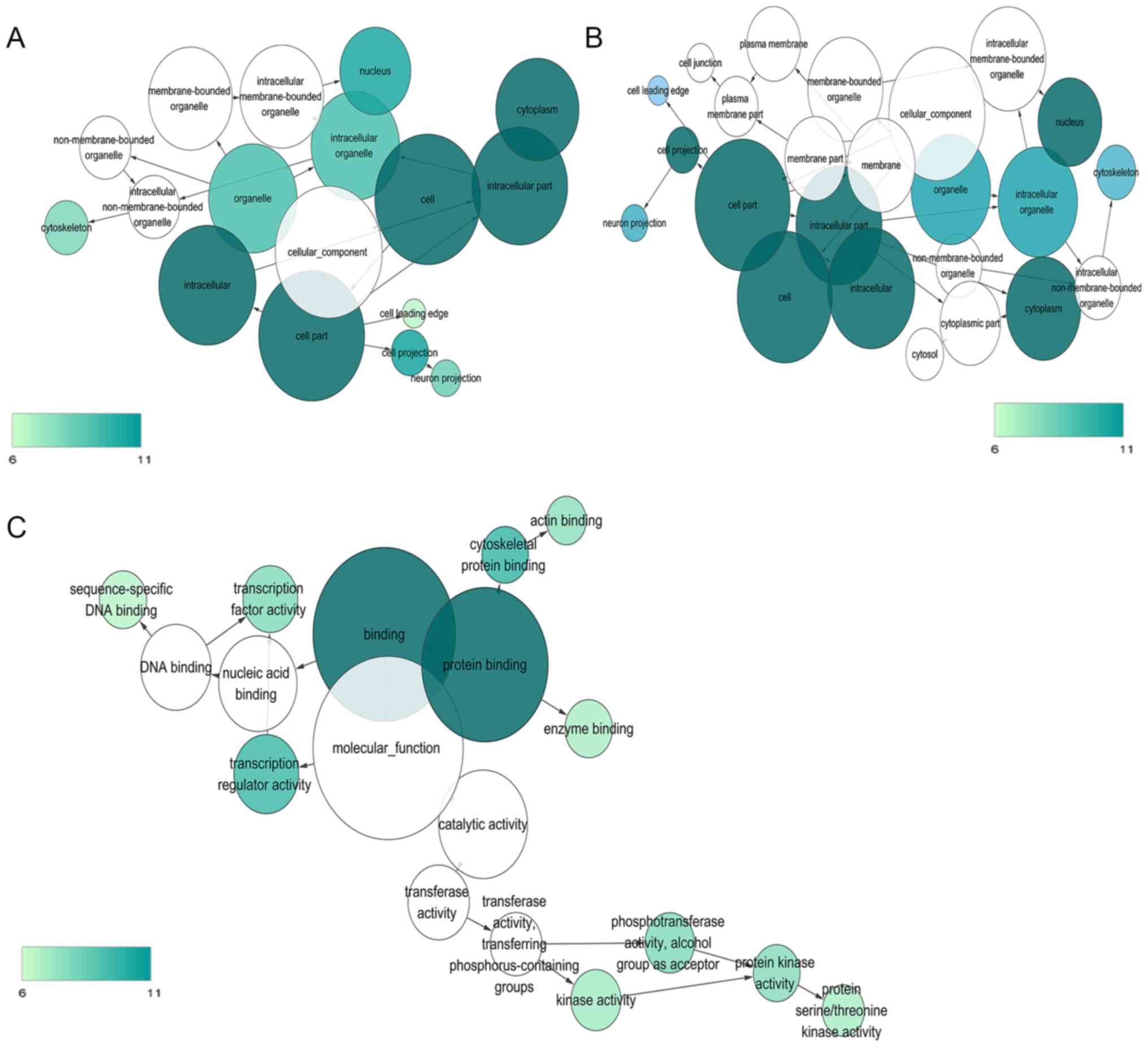
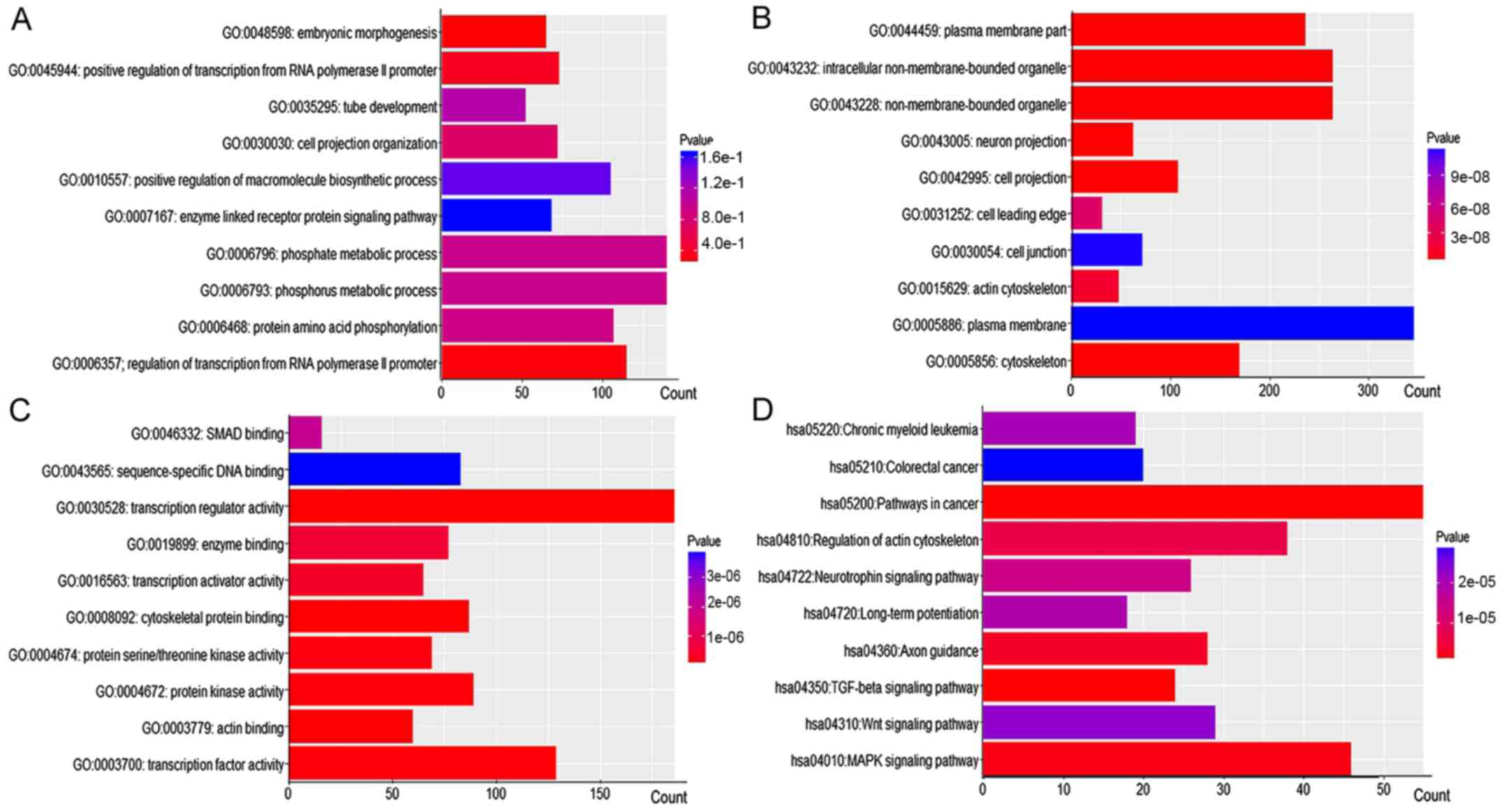
|
1
|
Schneider AP II, Zainer CM, Kubat CK,
Mullen NK and Windisch AK: The breast cancer epidemic: 10 facts.
Linacre Q. 81:244–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Donepudi MS, Kondapalli K, Amos SJ and
Venkanteshan P: Breast cancer statistics and markers. J Cancer Res
Ther. 10:506–511. 2014.PubMed/NCBI
|
|
4
|
Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y,
Kong R, Luo Y, Shi Y, Wang K, et al: miR-206 is down-regulated in
breast cancer and inhibits cell proliferation through the
up-regulation of cyclinD2. Biochem Biophys Res Commun. 433:207–212.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao Y, Cai Q, Huang Y, Li S, Yang H, Sun
L, Chen K and Wang Y: MicroRNA-21 as a potential diagnostic
biomarker for breast cancer patients: A pooled analysis of
individual studies. Oncotarget. 7:34498–34506. 2016.PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang P, Chen L, Zhang J, Chen H, Fan J,
Wang K, Luo J, Chen Z, Meng Z and Liu L: Methylation-mediated
silencing of the miR-124 genes facilitates pancreatic cancer
progression and metastasis by targeting Rac1. Oncogene. 33:514–524.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marquardt JU and Galle PR: Epigenetic
regulation of methionine adenosyltransferase 1A: A role for
MicroRNA-based treatment in liver cancer? Hepatology. 57:2081–2084.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia H, Ooi LL and Hui KM:
MicroRNA-216a/217-induced epithelial-mesenchymal transition targets
PTEN and SMAD7 to promote drug resistance and recurrence of liver
cancer. Hepatology. 58:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang F, Li L, Chen Z, Zhu M and Gu Y:
MicroRNA-214 acts as a potential oncogene in breast cancer by
targeting the PTEN-PI3K/Akt signaling pathway. Int J Mol Med.
37:1421–1428. 2016.PubMed/NCBI
|
|
12
|
Fu SW, Lee W, Coffey C, Lean A, Wu X, Tan
X, Man YG and Brem RF: miRNAs as potential biomarkers in early
breast cancer detection following mammography. Cell Biosci.
6:62016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi H, Chen J, Li Y, Li G, Zhong R, Du D,
Meng R, Kong W and Lu M: Identification of a six microRNA signature
as a novel potential prognostic biomarker in patients with head and
neck squamous cell carcinoma. Oncotarget. 7:21579–21590.
2016.PubMed/NCBI
|
|
14
|
Bland JM and Altman DG: Statistical
methods for assessing agreement between two methods of clinical
measurement. Lancet. 1:307–310. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Riecke L, Sack AT and Schroeder CE:
Endogenous delta/theta sound-brain phase entrainment accelerates
the buildup of auditory streaming. Curr Biol. 25:3196–3201. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng Q, Yi B, Wang A and Jiang X:
Exploring and exploiting the fundamental role of microRNAs in tumor
pathogenesis. Onco Targets Ther. 6:1675–1684. 2013.PubMed/NCBI
|
|
17
|
Tan YY, Xu XY, Wang JF, Zhang CW and Zhang
SC: MiR-654-5p attenuates breast cancer progression by targeting
EPSTI1. Am J Cancer Res. 6:522–532. 2016.PubMed/NCBI
|
|
18
|
Jacot W, Fiche M, Zaman K, Wolfer A and
Lamy PJ: The HER2 amplicon in breast cancer: Topoisomerase IIA and
beyond. Biochim Biophys Acta. 1836:146–157. 2013.PubMed/NCBI
|
|
19
|
Zaczek A, Markiewicz A, Supernat A,
Bednarz-Knoll N, Brandt B, Seroczyńska B, Skokowski J, Szade J,
Czapiewski P, Biernat W, et al: Prognostic value of TOP2A gene
amplification and chromosome 17 polysomy in early breast cancer.
Pathol Oncol Res. 18:885–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lambros MB, Natrajan R, Geyer FC,
Lopez-Garcia MA, Dedes KJ, Savage K, Lacroix-Triki M, Jones RL,
Lord CJ, Linardopoulos S, et al: PPM1D gene amplification and
overexpression in breast cancer: A qRT-PCR and chromogenic in situ
hybridization study. Mod Pathol. 23:1334–1345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mandal CC, Ghosh-Choudhury T, Dey N,
Choudhury GG and Ghosh-Choudhury N: miR-21 is targeted by omega-3
polyunsaturated fatty acid to regulate breast tumor CSF-1
expression. Carcinogenesis. 33:1897–1908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang F, Wang Y, Xue J, Ma Q, Zhang J, Chen
YF, Shang ZZ, Li QQ, Zhang SL and Zhao L: Effect of Corilagin on
the miR-21/smad7/ERK signaling pathway in a schistosomiasis-induced
hepatic fibrosis mouse model. Parasitol Int. 65:308–315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu F, Zheng S, Liu T, Liu Q, Liang M, Li
X, Sheyhidin I, Lu X and Liu W: MicroRNA-21 promotes the
proliferation and inhibits apoptosis in Eca109 via activating
ERK1/2/MAPK pathway. Mol Cell Biochem. 381:115–125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han M, Liu M, Wang Y, Mo Z, Bi X, Liu Z,
Fan Y, Chen X and Wu C: Re-expression of miR-21 contributes to
migration and invasion by inducing epithelial-mesenchymal
transition consistent with cancer stem cell characteristics in
MCF-7 cells. Mol Cell Biochem. 363:427–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han M, Wang F, Gu Y, Pei X, Guo G, Yu C,
Li L, Zhu M, Xiong Y and Wang Y: MicroRNA-21 induces breast cancer
cell invasion and migration by suppressing smad7 via EGF and TGF-β
pathways. Oncol Rep. 35:73–80. 2016.PubMed/NCBI
|
|
27
|
Marino AL, Evangelista AF, Vieira RA,
Macedo T, Kerr LM, Abrahão-Machado LF, Longatto-Filho A, Silveira
HC and Marques MM: MicroRNA expression as risk biomarker of breast
cancer metastasis: A pilot retrospective case-cohort study. BMC
Cancer. 14:7392014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gebremedhn S, Salilew-Wondim D, Hoelker M,
Rings F, Neuhoff C, Tholen E, Schellander K and Tesfaye D:
MicroRNA-183-96-182 cluster regulates bovine granulosa cell
proliferation and cell cycle transition by coordinately targeting
FOXO1. Biol Reprod. 94:1272016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang L, Guo F, Huo B, Lv Y, Wang Y and
Liu W: Expression and clinical significance of the microRNA-200
family in gastric cancer. Oncol Lett. 9:2317–2324. 2015.PubMed/NCBI
|
|
31
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tan H, Zhu Y, Zhang J, Peng L and Ji T:
miR141 expression is downregulated and negatively correlated with
STAT5 expression in esophageal squamous cell carcinoma. Exp Ther
Med. 11:1803–1808. 2016.PubMed/NCBI
|
|
33
|
Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di
GH, Wu J, Shen ZZ, Song HY and Shao ZM: MicroRNA-200a promotes
anoikis resistance and metastasis by targeting YAP1 in human breast
cancer. Clin Cancer Res. 19:1389–1399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Du L, Yang Y, Wang C, Liu H, Wang L,
Zhang X, Li W, Zheng G and Dong Z: MiR-429 is an independent
prognostic factor in colorectal cancer and exerts its
anti-apoptotic function by targeting SOX2. Cancer Lett. 329:84–90.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yoneyama K, Ishibashi O, Kawase R, Kurose
K and Takeshita T: miR-200a, miR-200b and miR-429 are onco-miRs
that target the PTEN gene in endometrioid endometrial carcinoma.
Anticancer Res. 35:1401–1410. 2015.PubMed/NCBI
|
|
36
|
Zhang X, Li P, Rong M, He R, Hou X, Xie Y
and Chen G: MicroRNA-141 is a biomarker for progression of squamous
cell carcinoma and adenocarcinoma of the lung: Clinical analysis of
125 patients. Tohoku J Exp Med. 235:161–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu CL, Ho JY, Chou SC and Yu DS: MiR-429
reverses epithelial-mesenchymal transition by restoring E-cadherin
expression in bladder cancer. Oncotarget. 7:26593–26603.
2016.PubMed/NCBI
|
|
38
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
miR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016.PubMed/NCBI
|
|
39
|
Asakura T, Yamaguchi N, Ohkawa K and
Yoshida K: Proteasome inhibitor-resistant cells cause EMT-induction
via suppression of E-cadherin by miR-200 and ZEB1. Int J Oncol.
46:2251–2260. 2015.PubMed/NCBI
|
|
40
|
Korpal M and Kang Y: The emerging role of
miR-200 family of microRNAs in epithelial-mesenchymal transition
and cancer metastasis. RNA Biol. 5:115–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee JW, Park YA, Choi JJ, Lee YY, Kim CJ,
Choi C, Kim TJ, Lee NW, Kim BG and Bae DS: The expression of the
miRNA-200 family in endometrial endometrioid carcinoma. Gynecol
Oncol. 120:56–62. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang HD, Sun DW, Mao L, Zhang J, Jiang
LH, Li J, Wu Y, Ji H, Chen W, Wang J, et al: MiR-139-5p inhibits
the biological function of breast cancer cells by targeting Notch1
and mediates chemosensitivity to docetaxel. Biochem Biophys Res
Commun. 465:702–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez
G, Bautista-Piña V, Arellano-Llamas R and Hidalgo-Miranda A:
Identification and pathway analysis of microRNAs with no previous
involvement in breast cancer. PLoS One. 7:e319042012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hua W, Sa KD, Zhang X, Jia LT, Zhao J,
Yang AG, Zhang R, Fan J and Bian K: MicroRNA-139 suppresses
proliferation in luminal type breast cancer cells by targeting
Topoisomerase II alpha. Biochem Biophys Res Commun. 463:1077–1083.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
MiR-145 promotes TNF-α-induced apoptosis
by facilitating the formation of RIP1-FADDcaspase-8 complex in
triple-negative breast cancer. Tumour Biol. 37:8599–8607. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng M, Sun X, Li Y and Zuo W:
MicroRNA-145 inhibits growth and migration of breast cancer cells
through targeting oncoprotein ROCK1. Tumour Biol. 37:8189–8196.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lv J, Xia K, Xu P, Sun E, Ma J, Gao S,
Zhou Q, Zhang M, Wang F, Chen F, et al: miRNA expression patterns
in chemoresistant breast cancer tissues. Biomed Pharmacother.
68:935–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sengupta S, Das JK and Gangopadhyay A:
Naevoid psoriasis and ILVEN: Same coin, two faces? Indian J
Dermatol. 57:489–491. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Min W, Wang B, Li J, Han J, Zhao Y, Su W,
Dai Z, Wang X and Ma Q: The expression and significance of five
types of miRNAs in breast cancer. Med Sci Monit Basic Res.
20:97–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hou Y, Wang J, Wang X, Shi S, Wang W and
Chen Z: Appraising microRNA-155 as a noninvasive diagnostic
biomarker for cancer detection: A meta-analysis. Medicine.
95:e24502016. View Article : Google Scholar : PubMed/NCBI
|